Transcriptome of Two-Hybrid Poplar (Populus alba × P. tomentiglandulosa) During Adventitious Root Formation After Stem Cutting
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site and Plant Materials
2.2. RNA Extraction and cDNA Library Construction
2.3. RNA-Seq Read Processing
2.4. Gene Ontology Enrichment Analysis
2.5. Validating RNA-Seq Results Using qRT-PCR
2.6. Data
3. Results
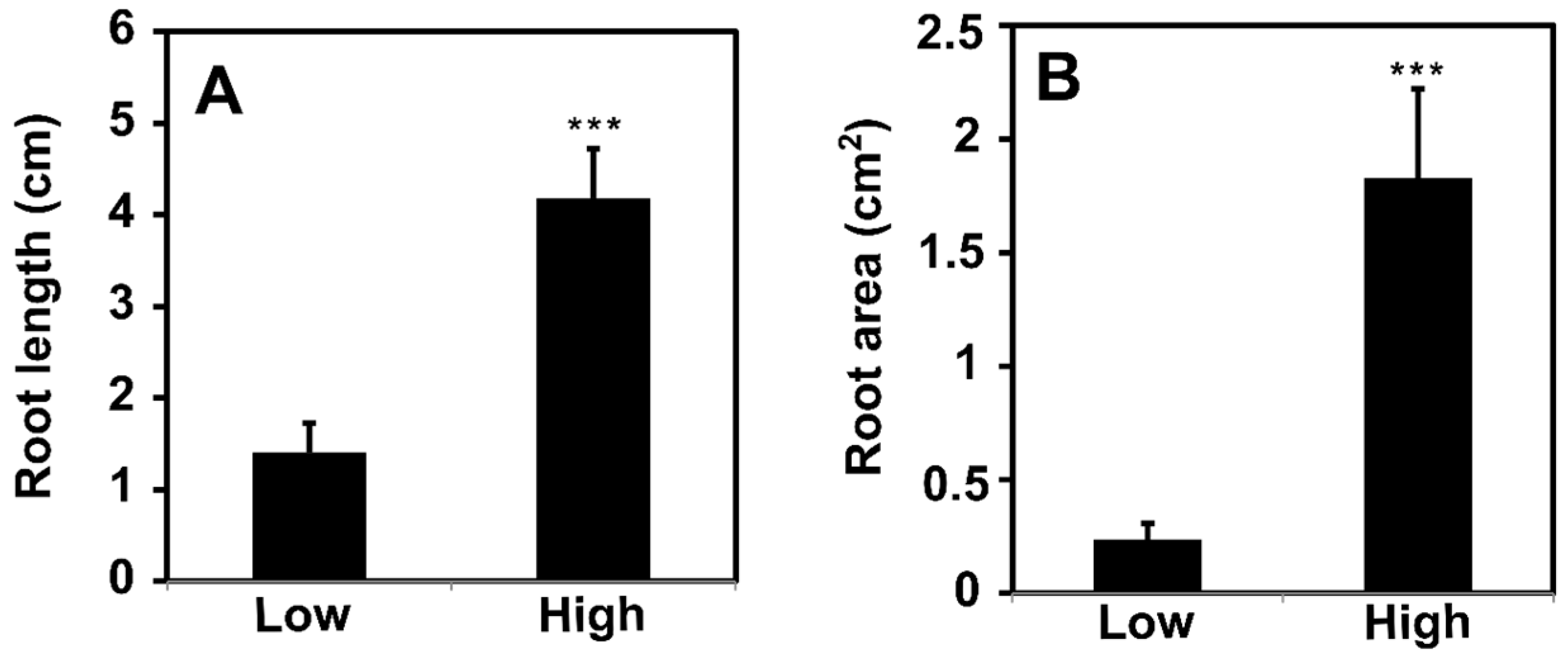
3.1. Phenotype of High- and Low-Rooting Ability Poplar Groups
3.2. De Novo Assembly of High- and Low-Rooting Ability Poplar Groups
3.3. Identifying DEGs Between High- and Low-Rooting Poplar Groups
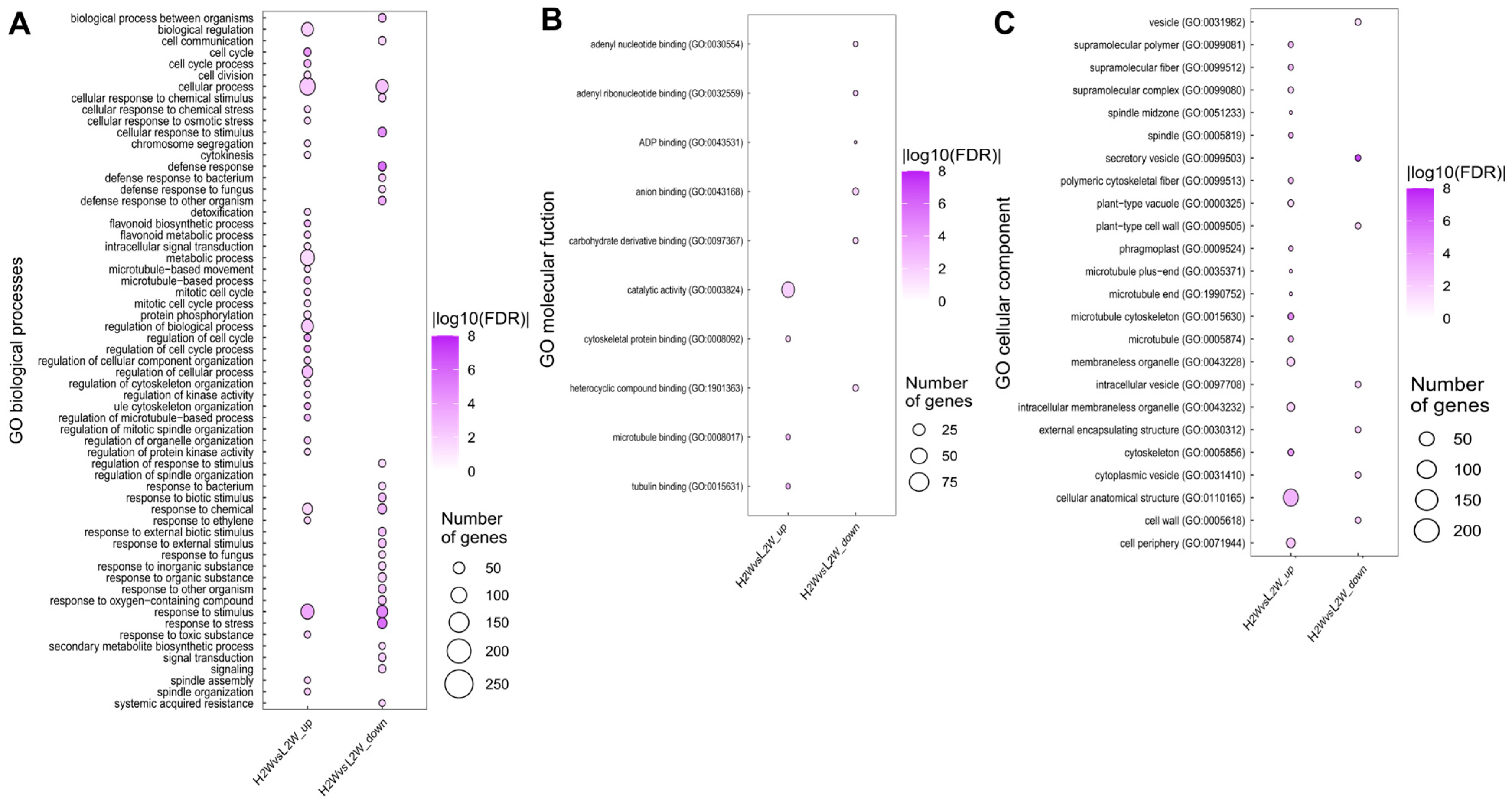
3.4. GO Enrichment Analysis of DEGs Between High- and Low-Rooting Poplar Groups
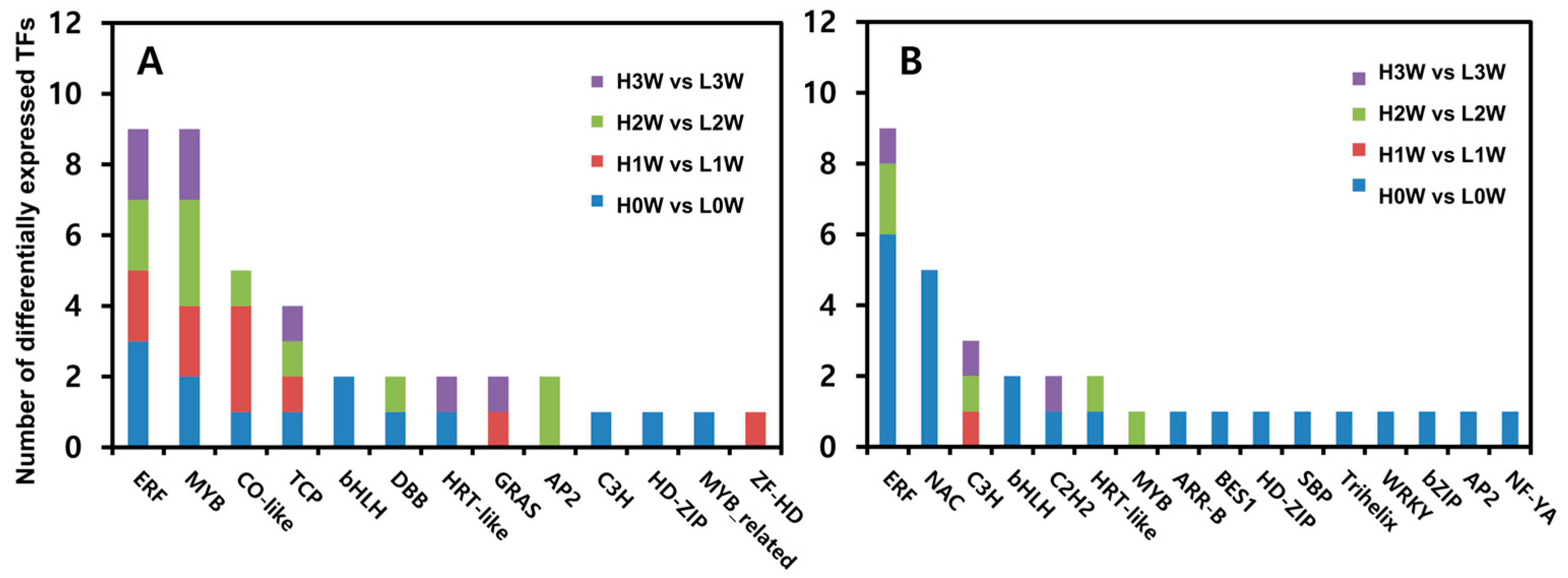
3.5. Rooting Ability-Related Transcription Factors
3.6. qRT-PCR Validation of DEG Expression Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jung, Y.H.; Cho, H.J.; Lee, J.-S.; Noh, E.W.; Park, O.K.; Kim, K.H. Evaluation of a transgenic poplar as a potential biomass crop for biofuel production. Bioresour. Technol. 2013, 129, 639–641. [Google Scholar] [CrossRef] [PubMed]
- Renninger, H. Aboveground woody biomass estimation of young bioenergy plantations of populus and its hybrids using LIDAR remote sensing. Trees For. People 2024, 18, 100665. [Google Scholar]
- Bae, E.-K.; Kang, M.-J.; Lee, S.-J.; Park, E.-J.; Kim, K.-T. Chromosome-level genome assembly of the Asian aspen Populus davidiana Dode. Sci. Data 2023, 10, 431. [Google Scholar] [CrossRef]
- Shi, T.; Zhang, X.; Hou, Y.; Jia, C.; Dan, X.; Zhang, Y.; Jiang, Y.; Lai, Q.; Feng, J.; Feng, J. The super-pangenome of Populus unveils genomic facets for its adaptation and diversification in widespread forest trees. Mol. Plant 2024, 17, 725–746. [Google Scholar] [CrossRef] [PubMed]
- Confalonieri, M.; Balestrazzi, A.; Bisoffi, S.; Carbonera, D. In vitro culture and genetic engineering of Populus spp.: Synergy for forest tree improvement. Plant Cell Tissue Organ Cult. 2003, 72, 109–138. [Google Scholar] [CrossRef]
