Amplification of Ultra-Trace DNA from Early Sheep Embryos Based on qPCR: Establishing a Gender Identification System
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experiment Location and Time
2.3. Extraction of Blood gDNA
2.4. Extraction of Embryo gDNA
2.4.1. Embryo Production System
2.4.2. Oocyte Maturation
2.4.3. Sperm Preparation
2.4.4. Embryo Culture
2.4.5. Embryo Biopsy Protocol
2.4.6. gDNA Extraction
2.4.7. Cryopreservation
2.5. Primer and Probe Design and Synthesis
2.6. Blood qPCR Reaction System
2.7. Embryo Ultra-Trace qPCR Reaction System
2.8. Agarose Gel Electrophoresis
2.9. Data Analysis
3. Results
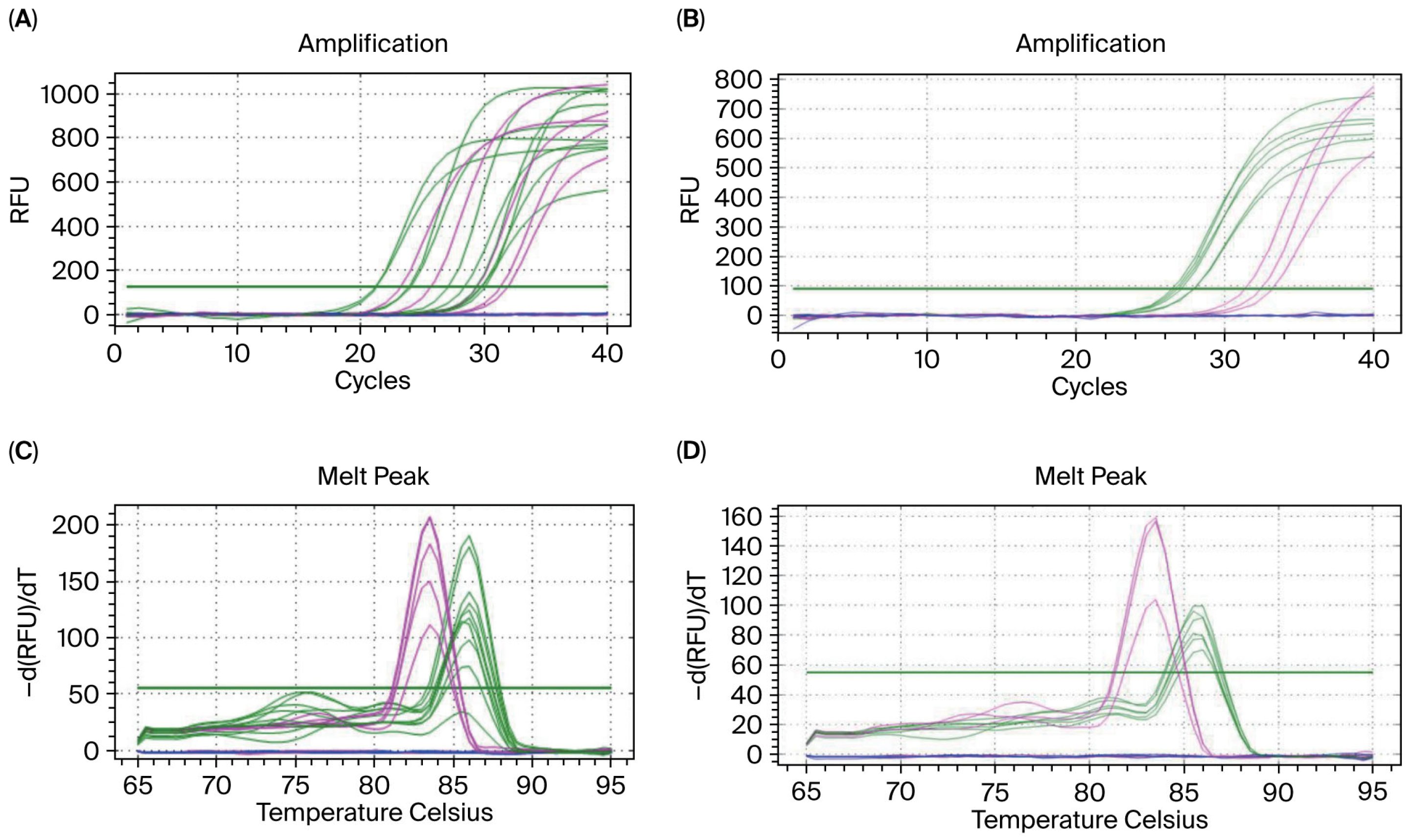
3.1. Establishing the Blood gDNA Ultra-Trace qPCR Amplification System
3.2. Verifying the Amplification Results for Blood gDNA Diluted 20,000 Times
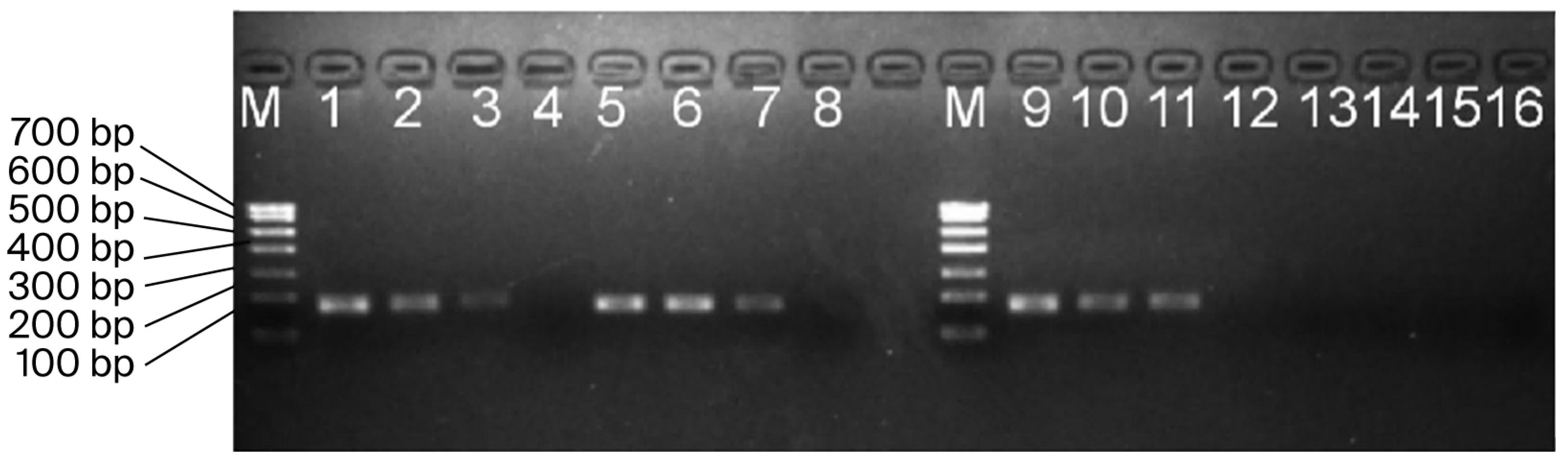
3.3. Serial Dilution of Blood gDNA for Electrophoresis Detection
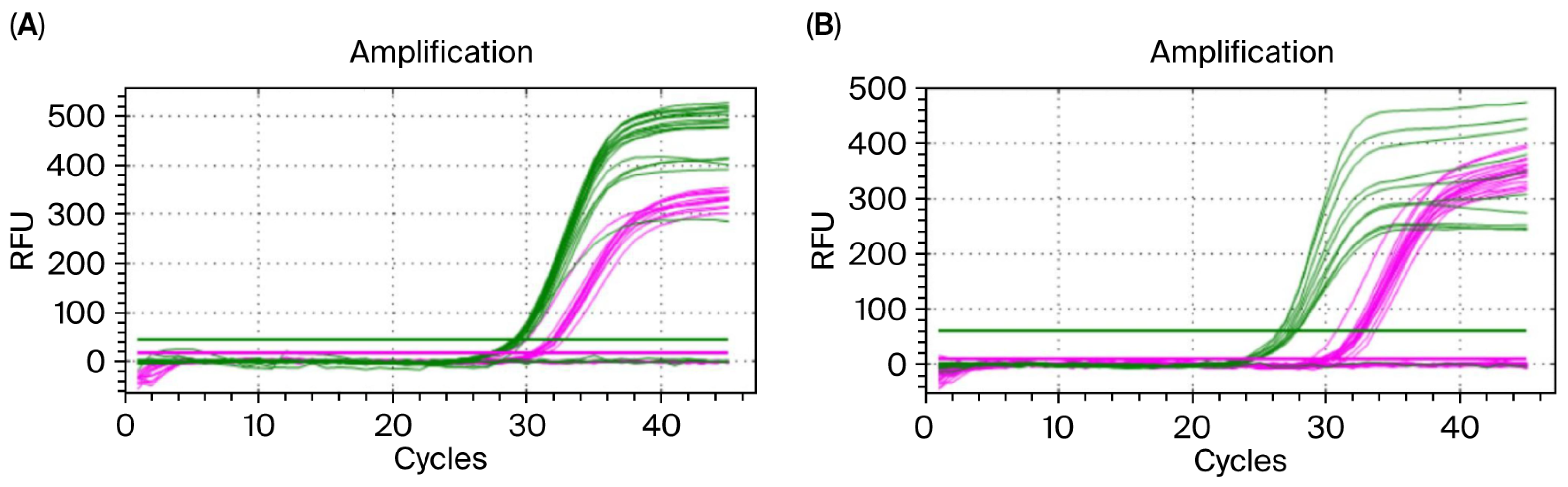
3.4. Gradient Concentration Amplification Results for Embryo gDNA
3.5. Embryo Sex Identification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xia, X.H.; Liang, N.; Ma, X.Y.; Lu, Q.M.; Wang, S.S.; Chang, Z.J. Inhibition of the NF-κB signaling pathway affects gonadal differentiation and leads to male bias in Paramisgurnus dabryanus. Theriogenology 2023, 207, 82–95. [Google Scholar] [CrossRef]
- Khalajzadeh, S.; Nejati-Javaremi, A.; Yeganeh, H. Effect of widespread and limited use of sexed semen on genetic progress and reproductive performance of dairy cows. Animal 2012, 6, 1398–1406. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khramtsova, E.A.; Wilson, M.A.; Martin, J.; Winham, S.J.; He, K.Y.; Davis, L.K.; Stranger, B.E. Quality control and analytic best practices for testing genetic models of sex differences in large populations. Cell 2023, 186, 2044–2061. [Google Scholar] [CrossRef] [PubMed]
- Cusanovich, D.A.; Reddington, J.P.; Garfield, D.A.; Daza, R.M.; Aghamirzaie, D.; Marco-Ferreres, R.; Pliner, H.A.; Christiansen, L.; Qiu, X.; Steemers, F.J.; et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 2018, 555, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Yang, A.; Li, G.; Wu, H.; Deng, S.; Yang, H.; Ma, W.; Lv, D.; Fu, Y.; Ji, P.; et al. Melatonin promotes the development of sheep transgenic cloned embryos by protecting donor and recipient cells. Cell Cycle 2022, 21, 1360–1375. [Google Scholar] [CrossRef]
