Identification of Self-Incompatibility Related Genes in Sweet Cherry Based on Transcriptomic Analysis
Simple Summary
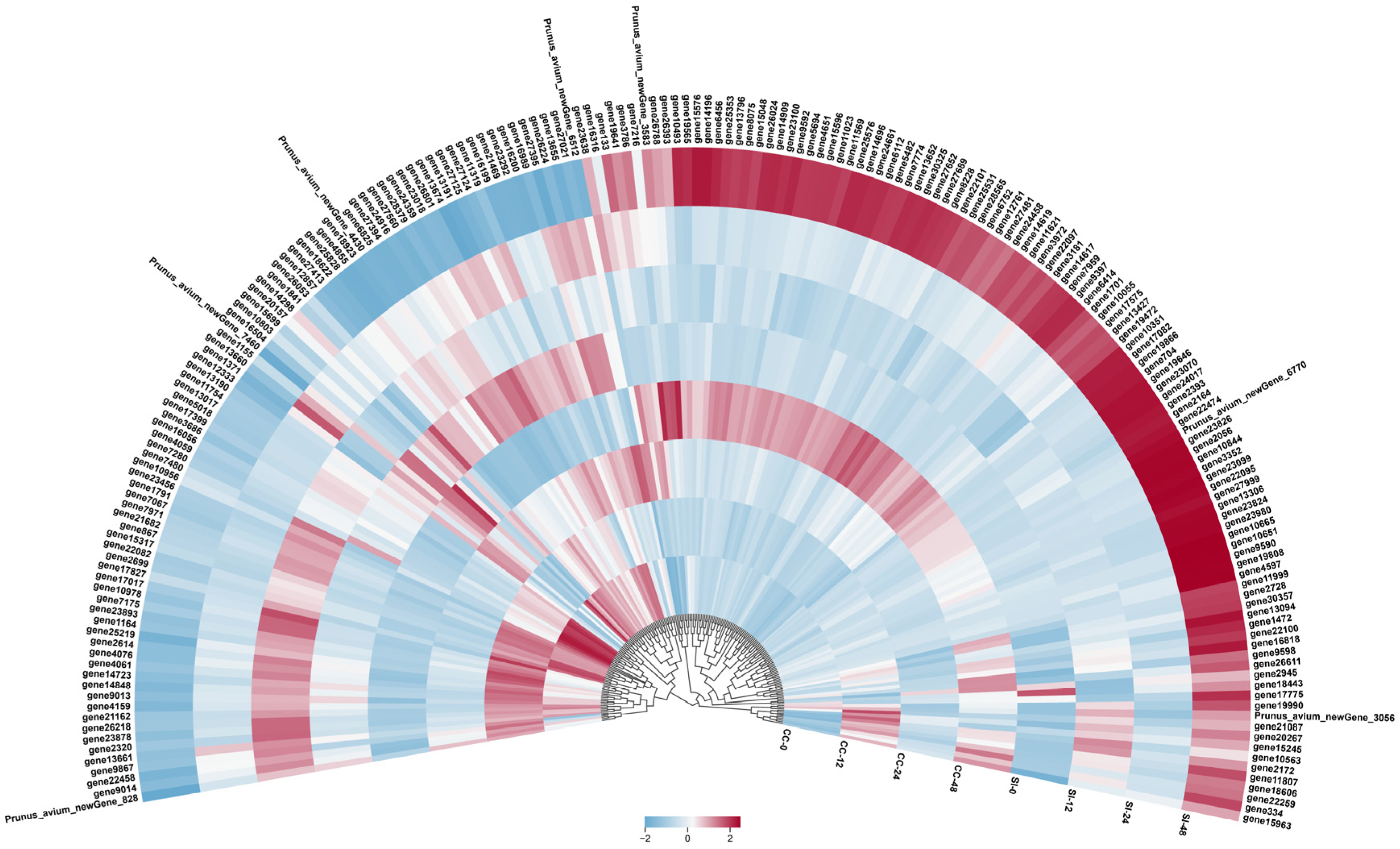
Abstract
1. Introduction
2. Results
2.1. Pollen Morphology and Activity Analysis of ‘Tieton’ and ‘Rainier’
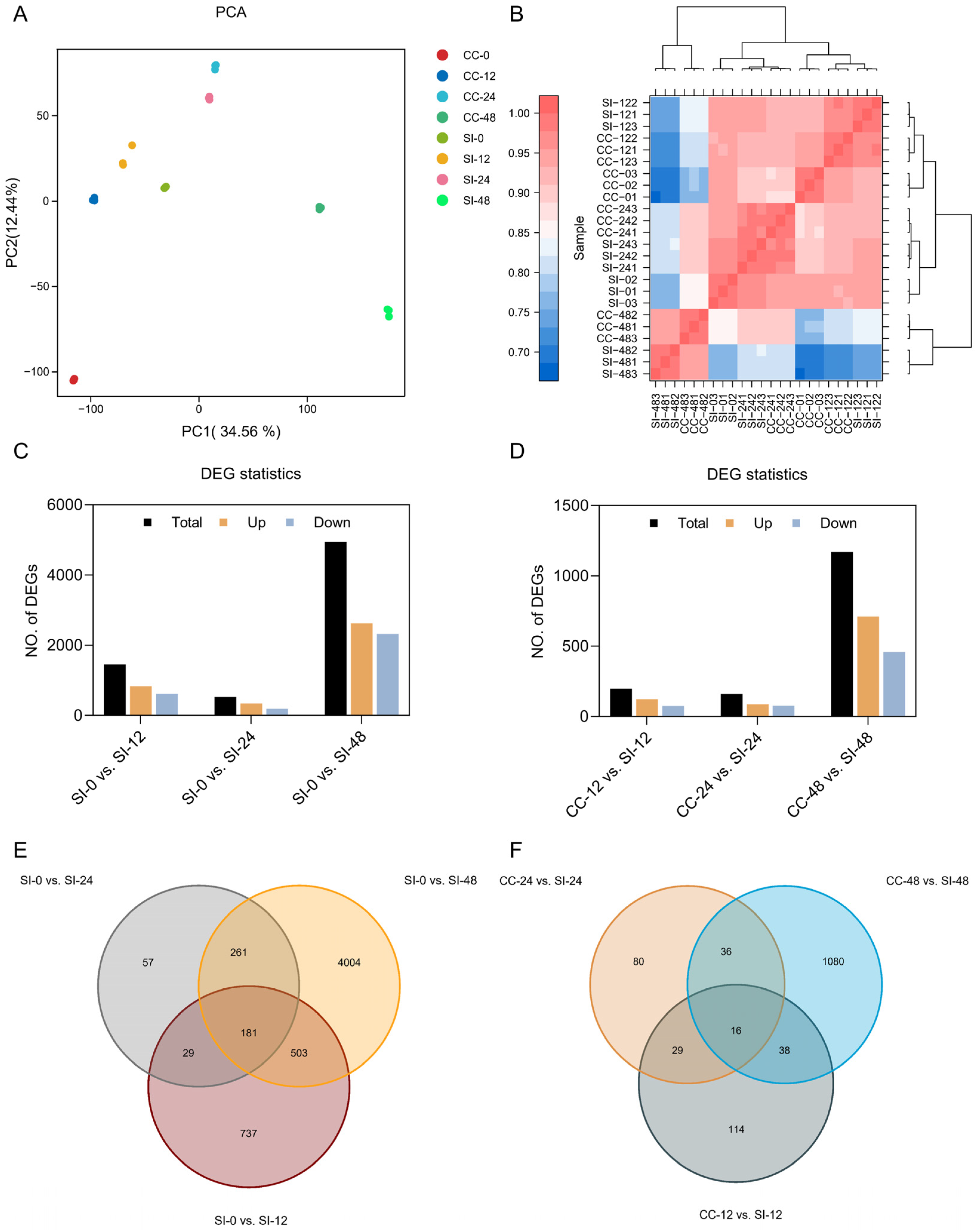
2.2. Transcriptome Analysis
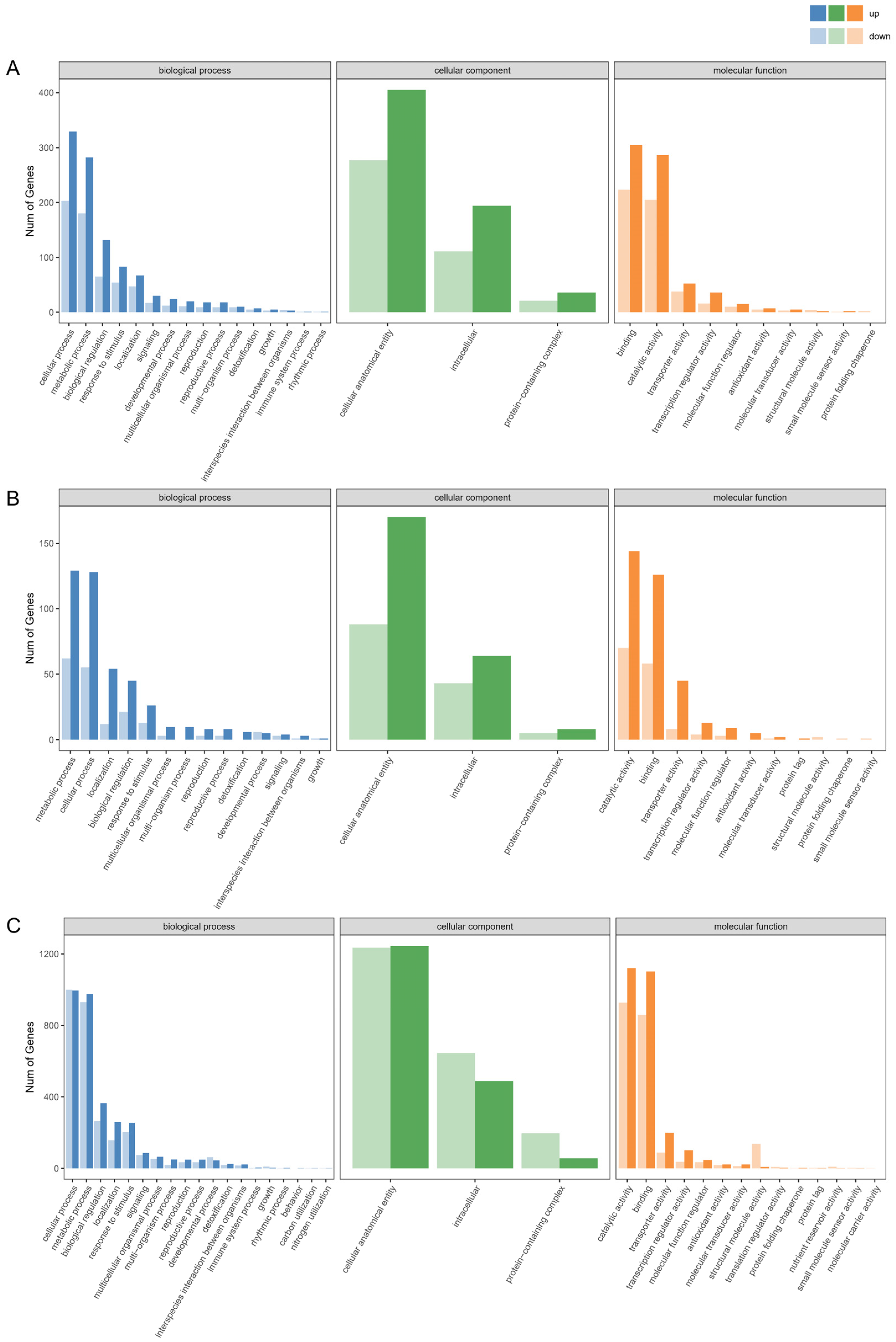
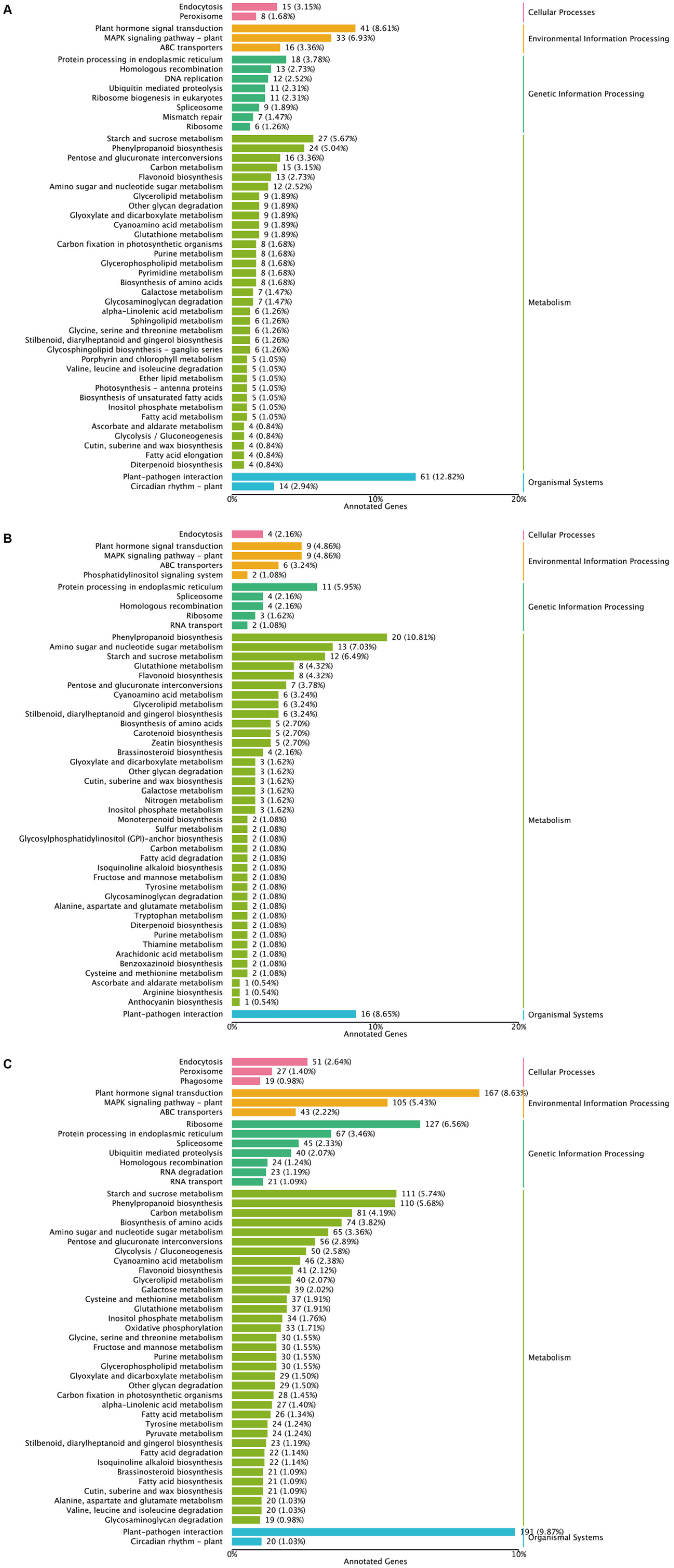
2.3. GO Term and KEGG Pathway Analysis of DEGs
2.4. Screening of Candidate Genes Related to SI in ‘Tieton’
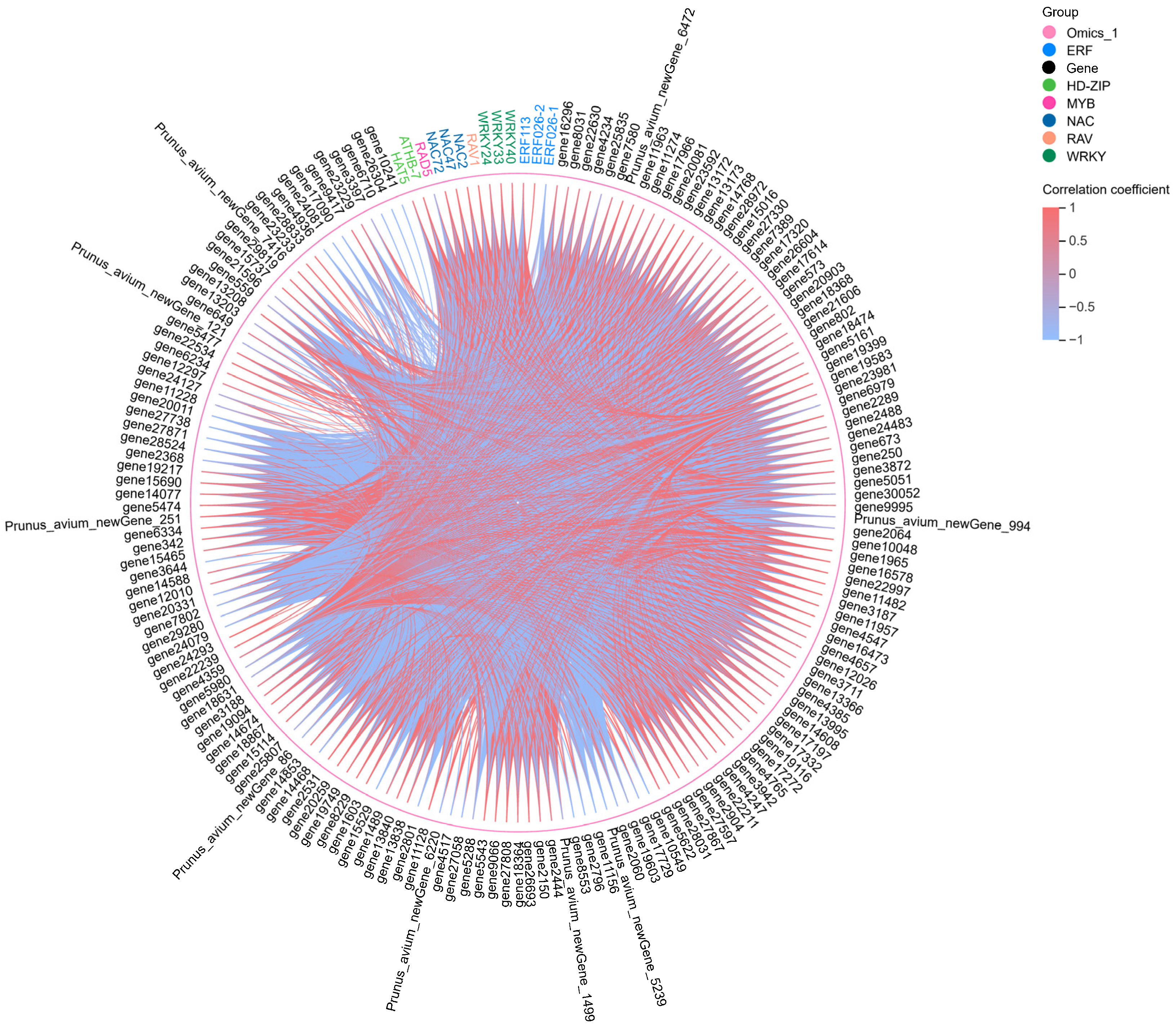
2.5. Transcription Factor Analysis
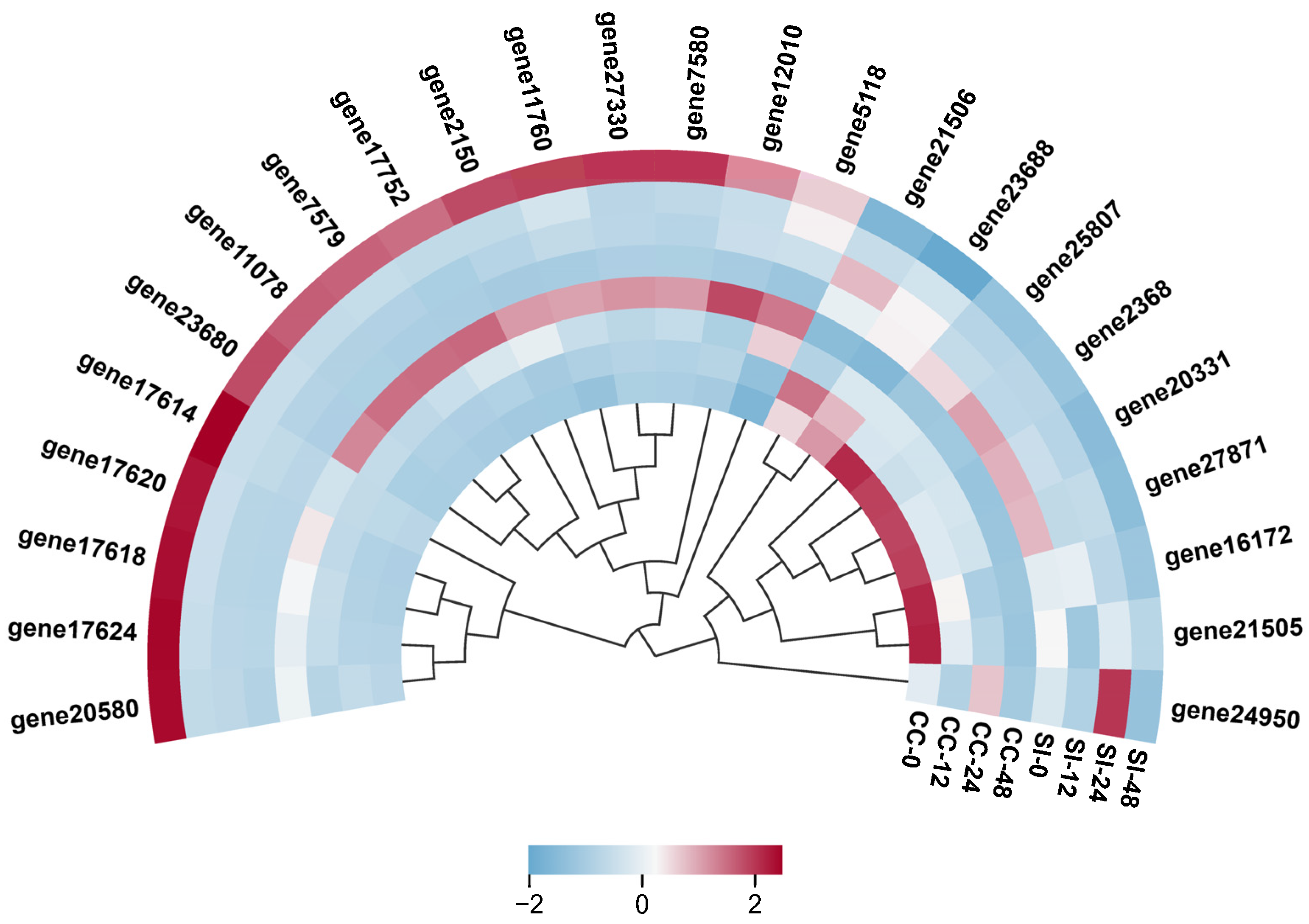
2.6. Co-Expression Network Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Pollen Vitality Identification
4.3. Field Pollination Treatments
4.4. RNA Extraction, Transcriptome Sequencing, and Real-Time Quantitative PCR
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Coulibaly, D.; Gao, F.; Bai, Y.; Ouma, K.O.; Antwi-Boasiako, A.; Zhou, P.; Iqbal, S.; Bah, A.A.; Huang, X.; Diarra, S.T.; et al. Molecular Research Progress on Gametophytic Self-Incompatibility in Rosaceae Species. Horticulturae 2024, 10, 1101. [Google Scholar] [CrossRef]
- Takayama, S.; Isogai, A. Self-incompatibility in plants. Annu. Rev. Plant Biol. 2005, 56, 467–489. [Google Scholar] [CrossRef]
- Li, W.; Meng, D.; Gu, Z.; Yang, Q.; Yuan, H.; Li, Y.; Chen, Q.; Yu, J.; Liu, C.; Li, T. Apple S-RNase triggers inhibition of tRNA aminoacylation by interacting with a soluble inorganic pyrophosphatase in growing self-pollen tubes in vitro. New Phytol. 2018, 218, 579–593. [Google Scholar] [CrossRef]
- Matsumoto, D.; Yamane, H.; Abe, K.; Tao, R. Identification of a Skp1-Like Protein Interacting with SFB, the Pollen S Determinant of the Gametophytic Self-Incompatibility in Prunus. Plant Physiol. 2012, 159, 1252–1262. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Esumi, T.; Hirakawa, H.; Tanaka, H.; Itai, A.; Ghelfi, A.; Nagasaki, H.; Isobe, S. Phased genome sequence of an interspecific hybrid flowering cherry, ‘Somei-Yoshino’ (Cerasus × yedoensis). DNA Res. 2019, 26, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Deng, C.H.; Cornille, A.; Marande, W.; López-Girona, E.; Foster, T.; Bowatte, D.; Chen, T.