- Petersen, L.A.; Phipps, H.M. Water soaking pretreatment improves rooting and early survival of hardwood cuttings of some populus clones. Tree Plant. Notes 1976, 27, 20–22. [Google Scholar]
- Bannoud, F.; Bellini, C. Adventitious rooting in Populus species: Update and perspectives. Front. Plant Sci. 2021, 12, 668837. [Google Scholar] [CrossRef]
- Ahkami, A.H. Systems biology of root development in Populus: Review and perspectives. Plant Sci. 2023, 335, 111818. [Google Scholar] [CrossRef]
- Zhao, X.; Zheng, H.; Li, S.; Yang, C.; Jiang, J.; Liu, G. The rooting of poplar cuttings: A review. New For. 2014, 45, 21–34. [Google Scholar] [CrossRef]
- Coffman, C.; Gentner, W. Greenhouse propagation of Cannabis sativa L. by vegetative cuttings. Econ. Bot. 1979, 33, 124–127. [Google Scholar] [CrossRef]
- LeBude, A.V.; Goldfarb, B.; Blazich, F.A. Mist, vapor pressure deficit, and cutting water potential influence rooting of stem cuttings of loblolly pine. HortScience 2004, 39, 890D–890. [Google Scholar] [CrossRef]
- Evlakov, P.; Tseplyaev, A.; Popova, A.; Zapletin, V.; Ryzhkova, V.; Repnikova, L.; Zhuzhukin, K. Influence of Container Volume and Cuttings Size on the Growth Parameters of Seedlings with a Closed Root System of Two Poplar Genotypes in the Voronezh Region. Int. J. Plant Biol. 2025, 16, 49. [Google Scholar] [CrossRef]
- Ye, X.; Busov, V.; Zhao, N.; Meilan, R.; McDonnell, L.M.; Coleman, H.D.; Mansfield, S.D.; Chen, F.; Li, Y.; Cheng, Z.-M. Transgenic Populus trees for forest products, bioenergy, and functional genomics. Crit. Rev. Plant Sci. 2011, 30, 415–434. [Google Scholar] [CrossRef]
- Xuemei, C.; Hongbing, G. Studies on endogenous hormone levels in cuttings of three poplar species during rooting process. Sci. Silvae Sin. 1994, 30, 1–7. [Google Scholar]
- Zalesny, J.; Ronald, S.; Riemenschneider, D.E.; Hall, R.B. Early rooting of dormant hardwood cuttings of Populus: Analysis of quantitative genetics and genotype× environment interactions. Can. J. For. Res. 2005, 35, 918–929. [Google Scholar] [CrossRef]
- Steffens, B.; Rasmussen, A. The physiology of adventitious roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef]
