Weak Genetic Isolation and Putative Phenotypic Selection in the Wild Carnation Dianthus virgineus (Caryophyllaceae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study System
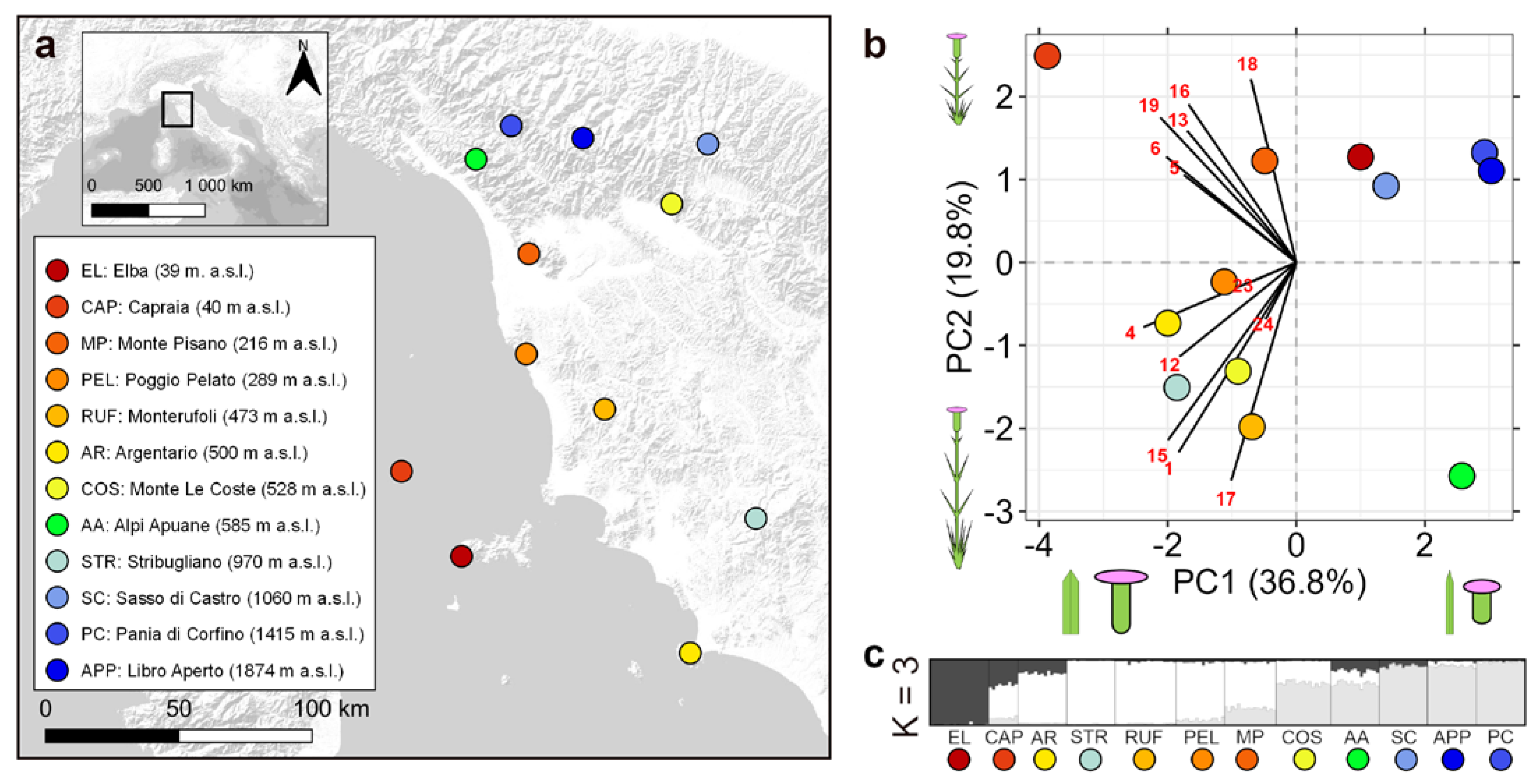
2.2. Study Area and Sampling
2.3. DNA Extraction, Libraries Preparation, and Sequencing
2.4. Reference Assembly, Mapping, SNPs Calling, and Filtering
2.5. Genetic Structure
2.6. Morphometry
| Character (Unit of Measure) | ID | Fresh (F)/Dried (D) Material |
|---|---|---|
| Plant height (cm) | 1 | D |
| Number of internodes | 2 | D |
| Lower internode length (mm) | 3 | D |
| Upper internode length (mm) | 4 | D |
| Basal leaf length (mm) | 5 | D |
| Basal leaf width (mm) | 6 | D |
| Upper stem leaf length (mm) | 7 | D |
| Upper stem leaf width (mm) | 8 | D |
| Lower stem leaf length (mm) | 9 | D |
| Lower stem leaf width (mm) | 10 | D |
| Number of flowers per stem | 11 | D |
| Number of epicalyx scales | 12 | D |
| Upper epicalyx scale length (mm) | 13 | D |
| Upper epicalyx scale mucro length (mm) | 14 | D |
| Upper epicalyx scale width (mm) | 15 | D |
| Calyx length (mm) | 16 | D |
| Calyx width (mm) | 17 | F |
| Calyx teeth length (mm) | 18 | D |
| Corolla diameter (mm) | 19 | F |
| Petal length (mm) | 20 | F |
| Petal limb length (mm) | 21 | F |
| Petal limb width (mm) | 22 | F |
| Ovary length (mm) | 23 | F |
| Anther length (mm) | 24 | F |
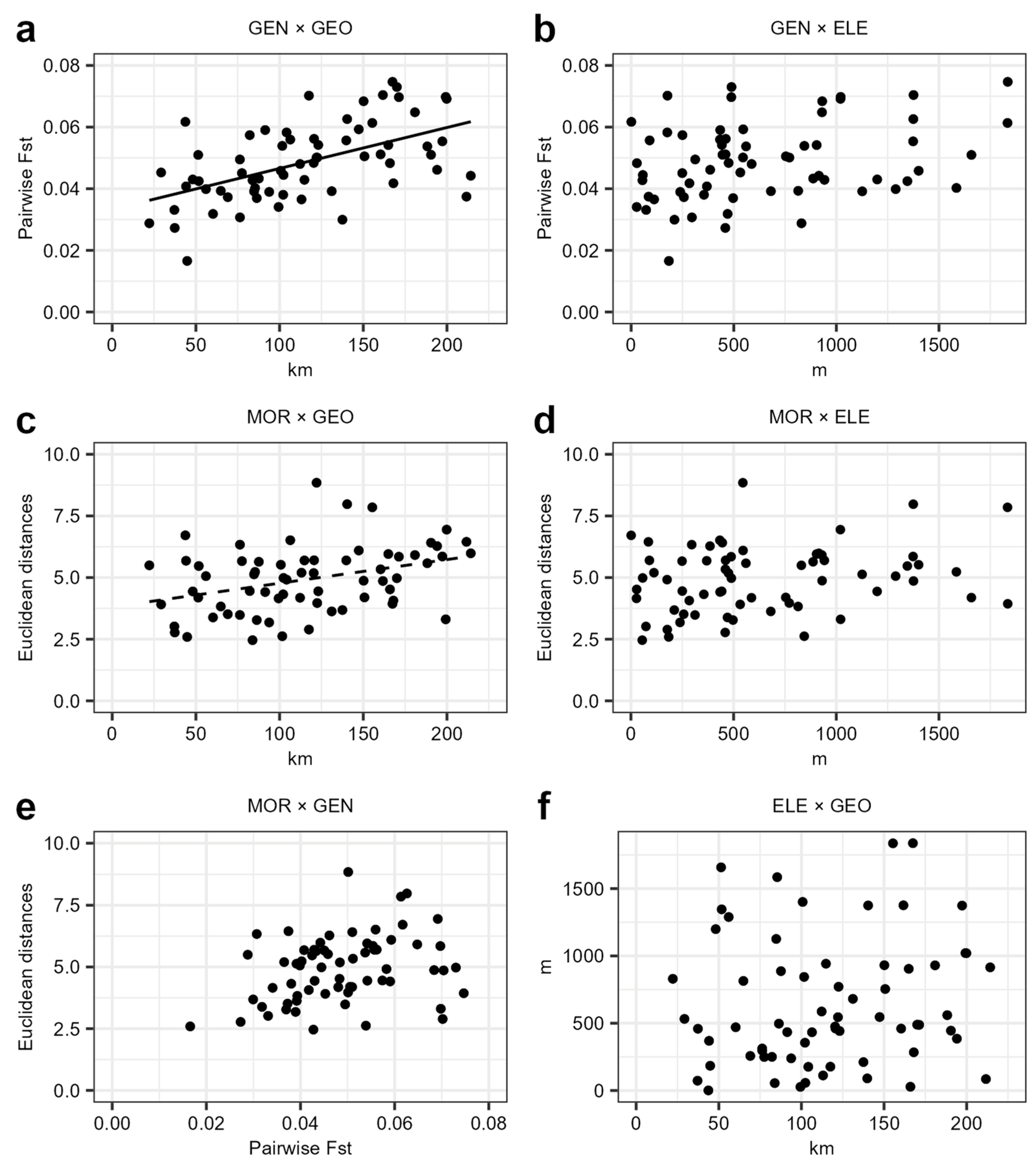
2.7. Mantel Tests
2.8. Pst-Fst Comparisons
3. Results
3.1. Genetic Analyses
3.2. Morphometric Analyses
3.3. Mantel Tests
3.4. Pst-Fst Comparison
4. Discussion
4.1. Patterns of Genetic Variation
4.2. Patterns of Phenotypic Variation
4.3. Scenarios of Intraspecific Differentiation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, Y.; Widmer, A.; Karrenberg, S. The Roles of Genetic Drift and Natural Selection in Quantitative Trait Divergence along an Altitudinal Gradient in Arabidopsis thaliana. Heredity 2015, 114, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. Gene Flow and the Geographic Structure of Natural Populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Nosil, P. Speciation with Gene Flow Could Be Common. Mol. Ecol. 2008, 17, 2103–2106. [Google Scholar] [CrossRef]