-H.; Chagné, D.; Schaffer, R.J.; et al. Characterisation of the Gillenia S-locus provides insight into evolution of the nonself-recognition self-incompatibility system in apple. Sci. Rep. 2025, 15, 14630. [Google Scholar] [CrossRef]
- De Franceschi, P.; Dondini, L.; Sanzol, J. Molecular bases and evolutionary dynamics of self-incompatibility in the Pyrinae (Rosaceae). J. Exp. Bot. 2012, 63, 4015–4032. [Google Scholar] [CrossRef]
- Feng, C.; Zhang, X.; Wang, J.; Wu, C.; Wang, W.; Yan, G.; Zhou, Y.; Zhang, K.; Duan, X. Characterization of volatile profiles in cherry fruits: Integration of E-nose and HS-SPME-GC–MS. Food Chem. X 2025, 28, 102632. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Duan, X.W.; Wang, J.; Yan, G.H.; Zhang, X.M.; Wu, C.B.; Zhou, Y.; Zhang, X.; Zhang, K.C. Pollen germination contributes to providing guidance for cross pollination of sweet cherry (Prunus avium L.). Acta Hortic. 2024, 1408, 463–468. [Google Scholar] [CrossRef]
- Schuster, M. Incompatible (S-) genotypes of sweet cherry cultivars (Prunus avium L.). Sci. Hortic. 2012, 148, 59–73. [Google Scholar] [CrossRef]
- Matsumoto, D.; Tao, R. Yeast Two-Hybrid Screening for the General Inhibitor Detoxifying S-Rnase in Prunus. Acta Hortic. 2012, 967, 167–170. [Google Scholar] [CrossRef]
- Sonneveld, T.; Tobutt, K.R.; Vaughan, S.P.; Robbins, T.P. Loss of Pollen-S Function in Two Self-Compatible Selections of Prunus avium Is Associated with Deletion/Mutation of an S Haplotype–Specific F-Box Gene. Plant Cell 2005, 17, 37–51. [Google Scholar] [CrossRef]
- Pratas, M.I.; Aguiar, B.; Vieira, J.; Nunes, V.; Teixeira, V.; Fonseca, N.A.; Iezzoni, A.; van Nocker, S.; Vieira, C.P. Inferences on specificity recognition at the Malus×domestica gametophytic self-incompatibility system. Sci. Rep. 2018, 8, 1717. [Google Scholar] [CrossRef]