- Jin, X.; Zhao, K.; Hu, J.; Gailing, O.; Zhou, L.; Du, S.; Han, Y.; Wang, S. PagMYB73A enhances poplar salt tolerance by facilitating adventitious roots elongation and stomata density. For. Res. 2023, 4, e003. [Google Scholar] [CrossRef]
- Mishra, P.; Roggen, A.; Ljung, K.; Albani, M.C.; Vayssières, A. Adventitious rooting in response to long-term cold: A possible mechanism of clonal growth in alpine perennials. Front. Plant Sci. 2024, 15, 1352830. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Subramanian, S.; Stacey, G.; Yu, O. Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J. 2009, 57, 171–183. [Google Scholar] [CrossRef]
- Ribeiro, C.L.; Silva, C.M.; Drost, D.R.; Novaes, E.; Novaes, C.R.; Dervinis, C.; Kirst, M. Integration of genetic, genomic and transcriptomic information identifies putative regulators of adventitious root formation in Populus. BMC Plant Biol. 2016, 16, 66. [Google Scholar] [CrossRef]
- Sun, P.; Jia, H.; Zhang, Y.; Li, J.; Lu, M.; Hu, J. Deciphering genetic architecture of adventitious root and related shoot traits in Populus using QTL mapping and RNA-Seq data. Int. J. Mol. Sci. 2019, 20, 6114. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Delaruelle, C.; Martin, D.; Encelot, N.; Martin, F. The poplar root transcriptome: Analysis of 7000 expressed sequence tags. FEBS Lett. 2003, 542, 37–41. [Google Scholar] [CrossRef]
- Islam, M.S.; Ghimire, A.; Lay, L.; Khan, W.; Lee, J.-D.; Song, Q.; Jo, H.; Kim, Y. Identification of quantitative trait loci controlling root morphological traits in an interspecific soybean population using 2D imagery data. Int. J. Mol. Sci. 2024, 25, 4687. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wei, J.; Tian, R.; Zeng, Z.; Tang, H.; Liu, Y.; Xu, Q.; Deng, M.; Jiang, Q.; Chen, G. A major quantitative trait locus for wheat total root length associated with precipitation distribution. Front. Plant Sci. 2022, 13, 995183. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Bernacki, M.J.; Czarnocka, W.; Witoń, D.; Rusaczonek, A.; Szechyńska-Hebda, M.; Ślesak, I.; Dąbrowska-Bronk, J.; Karpiński, S. ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) affects development, photosynthesis, and hormonal homeostasis in hybrid aspen (Populus tremula L.