- Orsini, L.; Vanoverbeke, J.; Swillen, I.; Mergeay, J.; De Meester, L. Drivers of Population Genetic Differentiation in the Wild: Isolation by Dispersal Limitation, Isolation by Adaptation and Isolation by Colonization. Mol. Ecol. 2013, 22, 5983–5999. [Google Scholar] [CrossRef]
- Wright, S. Isolation by Distance. Genetics 1943, 28, 139–156. [Google Scholar] [CrossRef]
- Sexton, J.P.; Hangartner, S.B.; Hoffmann, A.A. Genetic Isolation by Environment or Distance: Which Pattern of Gene Flow Is Most Common? Evolution 2014, 68, 1–15. [Google Scholar] [CrossRef]
- Barth, J.M.I.; Berg, P.R.; Jonsson, P.R.; Bonanomi, S.; Corell, H.; Hemmer-Hansen, J.; Jakobsen, K.S.; Johannesson, K.; Jorde, P.E.; Knutsen, H.; et al. Genome Architecture Enables Local Adaptation of Atlantic Cod despite High Connectivity. Mol. Ecol. 2017, 26, 4452–4466. [Google Scholar] [CrossRef]
- Farris, E.; Filigheddu, R.; Mameli, G.; Falanga, V.; Vanetti, I.; Rosati, L.; Binelli, G. Is Population Genetic Structure of Vascular Plants Shaped More by Ecological or Geographic Factors? A Study Case on the Mediterranean Endemic Centaurea filiformis (Asteraceae). Plant Biol. 2018, 20, 936–947. [Google Scholar] [CrossRef]
- Wang, I.J.; Bradburd, G.S. Isolation by Environment. Mol. Ecol. 2014, 23, 5649–5662. [Google Scholar] [CrossRef]
- Abratowska, A.; Wąsowicz, P.; Bednarek, P.T.; Telka, J.; Wierzbicka, M. Morphological and Genetic Distinctiveness of Metallicolous and Non-Metallicolous Populations of Armeria maritima s.l. (Plumbaginaceae) in Poland. Plant Biol. 2012, 14, 586–595. [Google Scholar] [CrossRef]