- Okada, K.; Tonaka, N.; Taguchi, T.; Ichikawa, T.; Sawamura, Y.; Nakanishi, T.; Takasaki-Yasuda, T. Related polymorphic F-box protein genes between haplotypes clustering in the BAC contig sequences around the S-RNase of Japanese pear. J. Exp. Bot. 2010, 62, 1887–1902. [Google Scholar] [CrossRef]
- Sonneveld, T.; Robbins, T.P.; Bošković, R.; Tobutt, K.R. Cloning of six cherry self-incompatibility alleles and development of allele-specific PCR detection. Theor. Appl. Genet. 2001, 102, 1046–1055. [Google Scholar] [CrossRef]
- Marchese, A.; Bošković, R.I.; Caruso, T.; Raimondo, A.; Cutuli, M.; Tobutt, K.R. A new self-compatibility haplotype in the sweet cherry ‘Kronio’, S5′, attributable to a pollen-part mutation in the SFB gene. J. Exp. Bot. 2007, 58, 4347–4356. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhang, X.; Zhang, K.; Jiang, L.; Zhang, L. Development of a simple molecular marker specific for detecting the self-compatibleS4′ haplotype in sweet cherry (Prunus avium L.). Plant Mol. Biol. Rep. 2004, 22, 387–398. [Google Scholar] [CrossRef]
- Muñoz-Espinoza, C.; Espinosa, E.; Bascuñán, R.; Tapia, S.; Meneses, C.; Almeida, A.M. Development of a molecular marker for self-compatible S4′ haplotype in sweet cherry (Prunus avium L.) using high-resolution melting. Plant Breed. 2017, 136, 987–993. [Google Scholar] [CrossRef]
- Marchese, A.; Tobutt, K.R.; Raimondo, A.; Motisi, A.; Bošković, R.I.; Clarke, J.; Caruso, T. Morphological characteristics, microsatellite fingerprinting and determination of incompatibility genotypes of Sicilian sweet cherry cultivars. J. Hortic. Sci. Biotechnol. 2007, 82, 41–48. [Google Scholar] [CrossRef]
- Ushijima, K.; Yamane, H.; Watari, A.; Kakehi, E.; Ikeda, K.; Hauck, N.R.; Iezzoni, A.F.; Tao, R. The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J. 2004, 39, 573–586. [Google Scholar] [CrossRef]
- Ushijima, K.; Sassa, H.; Dandekar, A.M.; Gradziel, T.M.; Tao, R.; Hirano, H. Structural and Transcriptional Analysis of the Self-Incompatibility Locus of Almond: Identification of a Pollen-Expressed F-Box Gene with Haplotype-Specific Polymorphism. Plant Cell 2003, 15, 771–781. [Google Scholar] [CrossRef]