× P. tremuloides). J. Plant Physiol. 2018, 226, 91–102. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Polle, A.; Klein, T.; Kettner, C. Impact of cadmium on young plants of Populus euphratica and P.× canescens, two poplar species that differ in stress tolerance. New For. 2013, 44, 13–22. [Google Scholar] [CrossRef]
- Adem, M.; Sharma, L.; Shekhawat, G.S.; Šafranek, M.; Jásik, J. Auxin Signaling Transportation and Regulation during Adventitious Root Formation. Curr. Plant Biol. 2024, 40, 100385. [Google Scholar] [CrossRef]
- Arya, A.; Husen, A. Role of various auxins in adventitious root formation. In Environmental, Physiological and Chemical Controls of Adventitious Rooting in Cuttings; Elsevier: Amsterdam, The Netherlands, 2022; pp. 213–238. [Google Scholar]
- Liu, S.; Li, X.; Xu, L.; Zhang, G. Hormone functions in adventitious root formation during cutting propagation of woody plants. J. Plant Res. 2024, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bustillo-Avendaño, E.; Ibáñez, S.; Sanz, O.; Sousa Barros, J.A.; Gude, I.; Perianez-Rodriguez, J.; Micol, J.L.; Del Pozo, J.C.; Moreno-Risueno, M.A.; Pérez-Pérez, J.M. Regulation of hormonal control, cell reprogramming, and patterning during de novo root organogenesis. Plant Physiol. 2018, 176, 1709–1727. [Google Scholar] [CrossRef]
- Bisgrove, S.R.; Lee, Y.-R.J.; Liu, B.; Peters, N.T.; Kropf, D.L. The microtubule plus-end binding protein EB1 functions in root responses to touch and gravity signals in Arabidopsis. Plant Cell 2008, 20, 396–410. [Google Scholar] [CrossRef]
- Molines, A.T.; Marion, J.; Chabout, S.; Besse, L.; Dompierre, J.P.; Mouille, G.g.; Coquelle, F.d.r.M. EB1 contributes to microtubule bundling and organization, along with root growth, in Arabidopsis thaliana. Biol. Open 2018, 7, bio030510. [Google Scholar] [CrossRef]
- Fukuda, Y.; Hirao, T.; Mishima, K.; Ohira, M.; Hiraoka, Y.; Takahashi, M.; Watanabe, A. Transcriptome dynamics of rooting zone and aboveground parts of cuttings during adventitious root formation in Cryptomeria japonica D. Don. BMC Plant Biol. 2018, 18, 201. [Google Scholar] [CrossRef]
