Regulation of Pol II Pausing during Daily Gene Transcription in Mouse Liver
Abstract
Simple Summary
Abstract
1. Introduction
2. Transcription Regulation Is the Main Driving Force for Gene Expression Rhythms
3. Both the Intrinsic Tissue Clock and Extrinsic Cues Can Regulate Gene Expression Rhythms
4. Clock Proteins Typically Collaborate with Other TFs to Regulate Transcription Rhythms
5. The Need to Study Pol II Pausing Regulation near the TSS
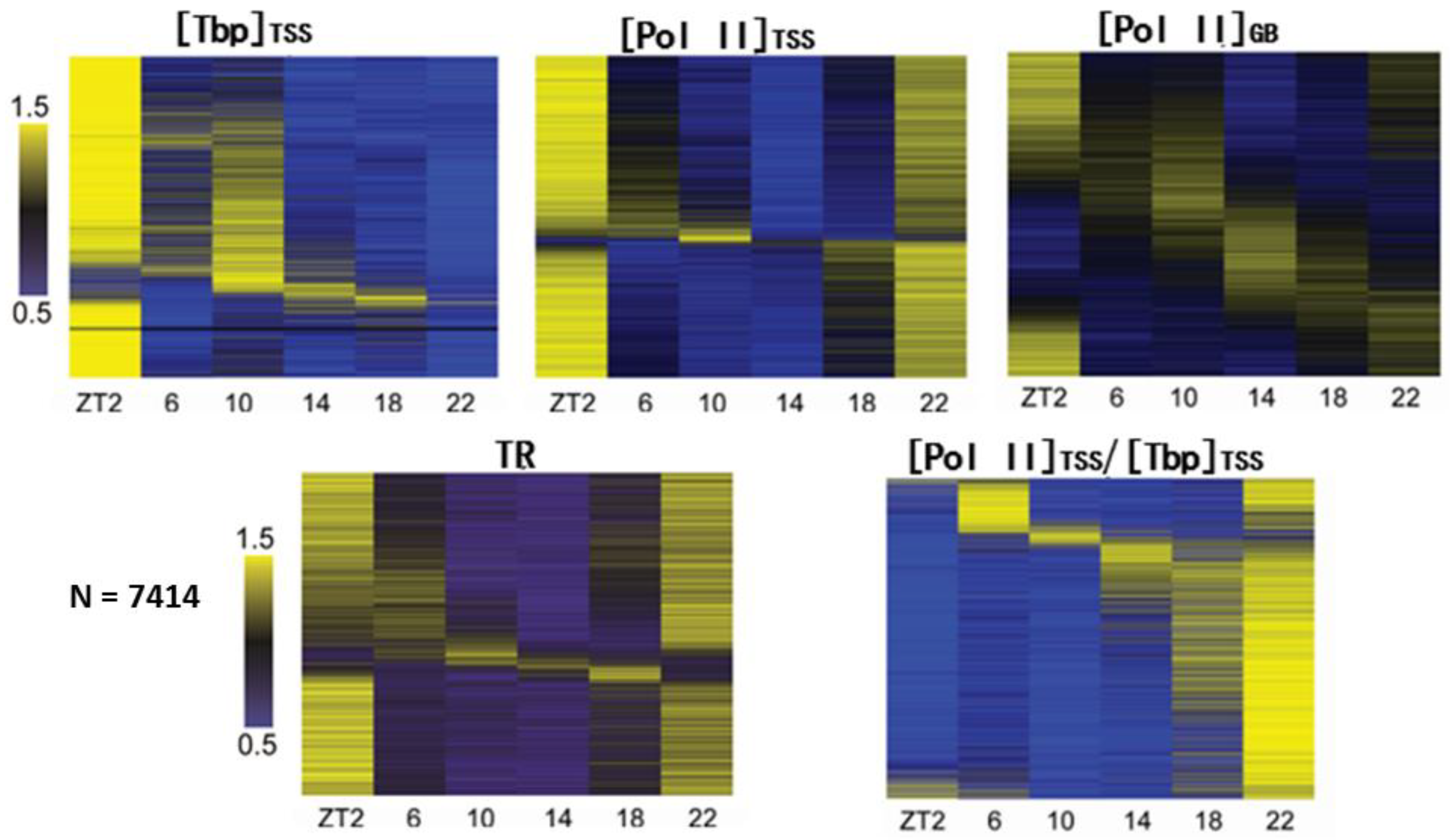
6. Global Characteristics of Pol II Recruitment and Pausing during Daily Transcription in Mouse Liver
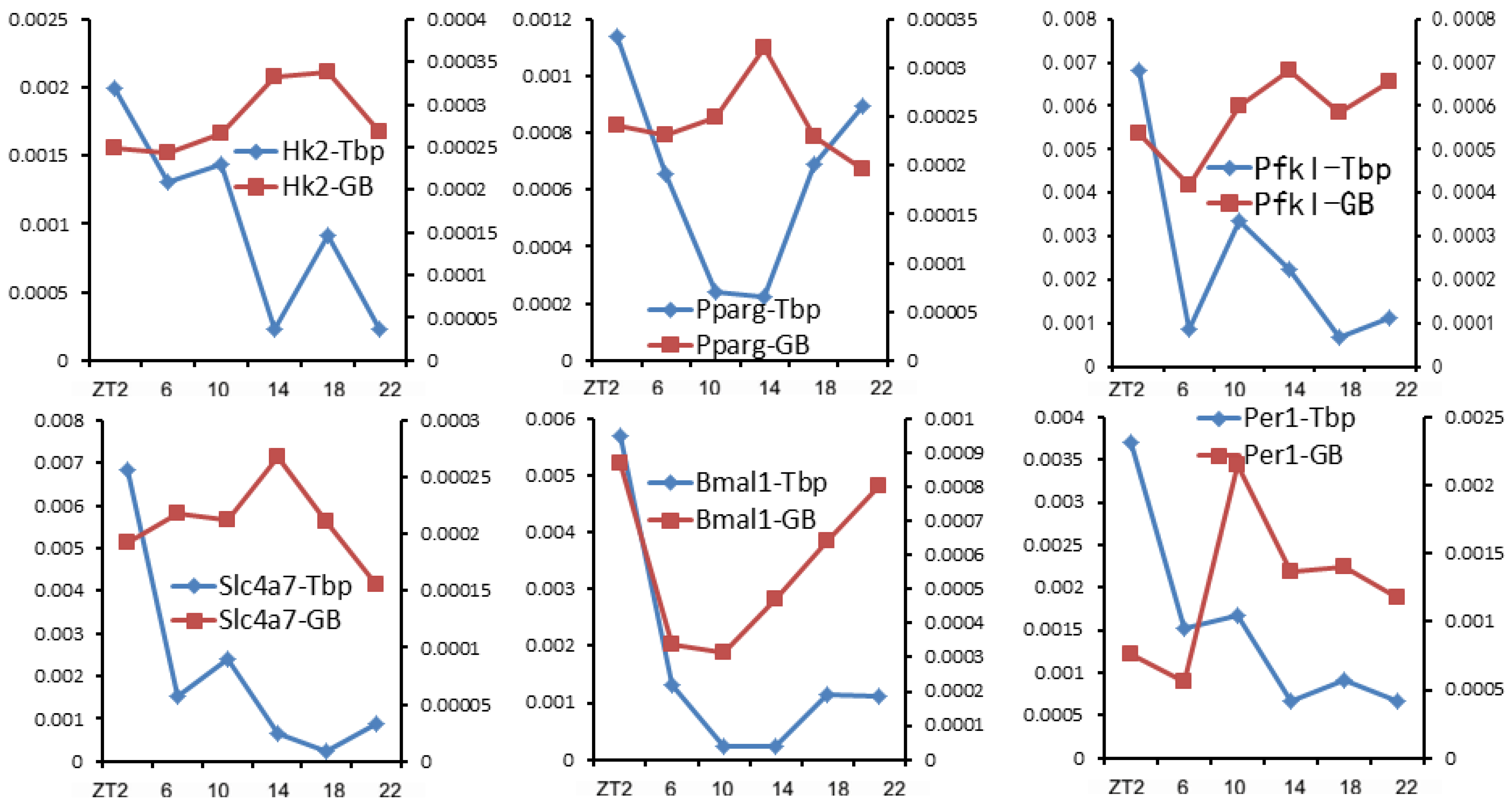
7. Pol II Recruitment Is Not a Direct Determinant of Gene Transcription Rate
8. Premature Transcription Termination at the 5′ End of Genes Contribute to the Regulation of Pol II Pausing
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosbash, M. The implications of multiple circadian clock origins. PLoS Biol. 2009, 7, e62. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Kim, J.K.; Eng, G.W.; Forger, D.B.; Virshup, D.M. A period2 phosphoswitch regulates and temperature compensates circadian period. Mol. Cell 2015, 60, 77–88. [Google Scholar] [CrossRef]
- Toh, K.L.; Jones, C.R.; He, Y.; Eide, E.J.; Hinz, W.A.; Virshup, D.M.; Ptacek, L.J.; Fu, Y.H. An hper2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 2001, 291, 1040–1043. [Google Scholar] [CrossRef] [PubMed]
- Montenegro-Montero, A.; Larrondo, L.F. In the driver’s seat: The case for transcriptional regulation and coupling as relevant determinants of the circadian transcriptome and proteome in eukaryotes. J. Biol. Rhythm. 2016, 31, 37–47. [Google Scholar] [CrossRef]
- Panda, S.; Antoch, M.P.; Miller, B.H.; Su, A.I.; Schook, A.B.; Straume, M.; Schultz, P.G.; Kay, S.A.; Takahashi, J.S.; Hogenesch, J.B. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 2002, 109, 307–320. [Google Scholar] [CrossRef]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [PubMed]
- Mure, L.S.; Le, H.D.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jillani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Cooper, H.M.; et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018, 359, eaao0318. [Google Scholar] [CrossRef]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef]
- Beytebiere, J.R.; Trott, A.J.; Greenwell, B.J.; Osborne, C.A.; Vitet, H.; Spence, J.; Yoo, S.H.; Chen, Z.; Takahashi, J.S.; Ghaffari, N.; et al. Tissue-specific bmal1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions. Genes Dev. 2019, 33, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Qu, H.; Jia, Z.; Kay, S.A. Hnf4a defines tissue-specific circadian rhythms by beaconing bmal1::Clock chromatin binding and shaping the rhythmic chromatin landscape. Nat. Commun. 2021, 12, 6350. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Duffy, T.; Hirota, T.; Kay, S.A. Nuclear receptor hnf4a transrepresses clock:Bmal1 and modulates tissue-specific circadian networks. Proc. Natl. Acad. Sci. USA 2018, 115, E12305–E12312. [Google Scholar] [CrossRef]
- Hong, H.K.; Maury, E.; Ramsey, K.M.; Perelis, M.; Marcheva, B.; Omura, C.; Kobayashi, Y.; Guttridge, D.C.; Barish, G.D.; Bass, J. Requirement for nf-kappab in maintenance of molecular and behavioral circadian rhythms in mice. Genes Dev. 2018, 32, 1367–1379. [Google Scholar] [CrossRef]
- Shen, Y.; Endale, M.; Wang, W.; Morris, A.R.; Francey, L.J.; Harold, R.L.; Hammers, D.W.; Huo, Z.; Partch, C.L.; Hogenesch, J.B.; et al. Nf-kappab modifies the mammalian circadian clock through interaction with the core clock protein bmal1. PLoS Genet. 2021, 17, e1009933. [Google Scholar] [CrossRef]