- Akagi, T.; Henry, I.M.; Morimoto, T.; Tao, R. Insights into the Prunus -Specific S-RNase-Based Self-Incompatibility System from a Genome-Wide Analysis of the Evolutionary Radiation of S Locus-Related F-box Genes. Plant Cell Physiol. 2016, 57, 1281–1294. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Iezzoni, A.F. The S-RNase-based gametophytic self-incompatibility system in Prunus exhibits distinct genetic and molecular features. Sci. Hortic. 2010, 124, 423–433. [Google Scholar] [CrossRef]
- Matsumoto, D.; Tao, R. Recognition of a wide-range of S-RNases by S locus F-box like 2, a general-inhibitor candidate in the Prunus-specific S-RNase-based self-incompatibility system. Plant Mol. Biol. 2016, 91, 459–469. [Google Scholar] [CrossRef]
- Li, Y.; Duan, X.; Wu, C.; Yu, J.; Liu, C.; Wang, J.; Zhang, X.; Yan, G.; Jiang, F.; Li, T.; et al. Ubiquitination of S4-RNase by S-LOCUS F-BOX LIKE2 Contributes to Self-Compatibility of Sweet Cherry ‘Lapins’. Plant Physiol. 2020, 184, 1702–1716. [Google Scholar] [CrossRef]
- Wünsch, A.; Tao, R.; Hormaza, J.I. Self-compatibility in ‘Cristobalina’ sweet cherry is not associated with duplications or modified transcription levels of S-locus genes. Plant Cell Rep. 2010, 29, 715–721. [Google Scholar] [CrossRef]
- Ono, K.; Akagi, T.; Morimoto, T.; Wünsch, A.; Tao, R. Genome Re-Sequencing of Diverse Sweet Cherry (Prunus avium) Individuals Reveals a Modifier Gene Mutation Conferring Pollen-Part Self-Compatibility. Plant Cell Physiol. 2018, 59, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Vasiliev, D.; Greenwood, S. The role of climate change in pollinator decline across the Northern Hemisphere is underestimated. Sci. Total Environ. 2021, 775, 145788. [Google Scholar] [CrossRef]
- Yu, J.; Wang, B.; Fan, W.; Fan, S.; Xu, Y.; Liu, C.; Lv, T.; Liu, W.; Wu, L.; Xian, L.; et al. Polyamines Involved in Regulating Self-Incompatibility in Apple. Genes 2021, 12, 1797. [Google Scholar] [CrossRef]
- Gradziel, T.M. Transfer of Self-Fruitfulness to Cultivated Almond from Peach and Wild Almond. Horticulturae 2022, 8, 965. [Google Scholar] [CrossRef]