- Dvoran, M.; Nemcova, L.; Kalous, J. An Interplay between Epigenetics and Translation in Oocyte Maturation and Embryo Development: Assisted Reproduction Perspective. Biomedicines 2022, 10, 1689. [Google Scholar] [CrossRef]
- Fraire-Zamora, J.; Martínez, M.F.; Torra-Massana, M.; Zamora, M.J.; Vassena, R.; Miguel-Escalada, I.; Rodríguez, A.; Popovic, M. P-135 No difference in morphokinetic patterns between male and female preimplantation embryos. Hum. Reprod. 2023, 38. [Google Scholar] [CrossRef]
- Herr, C.; Matthaei, K.I.; Steel, T.; Reed, K.C. Rapid Y-chromosome-assay sexing of peripheral blood lymphocytes from bovinae of known phenotypic sex. Theriogenology 1990, 33, 246. [Google Scholar] [CrossRef]
- Polisseni, J.; Sá, W.F.; Guerra, M.O.; Machado, M.A.; Serapião, R.V.; Carvalho, B.C.; Camargo, L.S.; Peters, V.M. Post-biopsy bovine embryo viability and whole genome amplification in preimplantation genetic diagnosis. Fertil. Steril. 2010, 93, 783–788. [Google Scholar] [CrossRef]
- Christian, L.; Timothy, L.B.; Peter, K. Switching on sex: Transcriptional regulation of the testis-determining gene Sry. Development 2014, 141, 2195–2205. [Google Scholar] [CrossRef]
- Vincent, R.H.; Michael, C.; Anthony, A. The Molecular Action and Regulation of the Testis-Determining Factors, SRY (Sex-Determining Region on the Y Chromosome) and SOX9 [SRY-Related High-Mobility Group (HMG) Box 9]. Endocr. Rev. 2003, 24, 466–487. [Google Scholar] [CrossRef]
- Wei, H.; Movahedi, A.; Yang, J.; Zhang, Y.Y.; Liu, G.Y.; Zhu, S.; Yu, C.M.; Chen, Y.H.; Zhong, F.; Zhang, J. Characteristics and molecular identification of glyceraldehyde-3-phosphate dehydrogenases in poplar. Int. J. Biol. Macromol. 2022, 219, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Rekawiecki, R.; Kowalik, M.K.; Kotwica, J. Validation of housekeeping genes for studying differential gene expression in the bovine myometrium. Acta Vet. Hung. 2013, 61, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Sonowal, J.; Lal, P.C.; Dev, K.; Singh, R.; Barkathullah, N.; Akram, M.W.; Kumar, G.R.; Kant, A.R.; Kumar, D.; Saxena, S.; et al. Selection and validation of suitable reference gene for qPCR gene expression analysis in lamb testis cells under Sheep pox virus infection. Gene 2022, 831, 146561. [Google Scholar] [CrossRef] [PubMed]