- Menet, J.S.; Pescatore, S.; Rosbash, M. Clock: Bmal1 is a pioneer-like transcription factor. Genes Dev. 2014, 28, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Lemon, B.; Tjian, R. Orchestrated response: A symphony of transcription factors for gene control. Genes Dev. 2000, 14, 2551–2569. [Google Scholar] [CrossRef] [PubMed]
- Aryal, R.P.; Kwak, P.B.; Tamayo, A.G.; Gebert, M.; Chiu, P.L.; Walz, T.; Weitz, C.J. Macromolecular assemblies of the mammalian circadian clock. Mol. Cell 2017, 67, 770–782.e776. [Google Scholar] [CrossRef] [PubMed]
- Mahesh, G.; Rivas, G.B.S.; Caster, C.; Ost, E.B.; Amunugama, R.; Jones, R.; Allen, D.L.; Hardin, P.E. Proteomic analysis of drosophila clock complexes identifies rhythmic interactions with saga and tip60 complex component nipped-a. Sci. Rep. 2020, 10, 17951. [Google Scholar] [CrossRef]
- Petkau, N.; Budak, H.; Zhou, X.; Oster, H.; Eichele, G. Acetylation of bmal1 by tip60 controls brd4-p-tefb recruitment to circadian promoters. Elife 2019, 8, e43235. [Google Scholar] [CrossRef]
- Ju, D.; Zhang, W.; Yan, J.; Zhao, H.; Li, W.; Wang, J.; Liao, M.; Xu, Z.; Wang, Z.; Zhou, G.; et al. Chemical perturbations reveal that ruvbl2 regulates the circadian phase in mammals. Sci. Transl. Med. 2020, 12, eaba0769. [Google Scholar] [CrossRef]
- Malik, S.; Roeder, R.G. The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010, 11, 761–772. [Google Scholar] [CrossRef]
- Thomas, M.C.; Chiang, C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 105–178. [Google Scholar] [CrossRef] [PubMed]
- Roeder, R.G. 50+ years of eukaryotic transcription: An expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol. 2019, 26, 783–791. [Google Scholar] [CrossRef]
- Hsieh, T.S.; Cattoglio, C.; Slobodyanyuk, E.; Hansen, A.S.; Darzacq, X.; Tjian, R. Enhancer-promoter interactions and transcription are largely maintained upon acute loss of ctcf, cohesin, wapl or yy1. Nat. Genet. 2022, 54, 1919–1932. [Google Scholar] [CrossRef]
- Richter, W.F.; Nayak, S.; Iwasa, J.; Taatjes, D.J. The mediator complex as a master regulator of transcription by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2022, 23, 732–749. [Google Scholar] [CrossRef]
- Suter, D.M.; Molina, N.; Gatfield, D.; Schneider, K.; Schibler, U.; Naef, F. Mammalian genes are transcribed with widely different bursting kinetics. Science 2011, 332, 472–474. [Google Scholar] [CrossRef]
- Rodriguez, J.; Larson, D.R. Transcription in living cells: Molecular mechanisms of bursting. Annu. Rev. Biochem. 2020, 89, 189–212. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Levine, M.S. Enhancer-promoter communication: Hubs or loops? Curr. Opin. Genet. Dev. 2021, 67, 5–9. [Google Scholar] [CrossRef]
- McSwiggen, D.T.; Mir, M.; Darzacq, X.; Tjian, R. Evaluating phase separation in live cells: Diagnosis, caveats, and functional consequences. Genes Dev. 2019, 33, 1619–1634. [Google Scholar] [CrossRef] [PubMed]
- Jonkers, I.; Lis, J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015, 16, 167–177. [Google Scholar] [CrossRef]
- Chiu, A.C.; Suzuki, H.I.; Wu, X.; Mahat, D.B.; Kriz, A.J.; Sharp, P.A. Transcriptional pause sites delineate stable nucleosome-associated premature polyadenylation suppressed by u1 snrnp. Mol. Cell 2018, 69, 648–663.e647. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Shibata, H.; Handa, H. Transcription elongation factors dsif and nelf: Promoter-proximal pausing and beyond. Biochim. Biophys. Acta 2013, 1829, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Lin, C.; Shilatifard, A. The super elongation complex (sec) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 2012, 13, 543–547. [Google Scholar] [CrossRef]