- Marić, S.; Radičević, S.; Sîrbu, S.; Malchev, S.; Filyova, P.; Milošević, N.; Glišić, I.; Đorđević, M.; Banović Đeri, B. Characterization of self-incompatibility genotypes in 48 sweet cherry cultivars and 21 promising hybrids bred in the Balkan region. Euphytica 2025, 221, 50. [Google Scholar] [CrossRef]
- Gu, C.; Xu, Y.; Wu, L.; Wang, X.; Qi, K.; Qiao, X.; Wang, Z.; Li, Q.; He, M.; Zhang, S. Long-read genome sequencing reveals the sequence characteristics of pear self-incompatibility locus. Mol. Hortic. 2025, 5, 13. [Google Scholar] [CrossRef]
- Abdallah, D.; Baraket, G.; Perez, V.; Salhi Hannachi, A.; Hormaza, J.I. Self-compatibility in peach [Prunus persica (L.) Batsch]: Patterns of diversity surrounding the S-locus and analysis of SFB alleles. Hortic. Res. 2020, 7, 170. [Google Scholar] [CrossRef]
- Li, K.; Wang, Y.; Qu, H. RNA-Seq analysis of compatible and incompatible styles of Pyrus species at the beginning of pollination. Plant Mol. Biol. 2020, 102, 287–306. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, M.; Meng, H.-L.; Yang, J.; Wei, X.; Yang, S.-C. Molecular cloning and expression analysis of SRLK1 gene in self-incompatible Asteraceae species Erigeron breviscapus. Mol. Biol. Rep. 2019, 46, 3157–3165. [Google Scholar] [CrossRef]
- Eschrig, S.; Schäffer, M.; Shu, L.-J.; Illig, T.; Eibel, S.; Fernandez, A.; Ranf, S. LORE receptor homomerization is required for 3-hydroxydecanoic acid-induced immune signaling and determines the natural variation of immunosensitivity within the Arabidopsis genus. New Phytol. 2024, 242, 2163–2179. [Google Scholar] [CrossRef]
- Morris, E.R.; Walker, J.C. Receptor-like protein kinases: The keys to response. Curr. Opin. Plant Biol. 2003, 6, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Zhao, T.; Yang, Z.; Liang, L.; Ma, W.; Wang, G.; Ma, Q. Stigmatic Transcriptome Analysis of Self-Incompatible and Compatible Pollination in Corylus heterophylla Fisch. × Corylus avellana L. Front. Plant Sci. 2022, 13, 2022. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Jia, J.; Huang, X.; Yan, X.; Cheng, L.; Chen, S.; Li, X.; Peng, X.; Liu, G. The large-scale investigation of gene expression in Leymus chinensis stigmas provides a valuable resource for understanding the mechanisms of poaceae self-incompatibility. BMC Genom. 2014, 15, 399. [Google Scholar] [CrossRef] [PubMed]