- Li, S.-W.; Shi, R.-F.; Leng, Y. De novo characterization of the mung bean transcriptome and transcriptomic analysis of adventitious rooting in seedlings using RNA-Seq. PLoS ONE 2015, 10, e0132969. [Google Scholar] [CrossRef]
- Brinker, M.; van Zyl, L.; Liu, W.; Craig, D.; Sederoff, R.R.; Clapham, D.H.; von Arnold, S. Microarray analyses of gene expression during adventitious root development in Pinus contorta. Plant Physiol. 2004, 135, 1526–1539. [Google Scholar] [CrossRef]
- Rahim, M.A.; Zhang, X.; Busatto, N. Phenylpropanoid biosynthesis in plants. Front. Plant Sci. 2023, 14, 1230664. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, Y.; Xu, C.; Mei, X. Contribution of phenylpropanoid metabolism to plant development and stress responses. J. Integr. Plant Biol. 2024, 15, 1456913. [Google Scholar] [CrossRef]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef]
- Barber, M.S.; Mitchell, H.J. Regulation of phenylpropanoid metabolism in relation to lignin biosynthesis in plants. In International Review of Cytology; Elsevier: Amsterdam, The Netherlands, 1997; Volume 172, pp. 243–293. [Google Scholar]
- Dos Santos, W.D.; Ferrarese, M.d.L.L.; Finger, A.; Teixeira, A.C.; Ferrarese-Filho, O. Lignification and related enzymes in Glycine max root growth-inhibition by ferulic acid. J. Chem. Ecol. 2004, 30, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Bergonci, T.; Fomsgaard, I.S.; Kjaer, K.H.; Paponov, I.A. Hormone–Flavonoid Patterns in Two Genotypes of Campanula portenschlagiana with Distinct Adventitious Rooting Competence. Horticulturae 2023, 9, 121. [Google Scholar] [CrossRef]
- Daryanavard, H.; Postiglione, A.E.; Mühlemann, J.K.; Muday, G.K. Flavonols modulate plant development, signaling, and stress responses. Curr. Opin. Plant Biol. 2023, 72, 102350. [Google Scholar] [CrossRef]
- Djordjevic, M.; Mathesius, U.; Arioli, T.; Weinman, J.; Gärtner, E. Chalcone synthase gene expression in transgenic subterranean clover correlates with localised accumulation of flavonoids. Funct. Plant Biol. 1997, 24, 119–132. [Google Scholar] [CrossRef]