- Chen, Y.; Vos, S.M.; Dienemann, C.; Ninov, M.; Urlaub, H.; Cramer, P. Allosteric transcription stimulation by RNA polymerase II super elongation complex. Mol. Cell 2021, 81, 3386–3399.e3310. [Google Scholar] [CrossRef] [PubMed]
- Price, D.H. Transient pausing by RNA polymerase II. Proc. Natl. Acad. Sci. USA 2018, 115, 4810–4812. [Google Scholar] [CrossRef]
- Kamieniarz-Gdula, K.; Proudfoot, N.J. Transcriptional control by premature termination: A forgotten mechanism. Trends Genet. 2019, 35, 553–564. [Google Scholar] [CrossRef]
- Beytebiere, J.R.; Greenwell, B.J.; Sahasrabudhe, A.; Menet, J.S. Clock-controlled rhythmic transcription: Is the clock enough and how does it work? Transcription 2019, 10, 212–221. [Google Scholar] [CrossRef]
- Zhu, J.; Li, C.; Gong, C.; Li, X. Regulation of pol II pausing is involved in daily gene transcription in the mouse liver. J. Biol. Rhythm. 2018, 33, 350–362. [Google Scholar] [CrossRef]
- Hardin, P.E.; Hall, J.C.; Rosbash, M. Feedback of the drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 1990, 343, 536–540. [Google Scholar] [CrossRef]
- Hardin, P.E.; Hall, J.C.; Rosbash, M. Circadian oscillations in period gene mrna levels are transcriptionally regulated. Proc. Natl. Acad. Sci. USA 1992, 89, 11711–11715. [Google Scholar] [CrossRef]
- Frisch, B.; Hardin, P.E.; Hamblen-Coyle, M.J.; Rosbash, M.; Hall, J.C. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the drosophila nervous system. Neuron 1994, 12, 555–570. [Google Scholar] [CrossRef] [PubMed]
- So, W.V.; Rosbash, M. Post-transcriptional regulation contributes to drosophila clock gene mrna cycling. EMBO J. 1997, 16, 7146–7155. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.S.; Albrecht, U.; Zhuchenko, O.; Bailey, J.; Eichele, G.; Lee, C.C. Rigui, a putative mammalian ortholog of the drosophila period gene. Cell 1997, 90, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Shearman, L.P.; Zylka, M.J.; Weaver, D.R.; Kolakowski, L.F., Jr.; Reppert, S.M. Two period homologs: Circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 1997, 19, 1261–1269. [Google Scholar] [CrossRef]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor rev-erbalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Atger, F.; Gobet, C.; Marquis, J.; Martin, E.; Wang, J.; Weger, B.; Lefebvre, G.; Descombes, P.; Naef, F.; Gachon, F. Circadian and feeding rhythms differentially affect rhythmic mrna transcription and translation in mouse liver. Proc. Natl. Acad. Sci. USA 2015, 112, E6579–E6588. [Google Scholar] [CrossRef]
- Luck, S.; Thurley, K.; Thaben, P.F.; Westermark, P.O. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep. 2014, 9, 741–751. [Google Scholar] [CrossRef]
- Wang, J.; Symul, L.; Yeung, J.; Gobet, C.; Sobel, J.; Luck, S.; Westermark, P.O.; Molina, N.; Naef, F. Circadian clock-dependent and -independent posttranscriptional regulation underlies temporal mrna accumulation in mouse liver. Proc. Natl. Acad. Sci. USA 2018, 115, E1916–E1925. [Google Scholar] [CrossRef]
- Hurni, C.; Weger, B.D.; Gobet, C.; Naef, F. Comprehensive analysis of the circadian nuclear and cytoplasmic transcriptome in mouse liver. PLoS Genet. 2022, 18, e1009903. [Google Scholar] [CrossRef]