- Andriamihaja, C.F.; Ramarosandratana, A.V.; Grisoni, M.; Jeannoda, V.H.; Besse, P. Drivers of Population Divergence and Species Differentiation in a Recent Group of Indigenous Orchids (Vanilla spp.) in Madagascar. Ecol. Evol. 2021, 11, 2681–2700. [Google Scholar] [CrossRef]
- Páez-Vacas, M.I.; Trumbo, D.R.; Funk, W.C. Contrasting Environmental Drivers of Genetic and Phenotypic Divergence in an Andean Poison Frog (Epipedobates anthonyi). Heredity 2021, 128, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Talavera, M.; Min, Y.; Flaven, E.; Imbert, E. Neutral Processes Contribute to Patterns of Spatial Variation for Flower Colour in the Mediterranean Iris lutescens (Iridaceae). Ann. Bot. 2016, 117, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Nosil, P.; Egan, S.P.; Funk, D.J. Heterogeneous Genomic Differentiation between Walking-Stick Ecotypes: “Isolation by Adaptation” and Multiple Roles for Divergent Selection. Evolution 2008, 62, 316–336. [Google Scholar] [CrossRef]
- Shafer, A.B.A.; Wolf, J.B.W. Widespread Evidence for Incipient Ecological Speciation: A Meta-Analysis of Isolation-by-Ecology. Ecol. Lett. 2013, 16, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Stojanova, B.; Šurinová, M.; Klápště, J.; Koláříková, V.; Hadincová, V.; Münzbergová, Z. Adaptive Differentiation of Festuca rubra along a Climate Gradient Revealed by Molecular Markers and Quantitative Traits. PLoS ONE 2018, 13, e0194670. [Google Scholar] [CrossRef]
- Filatov, D.A.; Osborne, O.G.; Papadopulos, A.S.T. Demographic History of Speciation in a Senecio Altitudinal Hybrid Zone on Mt. Etna. Mol. Ecol. 2016, 25, 2467–2481. [Google Scholar] [CrossRef]
- Gonzalo-Turpin, H.; Hazard, L. Local Adaptation Occurs along Altitudinal Gradient despite the Existence of Gene Flow in the Alpine Plant Species Festuca eskia. J. Ecol. 2009, 97, 742–751. [Google Scholar] [CrossRef]
- Hämälä, T.; Mattila, T.M.; Savolainen, O. Local Adaptation and Ecological Differentiation under Selection, Migration, and Drift in Arabidopsis lyrata: Local Adaptation under Gene Flow and Drift. Evolution 2018, 72, 1373–1386. [Google Scholar] [CrossRef]