- Morris, A.C.; Djordjevic, M.A. The Rhizobium leguminosarum biovar trifolii ANU794 induces novel developmental responses on the subterranean clover cultivar Woogenellup. Mol. Plant-Microbe Interact. 2006, 19, 471–479. [Google Scholar] [CrossRef]
- Lu, X.; Chen, X.; Liu, J.; Zheng, M.; Liang, H. Integrating histology and phytohormone/metabolite profiling to understand rooting in yellow camellia cuttings. Plant Sci. 2024, 346, 112160. [Google Scholar] [CrossRef]
- Santos-Rufo, A.; Rodríguez-Solana, R.; Fernández-Recamales, Á.; Sayago-Gómez, A.; Weiland-Ardáiz, C. Machine learning unveils the action of different endogenous phenolic compounds present or formed along the rooting development in olive stem cuttings. Sci. Hortic. 2024, 331, 113175. [Google Scholar] [CrossRef]
- Jin, Z.; Jiang, W.; Luo, Y.; Huang, H.; Yi, D.; Pang, Y. Analyses on flavonoids and transcriptome reveals key MYB gene for proanthocyanidins regulation in Onobrychis Viciifolia. Front. Plant Sci. 2022, 13, 941918. [Google Scholar] [CrossRef]
- Wang, W.-L.; Wang, Y.-X.; Li, H.; Liu, Z.-W.; Cui, X.; Zhuang, J. Two MYB transcription factors (CsMYB2 and CsMYB26) are involved in flavonoid biosynthesis in tea plant [Camellia sinensis (L.) O. Kuntze]. BMC Plant Biol. 2018, 18, 288. [Google Scholar] [CrossRef] [PubMed]
- Imin, N.; Nizamidin, M.; Wu, T.; Rolfe, B.G. Factors involved in root formation in Medicago truncatula. J. Exp. Bot. 2007, 58, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Liu, Y.; Wang, T.; Jiang, P.; Wen, W.; Nie, J. Combined transcriptomic and metabolomic analyses identifies CsERF003, a citrus ERF transcription factor, as flavonoid activator. Plant Sci. 2023, 334, 111762. [Google Scholar] [CrossRef]
- McKhann, H.I.; Hirsch, A.M. Isolation of chalcone synthase and chalcone isomerase cDNAs from alfalfa (Medicago sativa L.): Highest transcript levels occur in young roots and root tips. Plant Mol. Biol. 1994, 24, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Park, N.I.; Li, X.; Kim, Y.K.; Lee, S.Y.; Park, S.U. Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis. Bioresour. Technol. 2010, 101, 9715–9722. [Google Scholar] [CrossRef]