- Buhr, E.D.; Yoo, S.H.; Takahashi, J.S. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 2010, 330, 379–385. [Google Scholar] [CrossRef]
- Damiola, F.; Le Minh, N.; Preitner, N.; Kornmann, B.; Fleury-Olela, F.; Schibler, U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000, 14, 2950–2961. [Google Scholar] [CrossRef]
- Koronowski, K.B.; Sassone-Corsi, P. Communicating clocks shape circadian homeostasis. Science 2021, 371, eabd0951. [Google Scholar] [CrossRef] [PubMed]
- Reinke, H.; Saini, C.; Fleury-Olela, F.; Dibner, C.; Benjamin, I.J.; Schibler, U. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008, 22, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Gotic, I.; Schibler, U. Posttranscriptional mechanisms controlling diurnal gene expression cycles by body temperature rhythms. RNA Biol. 2017, 14, 1294–1298. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Marcacci, L.; Schibler, U. Multiple signaling pathways elicit circadian gene expression in cultured rat-1 fibroblasts. Curr. Biol. 2000, 10, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Damiola, F.; Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 1998, 93, 929–937. [Google Scholar] [CrossRef]
- Gerber, A.; Esnault, C.; Aubert, G.; Treisman, R.; Pralong, F.; Schibler, U. Blood-borne circadian signal stimulates daily oscillations in actin dynamics and srf activity. Cell 2013, 152, 492–503. [Google Scholar] [CrossRef]
- Pagani, L.; Schmitt, K.; Meier, F.; Izakovic, J.; Roemer, K.; Viola, A.; Cajochen, C.; Wirz-Justice, A.; Brown, S.A.; Eckert, A. Serum factors in older individuals change cellular clock properties. Proc. Natl. Acad. Sci. USA 2011, 108, 7218–7223. [Google Scholar] [CrossRef]
- Liu, S.; Brown, J.D.; Stanya, K.J.; Homan, E.; Leidl, M.; Inouye, K.; Bhargava, P.; Gangl, M.R.; Dai, L.; Hatano, B.; et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature 2013, 502, 550–554. [Google Scholar] [CrossRef]
- Koronowski, K.B.; Kinouchi, K.; Welz, P.S.; Smith, J.G.; Zinna, V.M.; Shi, J.; Samad, M.; Chen, S.; Magnan, C.N.; Kinchen, J.M.; et al. Defining the independence of the liver circadian clock. Cell 2019, 177, 1448–1462.e1414. [Google Scholar] [CrossRef]
- Welz, P.S.; Zinna, V.M.; Symeonidi, A.; Koronowski, K.B.; Kinouchi, K.; Smith, J.G.; Guillen, I.M.; Castellanos, A.; Furrow, S.; Aragon, F.; et al. Bmal1-driven tissue clocks respond independently to light to maintain homeostasis. Cell 2019, 177, 1436–1447.e1412. [Google Scholar] [CrossRef]
- Cho, H.; Zhao, X.; Hatori, M.; Yu, R.T.; Barish, G.D.; Lam, M.T.; Chong, L.W.; DiTacchio, L.; Atkins, A.R.; Glass, C.K.; et al. Regulation of circadian behaviour and metabolism by rev-erb-alpha and rev-erb-beta. Nature 2012, 485, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Kornmann, B.; Schaad, O.; Bujard, H.; Takahashi, J.S.; Schibler, U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007, 5, e34. [Google Scholar] [CrossRef] [PubMed]
- Greco, C.M.; Koronowski, K.B.; Smith, J.G.; Shi, J.; Kunderfranco, P.; Carriero, R.; Chen, S.; Samad, M.; Welz, P.S.; Zinna, V.M.; et al. Integration of feeding behavior by the liver circadian clock reveals network dependency of metabolic rhythms. Sci. Adv. 2021, 7, eabi7828. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Marhon, S.A.; Zhang, Y.; Steger, D.J.; Won, K.J.; Lazar, M.A. Rev-erbalpha dynamically modulates chromatin looping to control circadian gene transcription. Science 2018, 359, 1274–1277. [Google Scholar] [CrossRef]