- Denninger, P.; Bleckmann, A.; Lausser, A.; Vogler, F.; Ott, T.; Ehrhardt, D.W.; Frommer, W.B.; Sprunck, S.; Dresselhaus, T.; Grossmann, G. Male–female communication triggers calcium signatures during fertilization in Arabidopsis. Nat. Commun. 2014, 5, 4645. [Google Scholar] [CrossRef]
- Jiang, X.; Gao, Y.; Zhou, H.; Chen, J.; Wu, J.; Zhang, S. Apoplastic calmodulin promotes self-incompatibility pollen tube growth by enhancing calcium influx and reactive oxygen species concentration in Pyrus pyrifolia. Plant Cell Rep. 2014, 33, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, S.; Wang, Y.; Shang, Y.; Qi, Z.; Lin, H.; Niu, L. Transcriptomic Analysis of Self-Incompatibility in Alfalfa. Plants 2024, 13, 875. [Google Scholar] [CrossRef]
- Li, C.; Lu, M.; Zhou, J.; Wang, S.; Long, Y.; Xu, Y.; Tan, X. Transcriptome Analysis of the Late-Acting Self-Incompatibility Associated with RNase T2 Family in Camellia oleifera. Plants 2023, 12, 1932. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jia, J.; Cheng, L.; Zhao, P.; Qi, D.; Yang, W.; Liu, H.; Dong, X.; Li, X.; Liu, G. Transcriptomic Analysis Reveals a Comprehensive Calcium- and Phytohormone-Dominated Signaling Response in Leymus chinensis Self-Incompatibility. Int. J. Mol. Sci. 2019, 20, 2356. [Google Scholar] [CrossRef]
- Chen, D.; Zhao, J. Free IAA in stigmas and styles during pollen germination and pollen tube growth of Nicotiana tabacum. Physiol. Plant. 2008, 134, 202–215. [Google Scholar] [CrossRef]
- Wu, J.-Z.; Lin, Y.; Zhang, X.-L.; Pang, D.-W.; Zhao, J. IAA stimulates pollen tube growth and mediates the modification of its wall composition and structure in Torenia fournieri. J. Exp. Bot. 2008, 59, 2529–2543. [Google Scholar] [CrossRef]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis Enzyme Family That Conjugates Amino Acids to Indole-3-Acetic Acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef]