- Park, N.I.; Xu, H.; Li, X.; Kim, S.-J.; Park, S.U. Enhancement of flavone levels through overexpression of chalcone isomerase in hairy root cultures of Scutellaria baicalensis. Funct. Integr. Genom. 2011, 11, 491–496. [Google Scholar] [CrossRef]
- Zhang, H.-C.; Liu, J.-M.; Lu, H.-Y.; Gao, S.-L. Enhanced flavonoid production in hairy root cultures of Glycyrrhiza uralensis Fisch by combining the over-expression of chalcone isomerase gene with the elicitation treatment. Plant Cell Rep. 2009, 28, 1205–1213. [Google Scholar] [CrossRef]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.-S.; Amasino, R.; Scheres, B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef]
- Tao, G.-Y.; Xie, Y.-H.; Li, W.-F.; Li, K.-P.; Sun, C.; Wang, H.-M.; Sun, X.-M. LkARF7 and LkARF19 overexpression promote adventitious root formation in a heterologous poplar model by positively regulating LkBBM1. Commun. Biol. 2023, 6, 372. [Google Scholar] [CrossRef]
- Chen, J.; Tomes, S.; Gleave, A.P.; Hall, W.; Luo, Z.; Xu, J.; Yao, J.-L. Significant improvement of apple (Malus domestica Borkh.) transgenic plant production by pre-transformation with a Baby boom transcription factor. Hortic. Res. 2022, 9, uhab014. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.-M.; van Lammeren, A.A.; Miki, B.L. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef]
- Shu, W.; Zhou, H.; Jiang, C.; Zhao, S.; Wang, L.; Li, Q.; Yang, Z.; Groover, A.; Lu, M.Z. The auxin receptor TIR 1 homolog (Pag FBL 1) regulates adventitious rooting through interactions with Aux/IAA 28 in Populus. Plant Biotechnol. J. 2019, 17, 338–349. [Google Scholar] [CrossRef]
- Rigal, A.; Yordanov, Y.S.; Perrone, I.; Karlberg, A.; Tisserant, E.; Bellini, C.; Busov, V.B.; Martin, F.; Kohler, A.; Bhalerao, R. The AINTEGUMENTA LIKE1 homeotic transcription factor PtAIL1 controls the formation of adventitious root primordia in poplar. Plant Physiol. 2012, 160, 1996–2006. [Google Scholar] [CrossRef] [PubMed]
- Denaxa, N.-K.; Tsafouros, A.; Roussos, P.A. Role of phenolic compounds in adventitious root formation. In Environmental, Physiological and Chemical Controls of Adventitious Rooting in Cuttings; Elsevier: Amsterdam, The Netherlands, 2022; pp. 251–288. [Google Scholar]