- Abbott, R.J.; Brennan, A.C. Altitudinal Gradients, Plant Hybrid Zones and Evolutionary Novelty. Phil. Trans. R Soc. B 2014, 369, 20130346. [Google Scholar] [CrossRef]
- Körner, C. Alpine Plant Life: Functional Plant Ecology of High Mountain Ecosystems; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Pellissier, L.; Fournier, B.; Guisan, A.; Vittoz, P. Plant Traits Co-Vary with Altitude in Grasslands and Forests in the European Alps. Plant Ecol. 2010, 211, 351–365. [Google Scholar] [CrossRef]
- Halbritter, A.H.; Fior, S.; Keller, I.; Billeter, R.; Edwards, P.J.; Holderegger, R.; Karrenberg, S.; Pluess, A.R.; Widmer, A.; Alexander, J.M. Trait Differentiation and Adaptation of Plants along Elevation Gradients. J. Evol. Biol. 2018, 31, 784–800. [Google Scholar] [CrossRef] [PubMed]
- Merilä, J. Quantitative Trait and Allozyme Divergence in the Greenfinch (Carduelis chloris, Aves: Fringillidae). Biol. J. Linn. Soc. 1997, 61, 243–266. [Google Scholar] [CrossRef]
- Leinonen, T.; Cano, J.M.; Mäkinen, H.; Merilä, J. Contrasting Patterns of Body Shape and Neutral Genetic Divergence in Marine and Lake Populations of Threespine Sticklebacks. J. Evol. Biol. 2006, 19, 1803–1812. [Google Scholar] [CrossRef]
- Brommer, J.E. Whither Pst? The Approximation of Qst by Pst in Evolutionary and Conservation Biology: Whither Pst? J. Evol. Biol. 2011, 24, 1160–1168. [Google Scholar] [CrossRef]
- Spitze, K. Population Structure in Daphnia Obtusa: Quantitative Genetic and Allozymic Variation. Genetics 1993, 135, 367–374. [Google Scholar] [CrossRef]
- Leinonen, T.; McCairns, R.J.S.; O’Hara, R.B.; Merilä, J. QST–FST Comparisons: Evolutionary and Ecological Insights from Genomic Heterogeneity. Nat. Rev. Genet. 2013, 14, 179–190. [Google Scholar] [CrossRef]
- Marin, S.; Gibert, A.; Archambeau, J.; Bonhomme, V.; Lascoste, M.; Pujol, B. Potential Adaptive Divergence between Subspecies and Populations of Snapdragon Plants Inferred from QST—FST Comparisons. Mol. Ecol. 2020, 29, 3010–3021. [Google Scholar] [CrossRef]