- An, H.; Zhang, J.; Xu, F.; Jiang, S.; Zhang, X. Transcriptomic profiling and discovery of key genes involved in adventitious root formation from green cuttings of highbush blueberry (Vaccinium corymbosum L.). BMC Plant Biol. 2020, 20, 182. [Google Scholar] [CrossRef]
- Chai, L.; Tudor, R.L.; Poulter, N.S.; Wilkins, K.A.; Eaves, D.J.; Franklin, F.C.H.; Franklin-Tong, V.E. MAP Kinase PrMPK9-1 Contributes to the Self-Incompatibility Response. Plant Physiol. 2017, 174, 1226–1237. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lu, M.; Yu, S.; Liu, Y.; Yang, J.; Tan, X. In-Depth Understanding of Camellia oleifera Self-Incompatibility by Comparative Transcriptome, Proteome and Metabolome. Int. J. Mol. Sci. 2020, 21, 1600. [Google Scholar] [CrossRef]
- Hu, D.; Lin, D.; Yi, S.; Gao, S.; Lei, T.; Li, W.; Xu, T. Comparative stigmatic transcriptomics reveals self and cross pollination responses to heteromorphic incompatibility in Plumbago auriculata Lam. Front. Genet. 2024, 15, 1372644. [Google Scholar] [CrossRef] [PubMed]
- Lei, R.; Li, X.; Ma, Z.; Lv, Y.; Hu, Y.; Yu, D. Arabidopsis WRKY2 and WRKY34 transcription factors interact with VQ20 protein to modulate pollen development and function. Plant J. 2017, 91, 962–976. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Dai, H.; Wang, X.; Wang, C.; Zeng, W.; Huang, J.; Duan, Q. Ethylene negatively mediates self-incompatibility response in Brassica rapa. Biochem. Biophys. Res. Commun. 2020, 525, 600–606. [Google Scholar] [CrossRef]
- Feng, C.; Guo, Q.; Wu, C.; Wang, J.; Zhang, X.; Yan, G.; Zhou, Y.; Wang, W.; Xue, Z.; Zhang, K.; et al. Identification and characteristic analysis of PavUGT48 as a novel UDP-glycosyltransferase with dual functions on anthocyanin and amygdalin biosynthesis in sweet cherry. Int. J. Biol. Macromol. 2025, 309, 143062. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, Y.; Chen, H.; He, C.; Sun, L.; Zhang, X.; Xu, Z.; Liu, H. Integrated omics profiles for exploring the potential mechanism underlying aroma formation in the terpenoid-rich aromatic plant Opisthopappus taihangensis and the bioactivity of its leaf essential oil. Agric. Commun. 2024, 2, 100061. [Google Scholar] [CrossRef]