- Karr, J.P.; Ferrie, J.J.; Tjian, R.; Darzacq, X. The transcription factor activity gradient (tag) model: Contemplating a contact-independent mechanism for enhancer-promoter communication. Genes Dev. 2022, 36, 7–16. [Google Scholar] [CrossRef]
- Yudkovsky, N.; Ranish, J.A.; Hahn, S. A transcription reinitiation intermediate that is stabilized by activator. Nature 2000, 408, 225–229. [Google Scholar] [CrossRef]
- Hawley, D.K.; Roeder, R.G. Functional steps in transcription initiation and reinitiation from the major late promoter in a hela nuclear extract. J. Biol. Chem. 1987, 262, 3452–3461. [Google Scholar] [CrossRef] [PubMed]
- Zawel, L.; Kumar, K.P.; Reinberg, D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995, 9, 1479–1490. [Google Scholar] [CrossRef]
- Zhang, Z.; Boskovic, Z.; Hussain, M.M.; Hu, W.; Inouye, C.; Kim, H.J.; Abole, A.K.; Doud, M.K.; Lewis, T.A.; Koehler, A.N.; et al. Chemical perturbation of an intrinsically disordered region of tfiid distinguishes two modes of transcription initiation. Elife 2015, 4, e07777. [Google Scholar] [CrossRef]
- Shao, W.; Zeitlinger, J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 2017, 49, 1045–1051. [Google Scholar] [CrossRef]
- Gressel, S.; Schwalb, B.; Cramer, P. The pause-initiation limit restricts transcription activation in human cells. Nat. Commun. 2019, 10, 3603. [Google Scholar] [CrossRef] [PubMed]
- Tantale, K.; Mueller, F.; Kozulic-Pirher, A.; Lesne, A.; Victor, J.M.; Robert, M.C.; Capozi, S.; Chouaib, R.; Backer, V.; Mateos-Langerak, J.; et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 2016, 7, 12248. [Google Scholar] [CrossRef] [PubMed]
- Day, D.S.; Zhang, B.; Stevens, S.M.; Ferrari, F.; Larschan, E.N.; Park, P.J.; Pu, W.T. Comprehensive analysis of promoter-proximal RNA polymerase II pausing across mammalian cell types. Genome Biol. 2016, 17, 120. [Google Scholar] [CrossRef]
- Rahl, P.B.; Lin, C.Y.; Seila, A.C.; Flynn, R.A.; McCuine, S.; Burge, C.B.; Sharp, P.A.; Young, R.A. C-myc regulates transcriptional pause release. Cell 2010, 141, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Schade, A.E.; Branigan, T.B.; Muller, G.A.; DeCaprio, J.A. Coordinating gene expression during the cell cycle. Trends Biochem. Sci. 2022, 47, 1009–1022. [Google Scholar] [CrossRef] [PubMed]
- Hsiung, C.C.; Bartman, C.R.; Huang, P.; Ginart, P.; Stonestrom, A.J.; Keller, C.A.; Face, C.; Jahn, K.S.; Evans, P.; Sankaranarayanan, L.; et al. A hyperactive transcriptional state marks genome reactivation at the mitosis-g1 transition. Genes Dev. 2016, 30, 1423–1439. [Google Scholar] [CrossRef]
- Palozola, K.C.; Donahue, G.; Liu, H.; Grant, G.R.; Becker, J.S.; Cote, A.; Yu, H.; Raj, A.; Zaret, K.S. Mitotic transcription and waves of gene reactivation during mitotic exit. Science 2017, 358, 119–122. [Google Scholar] [CrossRef]
- Gaucher, J.; Montellier, E.; Sassone-Corsi, P. Molecular cogs: Interplay between circadian clock and cell cycle. Trends Cell Biol. 2018, 28, 368–379. [Google Scholar] [CrossRef]
- Bieler, J.; Cannavo, R.; Gustafson, K.; Gobet, C.; Gatfield, D.; Naef, F. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol. Syst. Biol. 2014, 10, 739. [Google Scholar] [CrossRef]
- Matsuo, T.; Yamaguchi, S.; Mitsui, S.; Emi, A.; Shimoda, F.; Okamura, H. Control mechanism of the circadian clock for timing of cell division in vivo. Science 2003, 302, 255–259. [Google Scholar] [CrossRef]