- Merilä, J.; Crnokrak, P. Comparison of Genetic Differentiation at Marker Loci and Quantitative Traits: Natural Selection and Genetic Differentiation. J. Evol. Biol. 2001, 14, 892–903. [Google Scholar] [CrossRef]
- Pujol, B.; Wilson, A.J.; Ross, R.I.C.; Pannell, J.R. Are QST—FST Comparisons for Natural Populations Meaningful? Mol. Ecol. 2008, 17, 4782–4785. [Google Scholar] [CrossRef]
- Bajpai, P.K.; Weiss, H.; Dvir, G.; Hanin, N.; Wasserstrom, H.; Barazani, O. Phenotypic Differentiation and Diversifying Selection in Populations of Eruca sativa along an Aridity Gradient. BMC Ecol. Evol. 2022, 22, 40. [Google Scholar] [CrossRef] [PubMed]
- Vitek, N.S.; McDaniel, S.F.; Bloch, J.I. Microevolutionary Variation in Molar Morphology of Onychomys leucogaster Decoupled from Genetic Structure. Evolution 2022, 76, 2032–2048. [Google Scholar] [CrossRef]
- Castellani, M.B.; Dalla Vecchia, A.; Bolpagni, R.; Natale, R.; Piaser, E.; Lastrucci, L.; Coppi, A.; Villa, P. Genetic Drift versus Natural Selection Affecting the Evolution of Spectral and Functional Traits of Two Key Macrophytes: Phragmites australis and Nuphar lutea. Freshw. Biol. 2023, 68, 1739–1750. [Google Scholar] [CrossRef]
- Salloum, P.M.; Lavery, S.D.; de Villemereuil, P.; Santure, A.W. Local Adaptation in Shell Shape Traits of a Brooding Chiton with Strong Population Genomic Differentiation. Evolution 2023, 77, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Domina, G.; Astuti, G.; Barone, G.; Gargano, D.; Minuto, L.; Varaldo, L.; Peruzzi, L. Lectotypification of the Linnaean Name Dianthus virgineus (Caryophyllaceae) and Its Taxonomic Consequences. Taxon 2021, 70, 1096–1100. [Google Scholar] [CrossRef]
- Tison, J.-M.; de Foucault, B. Flora Gallica: Flore de France; Biotope Éditions: Mèze, France, 2014. [Google Scholar]