- Feng, C.; Zhang, X.; Du, B.; Xiao, Y.; Wang, Y.; Sun, Y.; Zhou, X.; Wang, C.; Liu, Y.; Li, T.-H. MicroRNA156ab regulates apple plant growth and drought tolerance by targeting transcription factor MsSPL13. Plant Physiol. 2023, 192, 1836–1857. [Google Scholar] [CrossRef]
- Yang, J.; Ma, Y.; Li, Z.; Zhong, H.; Wang, X.; Zeng, T. Mechanistic elucidation of the role of Salicylhydroxamic acid methyl ester in resistance of kiwifruit to Botrytis cinerea. Technol. Hortic. 2024, 4, e026. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Pollination Combination | No. of Flowers | No. of Fruits | Fruit Set Rate |
|---|---|---|---|
| ‘Tieton’ × ‘Tieton’ | 50 | 1 | 2% |
| ‘Tieton’ × ‘Rainier’ | 50 | 28 | 56% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, C.; Wu, C.; Wang, J.; Wang, W.; Yan, G.; Zhou, Y.; Zhang, K.; Zhang, X.; Duan, X. Identification of Self-Incompatibility Related Genes in Sweet Cherry Based on Transcriptomic Analysis. Biology 2025, 14, 1125. https://doi.org/10.3390/biology14091125
Feng C, Wu C, Wang J, Wang W, Yan G, Zhou Y, Zhang K, Zhang X, Duan X. Identification of Self-Incompatibility Related Genes in Sweet Cherry Based on Transcriptomic Analysis. Biology. 2025; 14(9):1125. https://doi.org/10.3390/biology14091125
Chicago/Turabian StyleFeng, Chen, Chuanbao Wu, Jing Wang, Wei Wang, Guohua Yan, Yu Zhou, Kaichun Zhang, Xiaoming Zhang, and Xuwei Duan. 2025. "Identification of Self-Incompatibility Related Genes in Sweet Cherry Based on Transcriptomic Analysis" Biology 14, no. 9: 1125. https://doi.org/10.3390/biology14091125
APA StyleFeng, C., Wu, C., Wang, J., Wang, W., Yan, G., Zhou, Y., Zhang, K., Zhang, X., & Duan, X. (2025). Identification of Self-Incompatibility Related Genes in Sweet Cherry Based on Transcriptomic Analysis. Biology, 14(9), 1125. https://doi.org/10.3390/biology14091125