- Azarpeykan, S.; Dittmer, K.E. Evaluation of housekeeping genes for quantitative gene expression analysis in the equine kidney. J. Equine Sci. 2016, 27, 165–168. [Google Scholar] [CrossRef][Green Version]
- Mogilicherla, K.; Athe, R.P.; Chatterjee, R.N.; Bhattacharya, T.K. Identification of suitable reference genes for normalization of quantitative real-time PCR-based gene expression in chicken (Gallus gallus). Anim. Genet. 2022, 53, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Park, J.H.; Lee, S.H.; Park, C.S.; Jin, D.I. Sexing using single blastomere derived from IVF bovine embryos by fluorescence in situ hybridization (FISH). Theriogenology 2004, 62, 1452–1458. [Google Scholar] [CrossRef]
- Mishra, A.; Dhali, A.; Reddy, I.J.; Kolte, A.P. Sexing of pre-implantation ovine embryos through polymerase chain reaction-based amplification of GAPDH, SRY and AMEL genes. Reprod. Domest. Anim. 2020, 55, 885–892. [Google Scholar] [CrossRef]
- Nix, J.L.; Marrella, M.A.; Oliver, M.A.; Ealy, A.D.; Biase, F.H. Cleavage kinetics is a better indicator of embryonic developmental competency than brilliant cresyl blue staining of oocytes. Anim. Reprod. 2023, 248, 107174. [Google Scholar] [CrossRef]
- He, Q.F.; Huang, M.; Cao, X.Y.; Zhang, K.; Li, J.; Quan, F.S. Advancements in mammalian X and Y sperm differences and sex control technology. Zygote 2022, 30, 423–430. [Google Scholar] [CrossRef]
- Nix, J.L.; Schettini, G.; Biase, F.H. Sexing of cattle embryos using RNA-sequencing data or polymerase chain reaction based on a complete sequence of cattle chromosome Y. Front. Genet. 2023, 14, 1038291. [Google Scholar] [CrossRef] [PubMed]
- López-Gatius, F.; García-Ispierto, I. Sexing of Embryos at the Time of Twin Reduction: A Clinical Approach. Animals 2023, 13, 1326. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, C.D.; Gonceer, N.; Dorus, S.; Crill, J.E.; Moshayoff, V.; Lachman, A.; Moran, A.P.; Vilenchik, D.; Fedida-Metula, S. Fast, accurate, and cost-effective poultry sex genotyping using real-time polymerase chain reaction. Front. Vet. Sci. 2023, 10, 1196755. [Google Scholar] [CrossRef] [PubMed]