- Zhao, X.; Hirota, T.; Han, X.; Cho, H.; Chong, L.W.; Lamia, K.; Liu, S.; Atkins, A.R.; Banayo, E.; Liddle, C.; et al. Circadian amplitude regulation via fbxw7-targeted rev-erbalpha degradation. Cell 2016, 165, 1644–1657. [Google Scholar] [CrossRef]
- Wang, J.; Mauvoisin, D.; Martin, E.; Atger, F.; Galindo, A.N.; Dayon, L.; Sizzano, F.; Palini, A.; Kussmann, M.; Waridel, P.; et al. Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 2017, 25, 102–117. [Google Scholar] [CrossRef]
- Segil, N.; Guermah, M.; Hoffmann, A.; Roeder, R.G.; Heintz, N. Mitotic regulation of tfiid: Inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996, 10, 2389–2400. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Yang, W.; Ni, T.; Tang, Z.; Nakadai, T.; Zhu, J.; Roeder, R.G. RNA polymerase ii-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 2015, 350, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Guermah, M.; McGinty, R.K.; Lee, J.S.; Tang, Z.; Milne, T.A.; Shilatifard, A.; Muir, T.W.; Roeder, R.G. Rad6-mediated transcription-coupled h2b ubiquitylation directly stimulates h3k4 methylation in human cells. Cell 2009, 137, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, J.A.; McGinty, R.K.; Nguyen, U.T.; Muir, T.W.; Allis, C.D.; Roeder, R.G. The n-set domain of set1 regulates h2b ubiquitylation-dependent h3k4 methylation. Mol. Cell 2013, 49, 1121–1133. [Google Scholar] [CrossRef]
- Valekunja, U.K.; Edgar, R.S.; Oklejewicz, M.; van der Horst, G.T.; O’Neill, J.S.; Tamanini, F.; Turner, D.J.; Reddy, A.B. Histone methyltransferase mll3 contributes to genome-scale circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 1554–1559. [Google Scholar] [CrossRef]
- Baxter, M.; Poolman, T.; Cunningham, P.; Hunter, L.; Voronkov, M.; Kitchen, G.B.; Goosey, L.; Begley, N.; Kay, D.; Hespe, A.; et al. Circadian clock function does not require the histone methyltransferase mll3. FASEB J. 2022, 36, e22356. [Google Scholar] [CrossRef]
- Li, Y.; Shan, Y.; Desai, R.V.; Cox, K.H.; Weinberger, L.S.; Takahashi, J.S. Noise-driven cellular heterogeneity in circadian periodicity. Proc. Natl. Acad. Sci. USA 2020, 117, 10350–10356. [Google Scholar] [CrossRef]
- Li, Y.; Shan, Y.; Kilaru, G.K.; Berto, S.; Wang, G.Z.; Cox, K.H.; Yoo, S.H.; Yang, S.; Konopka, G.; Takahashi, J.S. Epigenetic inheritance of circadian period in clonal cells. Elife 2020, 9, e54186. [Google Scholar] [CrossRef] [PubMed]
- Nikhil, K.L.; Korge, S.; Kramer, A. Heritable gene expression variability and stochasticity govern clonal heterogeneity in circadian period. PLoS Biol. 2020, 18, e3000792. [Google Scholar] [CrossRef] [PubMed]
- Bartman, C.R.; Hamagami, N.; Keller, C.A.; Giardine, B.; Hardison, R.C.; Blobel, G.A.; Raj, A. Transcriptional burst initiation and polymerase pause release are key control points of transcriptional regulation. Mol. Cell 2019, 73, 519–532.e514. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, M.G.; Schwalb, B.; Mackowiak, S.D.; Velychko, T.; Hanzl, A.; Imrichova, H.; Brand, M.; Agerer, B.; Chorn, S.; Nabet, B.; et al. Selective mediator dependence of cell-type-specifying transcription. Nat. Genet. 2020, 52, 719–727. [Google Scholar] [CrossRef]
- Nicolas, D.; Phillips, N.E.; Naef, F. What shapes eukaryotic transcriptional bursting? Mol. Biosyst. 2017, 13, 1280–1290. [Google Scholar] [CrossRef]
- Fukaya, T.; Lim, B.; Levine, M. Enhancer control of transcriptional bursting. Cell 2016, 166, 358–368. [Google Scholar] [CrossRef]
- Yang, Z.; Yik, J.H.; Chen, R.; He, N.; Jang, M.K.; Ozato, K.; Zhou, Q. Recruitment of p-tefb for stimulation of transcriptional elongation by the bromodomain protein brd4. Mol. Cell 2005, 19, 535–545. [Google Scholar] [CrossRef]