- Brullo, S.; Guarino, R. Complesso Di Dianthus Sylvestris. In Flora d’Italia; Pignatti, S., Ed.; New Business Media: Milan, Italy, 2017; Volume 2, pp. 200–205. [Google Scholar]
- Gargano, D.; Franzoni, J.; Luqman, H.; Fior, S.; Rovito, S.; Peruzzi, L. Phenotypic Correlates of Genetic Divergence Suggest at Least Three Species in the Complex of Dianthus virgineus (Caryophyllaceae). Taxon 2023. [Google Scholar] [CrossRef]
- Luqman, H.; Wegmann, D.; Fior, S.; Widmer, A. Climate-Induced Range Shifts Drive Adaptive Response via Spatio-Temporal Sieving of Alleles. Nat. Commun. 2023, 14, 1080. [Google Scholar] [CrossRef]
- Caruel, T. Prodromo Della Flora Toscana; Felice Le Monnier: Firenze, Italy, 1860. [Google Scholar]
- Bacchetta, G.; Brullo, S.; Casti, M.; Giusso del Galdo, G.P. Taxonomic Revision of the Dianthus sylvestris Group (Caryophyllaceae) in Central-Southern Italy, Sicily and Sardinia. Nord. J. Bot. 2010, 28, 137–173. [Google Scholar] [CrossRef]
- Tutin, T.G. Dianthus L. In Flora Europaea; Tutin, T.G., Burges, N.A., Chater, A.D., Edmondson, J.R., Heywood, V.H., Moore, D.M., Valentine, D.H., Walters, S.M., Webb, D.A., Eds.; Cambridge University Press: Cambridge, UK, 1993; Volume 1, pp. 227–246. [Google Scholar]
- Erhardt, A. Pollination and Reproduction in Dianthus silvester Wulf. In Sexual Reproduction in Higher Plants; Cresti, M., Gori, P., Pacini, E., Eds.; Springer: Berlin/Heidelberg, Germany, 1988; pp. 351–356. [Google Scholar]
- Collin, C.L.; Shykoff, J.A. Outcrossing Rates in the Gynomonoecious-Gynodioecious Species Dianthus sylvestris (Caryophyllaceae). Am. J. Bot. 2003, 90, 579–585. [Google Scholar] [CrossRef]
- Nebot, A.; Cogoni, D.; Fenu, G.; Bacchetta, G. Floral Biology and Breeding System of the Narrow Endemic Dianthus morisianus Vals. (Caryophyllaceae). Flora 2016, 219, 1–7. [Google Scholar] [CrossRef]
- Peruzzi, L.; Bedini, G. Wikiplantbase #Toscana v2.1. Available online: http://bot.biologia.unipi.it/wpb/toscana/index.html (accessed on 5 October 2023).