- Camargo, G.S.; Meira, C.D.; Bergfelt, D.R.; Oliveira, J.P.; Bromberger, C.R.; Barros, L.D.; Canesin, H.S.; Lopes, G.; Monteiro, G.A.; Ignácio, F.S. Equine blastocyst re-expansion rate, quality, and sex following embryonic collapse using needle perforation of blastocoel and subsequent PCR amplification of blastocoel DNA. J. Equine Vet. Sci. 2023, 118, 269. [Google Scholar] [CrossRef]
- Ridnik, M.; Abberbock, E.; Alipov, V.; Lhermann, S.Z.; Kaufman, S.; Lubman, M.; Poulat, F.; Gonen, N. Two redundant transcription factor binding sites in a single enhancer are essential for mammalian sex determination. Nucleic Acids Res. 2024, 52, 5514–5528. [Google Scholar] [CrossRef]
- Pathak, D.; Baksi, A.; Vasan, S.S.; Dighe, R.R. Molecular and Functional Characterization of Human Sex-Determining Region on the Y Chromosome Variants Using Protamine 1 Promoter. DNA Cell Biol. 2024, 43, 12–25. [Google Scholar] [CrossRef]
- Angove, M.L.; Turner, Z.; Conley, A.J.; McNabb, B.R.; Eenennaam, A.L. PSI-4 Investigation of a Naturally Occurring XX Male Bull. J. Anim. Sci. 2022, 100, 22–23. [Google Scholar] [CrossRef]
- Dujardin, É.; André, M.; Dewaele, A.; Mandon-Pepin, B.; Poulat, F.; Frambourg, A.; Thépot, D.; Jouneau, L.; Jolivet, G.; Pailhoux, É.; et al. DMRT1 is a testis-determining gene in rabbits and is also essential for female fertility. eLife 2023, 12, RP89284. [Google Scholar] [CrossRef]
- Li, N.; Zhou, J.; Zhang, W.Q.; Liu, W.J.; Wang, B.X.; She, H.B.; Mirbahar, A.A.; Li, S.F.; Zhang, Y.L.; Gao, W.J.; et al. A rapid method for assembly of single chromosome and identification of sex determination region based on single-chromosome sequencing. New Phytol. 2023, 240, 892–903. [Google Scholar] [CrossRef]
- Wu, L.R.; Chen, S.X.; Wu, Y.L.; Patel, A.A.; Zhang, D.Y. Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification. Nat. Biomed. Eng. 2017, 1, 714–723. [Google Scholar] [CrossRef]