- Jang, M.K.; Mochizuki, K.; Zhou, M.; Jeong, H.S.; Brady, J.N.; Ozato, K. The bromodomain protein brd4 is a positive regulatory component of p-tefb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 2005, 19, 523–534. [Google Scholar] [CrossRef]
- Conaway, R.C.; Conaway, J.W. The mediator complex and transcription elongation. Biochim. Biophys. Acta 2013, 1829, 69–75. [Google Scholar] [CrossRef]
- Luyties, O.; Taatjes, D.J. The mediator kinase module: An interface between cell signaling and transcription. Trends Biochem. Sci. 2022, 47, 314–327. [Google Scholar] [CrossRef]
- Li, Y.; Liu, M.; Chen, L.F.; Chen, R. P-tefb: Finding its ways to release promoter-proximally paused RNA polymerase II. Transcription 2018, 9, 88–94. [Google Scholar] [CrossRef]
- Contreras, X.; Barboric, M.; Lenasi, T.; Peterlin, B.M. Hmba releases p-tefb from hexim1 and 7sk snrna via pi3k/akt and activates hiv transcription. PLoS Pathog. 2007, 3, 1459–1469. [Google Scholar] [CrossRef]
- Krebs, A.R.; Imanci, D.; Hoerner, L.; Gaidatzis, D.; Burger, L.; Schubeler, D. Genome-wide single-molecule footprinting reveals high RNA polymerase II turnover at paused promoters. Mol. Cell 2017, 67, 411–422.e414. [Google Scholar] [CrossRef]
- Steurer, B.; Janssens, R.C.; Geverts, B.; Geijer, M.E.; Wienholz, F.; Theil, A.F.; Chang, J.; Dealy, S.; Pothof, J.; van Cappellen, W.A.; et al. Live-cell analysis of endogenous gfp-rpb1 uncovers rapid turnover of initiating and promoter-paused RNA polymerase II. Proc. Natl. Acad. Sci. USA 2018, 115, E4368–E4376. [Google Scholar] [CrossRef]
- Buckley, M.S.; Kwak, H.; Zipfel, W.R.; Lis, J.T. Kinetics of promoter pol II on hsp70 reveal stable pausing and key insights into its regulation. Genes Dev. 2014, 28, 14–19. [Google Scholar] [CrossRef]
- Cortazar, M.A.; Erickson, B.; Fong, N.; Pradhan, S.J.; Ntini, E.; Bentley, D.L. Xrn2 substrate mapping identifies torpedo loading sites and extensive premature termination of RNA pol II transcription. Genes Dev. 2022, 36, 1062–1078. [Google Scholar] [CrossRef]
- Proudfoot, N.J. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 2016, 352, aad9926. [Google Scholar] [CrossRef]
- Padmanabhan, K.; Robles, M.S.; Westerling, T.; Weitz, C.J. Feedback regulation of transcriptional termination by the mammalian circadian clock period complex. Science 2012, 337, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Ehrensberger, A.H.; Kelly, G.P.; Svejstrup, J.Q. Mechanistic interpretation of promoter-proximal peaks and rnapii density maps. Cell 2013, 154, 713–715. [Google Scholar] [CrossRef] [PubMed]
- Quinodoz, M.; Gobet, C.; Naef, F.; Gustafson, K.B. Characteristic bimodal profiles of RNA polymerase II at thousands of active mammalian promoters. Genome Biol. 2014, 15, R85. [Google Scholar] [CrossRef] [PubMed]
- Jonkers, I.; Kwak, H.; Lis, J.T. Genome-wide dynamics of pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife 2014, 3, e02407. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Gao, X.; Shilatifard, A. Stably paused genes revealed through inhibition of transcription initiation by the tfiih inhibitor triptolide. Genes Dev. 2015, 29, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Noe Gonzalez, M.; Sato, S.; Tomomori-Sato, C.; Conaway, J.W.; Conaway, R.C. Ctd-dependent and -independent mechanisms govern co-transcriptional capping of pol II transcripts. Nat. Commun. 2018, 9, 3392. [Google Scholar] [CrossRef] [PubMed]