- Deng, Y.; Wang, C.; Wang, N.; Wei, L.; Li, W.; Yao, Y.; Liao, W. Roles of small-molecule compounds in plant adventitious root development. Biomolecules 2019, 9, 420. [Google Scholar] [CrossRef] [PubMed]
- Nag, S.; Saha, K.; Choudhuri, M. Role of auxin and polyamines in adventitious root formation in relation to changes in compounds involved in rooting. J. Plant Growth Regul. 2001, 20, 182–194. [Google Scholar] [CrossRef]
- Bojarczuk, T.; Jankiewicz, L. Influence of phenolic substances on rooting of softtrood cuttings of Populus alba L. and P. canescens Sm. Acta Agrobot. 1975, 28, 121–129. [Google Scholar] [CrossRef][Green Version]



| Assembled Contigs | No. |
|---|---|
| Total Trinity genes (n) | 379,983 |
| Total Trinity transcripts (n) | 589,964 |
| GC content (%) | 43.44 |
| Contig N50 length (bp) | 1362 |
| Average contig length (bp) | 538.96 |
| Total assembled bases | 204,796,926 |
| KEGG Pathway | Gene | FDR | Rich Factor (%) | |
|---|---|---|---|---|
| H0W vs. L0W | Fatty acid degradation | 7 | 0.018 | 1.84 |
| H1W vs. L1W | n.s. | |||
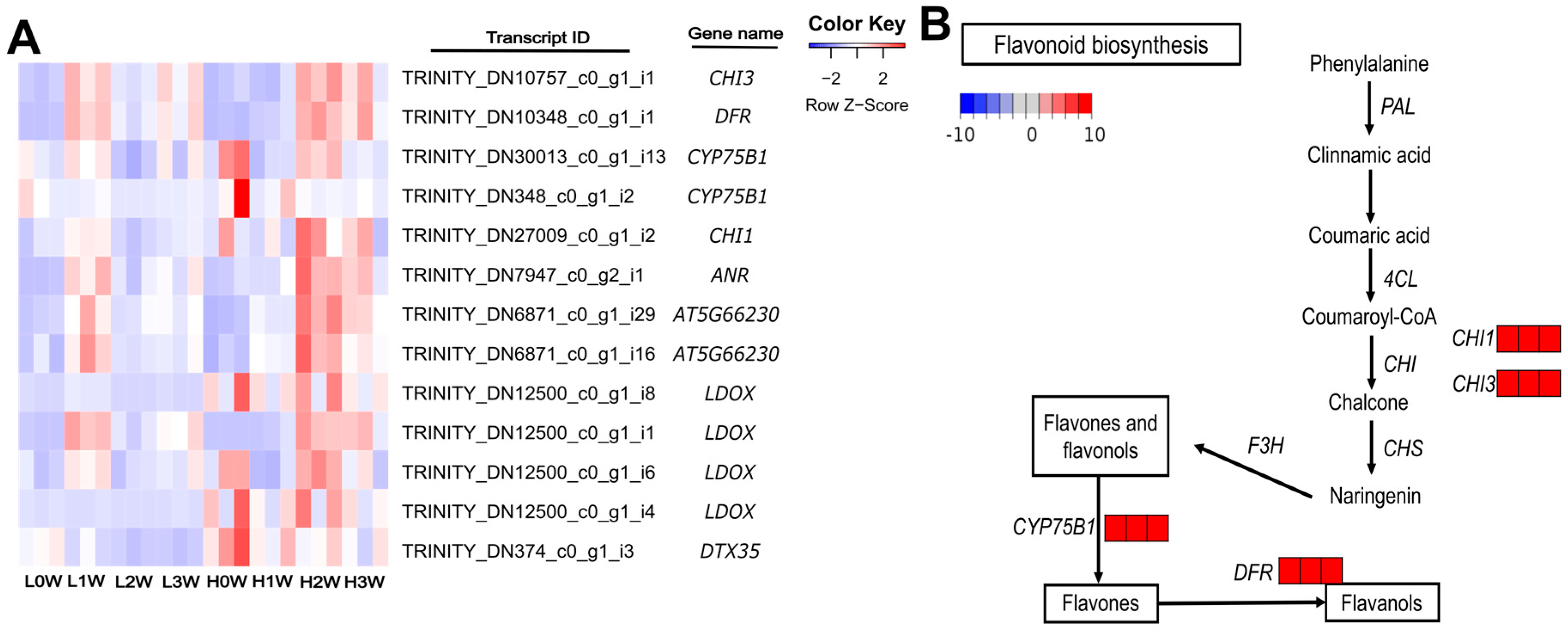
| H2W vs. L2W | Flavonoid biosynthesis | 6 | <0.001 | 2.75 |
| H3W vs. L3W | n.s. | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Byeon, S.; Lee, I.H. Transcriptome of Two-Hybrid Poplar (Populus alba × P. tomentiglandulosa) During Adventitious Root Formation After Stem Cutting. Biology 2025, 14, 751. https://doi.org/10.3390/biology14070751
Byeon S, Lee IH. Transcriptome of Two-Hybrid Poplar (Populus alba × P. tomentiglandulosa) During Adventitious Root Formation After Stem Cutting. Biology. 2025; 14(7):751. https://doi.org/10.3390/biology14070751
Chicago/Turabian StyleByeon, Siyeon, and Il Hwan Lee. 2025. "Transcriptome of Two-Hybrid Poplar (Populus alba × P. tomentiglandulosa) During Adventitious Root Formation After Stem Cutting" Biology 14, no. 7: 751. https://doi.org/10.3390/biology14070751
APA StyleByeon, S., & Lee, I. H. (2025). Transcriptome of Two-Hybrid Poplar (Populus alba × P. tomentiglandulosa) During Adventitious Root Formation After Stem Cutting. Biology, 14(7), 751. https://doi.org/10.3390/biology14070751