- Coster, T.D.; Poucke, M.V.; Pascottini, O.B.; Angel-Velez, D.; Branden, E.V.; Peere, S.; Papas, M.; Gerits, I.; Govaere, J.; Peelman, L.; et al. Single closed-tube quantitative real-time PCR assay with dual-labelled probes for improved sex determination of equine embryos. Animal 2023, 17, 100952. [Google Scholar] [CrossRef]
- Rogers, F.; Rhoades, J.R.; Quail, L.K.; Rich, J.J. 113 Investigation of differences in pregnancy associated glycoproteins due to different fetal parameters (sex and number) using commercial tests in multiparous ewes. J. Anim. Sci. 2024, 102, 104–105. [Google Scholar] [CrossRef]
- Sánchez, J.M.; Gómez-Redondo, I.; Browne, J.A.; Planells, B.; Gutiérrez-Adán, A.; Lonergan, P. MicroRNAs in amniotic fluid and maternal blood plasma associated with sex determination and early gonad differentiation in cattle. Biol. Reprod. 2021, 105, 345–358. [Google Scholar] [CrossRef]
| Primers | Primer Sequences | Size of Products (bp) | Melting Temperature (°C) |
|---|---|---|---|
| GAPDH SRY SRY-Probe | Forward: GGTCCACATGGCCTCCAAG Reverse: TCCATTTGTGAGTGTGTGGTCTT Forward: CTATACACCGAGACAAATACCCG Reverse: AATCGTCCCTGTATGTGAAGG Forward: AM-AAGAGGCCACAGAAATCCCTTGCT-MGB (Minor Groove Binder) | 167 148 | 65 60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, P.; Tao, W.; Huang, F.; Li, X.; Wang, X.; Wang, J.; Gao, Q.; Fang, D. Amplification of Ultra-Trace DNA from Early Sheep Embryos Based on qPCR: Establishing a Gender Identification System. Biology 2025, 14, 1144. https://doi.org/10.3390/biology14091144
Niu P, Tao W, Huang F, Li X, Wang X, Wang J, Gao Q, Fang D. Amplification of Ultra-Trace DNA from Early Sheep Embryos Based on qPCR: Establishing a Gender Identification System. Biology. 2025; 14(9):1144. https://doi.org/10.3390/biology14091144
Chicago/Turabian StyleNiu, Peng, Weikun Tao, Fei Huang, Xiaopeng Li, Xueyan Wang, Jie Wang, Qinghua Gao, and Di Fang. 2025. "Amplification of Ultra-Trace DNA from Early Sheep Embryos Based on qPCR: Establishing a Gender Identification System" Biology 14, no. 9: 1144. https://doi.org/10.3390/biology14091144
APA StyleNiu, P., Tao, W., Huang, F., Li, X., Wang, X., Wang, J., Gao, Q., & Fang, D. (2025). Amplification of Ultra-Trace DNA from Early Sheep Embryos Based on qPCR: Establishing a Gender Identification System. Biology, 14(9), 1144. https://doi.org/10.3390/biology14091144