- Henriques, T.; Gilchrist, D.A.; Nechaev, S.; Bern, M.; Muse, G.W.; Burkholder, A.; Fargo, D.C.; Adelman, K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol. Cell 2013, 52, 517–528. [Google Scholar] [CrossRef]
- Aoi, Y.; Smith, E.R.; Shah, A.P.; Rendleman, E.J.; Marshall, S.A.; Woodfin, A.R.; Chen, F.X.; Shiekhattar, R.; Shilatifard, A. Nelf regulates a promoter-proximal step distinct from RNA pol II pause-release. Mol. Cell 2020, 78, 261–274.e265. [Google Scholar] [CrossRef] [PubMed]
- Lidschreiber, M.; Leike, K.; Cramer, P. Cap completion and c-terminal repeat domain kinase recruitment underlie the initiation-elongation transition of RNA polymerase II. Mol. Cell Biol. 2013, 33, 3805–3816. [Google Scholar] [CrossRef]
- Ramanathan, A.; Robb, G.B.; Chan, S.H. Mrna capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Topisirovic, I.; Svitkin, Y.V.; Sonenberg, N.; Shatkin, A.J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev. RNA 2011, 2, 277–298. [Google Scholar] [CrossRef]
- Jiao, X.; Chang, J.H.; Kilic, T.; Tong, L.; Kiledjian, M. A mammalian pre-mrna 5’ end capping quality control mechanism and an unexpected link of capping to pre-mrna processing. Mol. Cell 2013, 50, 104–115. [Google Scholar] [CrossRef]
- Wagner, E.J.; Tong, L.; Adelman, K. Integrator is a global promoter-proximal termination complex. Mol. Cell 2023, 83, 416–427. [Google Scholar] [CrossRef]
- Lykke-Andersen, S.; Zumer, K.; Molska, E.S.; Rouviere, J.O.; Wu, G.; Demel, C.; Schwalb, B.; Schmid, M.; Cramer, P.; Jensen, T.H. Integrator is a genome-wide attenuator of non-productive transcription. Mol. Cell 2021, 81, 514–529.e516. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.B.; Field, A.R.; Mimoso, C.A.; Zhao, C.; Huang, K.L.; Wagner, E.J.; Adelman, K. Integrator endonuclease drives promoter-proximal termination at all RNA polymerase II-transcribed loci. Mol. Cell 2022, 82, 4232–4245.e4211. [Google Scholar] [CrossRef] [PubMed]
- Rambout, X.; Cho, H.; Blanc, R.; Lyu, Q.; Miano, J.M.; Chakkalakal, J.V.; Nelson, G.M.; Yalamanchili, H.K.; Adelman, K.; Maquat, L.E. Pgc-1alpha senses the cbc of pre-mrna to dictate the fate of promoter-proximally paused rnapii. Mol. Cell 2023, 83, 186–202.e111. [Google Scholar] [CrossRef] [PubMed]
- Lenasi, T.; Peterlin, B.M.; Barboric, M. Cap-binding protein complex links pre-mrna capping to transcription elongation and alternative splicing through positive transcription elongation factor b (p-tefb). J. Biol. Chem. 2011, 286, 22758–22768. [Google Scholar] [CrossRef] [PubMed]
- Nojima, T.; Gomes, T.; Grosso, A.R.F.; Kimura, H.; Dye, M.J.; Dhir, S.; Carmo-Fonseca, M.; Proudfoot, N.J. Mammalian net-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 2015, 161, 526–540. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; Li, X. Regulation of Pol II Pausing during Daily Gene Transcription in Mouse Liver. Biology 2023, 12, 1107. https://doi.org/10.3390/biology12081107
Xu W, Li X. Regulation of Pol II Pausing during Daily Gene Transcription in Mouse Liver. Biology. 2023; 12(8):1107. https://doi.org/10.3390/biology12081107
Chicago/Turabian StyleXu, Wei, and Xiaodong Li. 2023. "Regulation of Pol II Pausing during Daily Gene Transcription in Mouse Liver" Biology 12, no. 8: 1107. https://doi.org/10.3390/biology12081107
APA StyleXu, W., & Li, X. (2023). Regulation of Pol II Pausing during Daily Gene Transcription in Mouse Liver. Biology, 12(8), 1107. https://doi.org/10.3390/biology12081107