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km Spatial Resolution Climate Surfaces for Global Land Areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Franzoni, J.; Astuti, G.; Bacchetta, G.; Barone, G.; Bartolucci, F.; Bernardo, L.; Carta, A.; Conti, F.; Domina, G.; Frajman, B.; et al. A Cytosystematic Study of the Dianthus virgineus Complex (Caryophyllaceae) in the Central Mediterranean. J. Syst. Evol. 2023, in press. [Google Scholar]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double Digest RADseq: An Inexpensive Method for de Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Westergaard, K.B.; Zemp, N.; Bruederle, L.P.; Stenøien, H.K.; Widmer, A.; Fior, S. Population Genomic Evidence for Plant Glacial Survival in Scandinavia. Mol. Ecol. 2019, 28, 818–832. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An Analysis Tool Set for Population Genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef]
- Puritz, J.B.; Hollenbeck, C.M.; Gold, J.R. dDocent: A RADseq, Variant-Calling Pipeline Designed for Population Genomics of Non-Model Organisms. PeerJ 2014, 2, e431. [Google Scholar] [CrossRef]
- Chong, Z.; Ruan, J.; Wu, C.-I. Rainbow: An Integrated Tool for Efficient Clustering and Assembling RAD-Seq Reads. Bioinformatics 2012, 28, 2732–2737. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997v2. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup the Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based Variant Detection from Short-read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Wigginton, J.E.; Cutler, D.J.; Abecasis, G.R. A Note on Exact Tests of Hardy-Weinberg Equilibrium. Am. J. Hum. Genet. 2005, 76, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Bresadola, L.; Link, V.; Buerkle, C.A.; Lexer, C.; Wegmann, D. Estimating and Accounting for Genotyping Errors in RAD-seq Experiments. Mol. Ecol. Resour. 2020, 20, 856–870. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Dominant Markers and Null Alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring Weak Population Structure with the Assistance of Sample Group Information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Wang, J. The Computer Program STRUCTURE for Assigning Individuals to Populations: Easy to Use but Easier to Misuse. Mol. Ecol. Resour. 2017, 17, 981–990. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A Program for Identifying Clustering Modes and Packaging Population Structure Inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Goudet, J. Hierfstat, a Package for r to Compute and Test Hierarchical F-Statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Terlević, A.; Temunović, M.; Bogdanović, S.; Grgurev, M.; Ljubičić, I.; Rešetnik, I. Morphological and Environmental Variability of Dianthus sylvestris (Caryophyllaceae) in the Balkan Peninsula. Bot. J. Linn. Soc. 2023, 201, 377–389. [Google Scholar] [CrossRef]
- Castro, I.; Rocha, J.; Martins, M.; Carnide, V.; Martín, J.P.; Veiga, P.; Serafim, A.B.; Amich, F.; Ramírez-Rodríguez, R.; Colombo, G.; et al. The Redundancy Effect under Morphogenetic and Environmental Fluctuations. The Case of the Dianthus pungens Group. Plant Biosyst. 2022, 156, 292–306. [Google Scholar] [CrossRef]
- De Vries, J.; Fior, S.; Pålsson, A.; Widmer, A.; Alexander, J.M. Unravelling Drivers of Local Adaptation through Evolutionary Functional-Structural Plant Modelling. bioRxiv 2022. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 1 September 2023).
- Legendre, P.; Fortin, M.-J. Comparison of the Mantel Test and Alternative Approaches for Detecting Complex Multivariate Relationships in the Spatial Analysis of Genetic Data: Spatial Analysis of Genetic Data. Mol. Ecol. Resour. 2010, 10, 831–844. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A. The ade4 Package: Implementing the Duality Diagram for Ecologists. J. Stat. Soft 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Hijmans, R.J. Geosphere: Spherical Trigonometry. Available online: https://CRAN.R-project.org/package=geosphere (accessed on 5 October 2023).
- Da Silva, S.B.; Da Silva, A. Pstat: An R Package to Assess Population Differentiation in Phenotypic Traits. R J. 2018, 10, 447. [Google Scholar] [CrossRef]
- Bradburd, G.S.; Coop, G.M.; Ralph, P.L. Inferring Continuous and Discrete Population Genetic Structure across Space. Genetics 2018, 210, 33–52. [Google Scholar] [CrossRef]
- Jaros, U.; Tribsch, A.; Comes, H.P. Diversification in Continental Island Archipelagos: New Evidence on the Roles of Fragmentation, Colonization and Gene Flow on the Genetic Divergence of Aegean Nigella (Ranunculaceae). Ann. Bot. 2018, 121, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Volkova, P.A.; Bog, M.; Zablocka, B.; Oberprieler, C. Elevation Does Not Matter? Genome Screening Using AFLP Fails to Reveal Selection along Elevational Transects: A Case Study of Caucasian Primula vulgaris Huds. (Primulaceae). Flora 2021, 274, 151726. [Google Scholar] [CrossRef]
- Jürgens, A.; Witt, T.; Gottsberger, G. Flower Scent Composition in Dianthus and Saponaria Species (Caryophyllaceae) and Its Relevance for Pollination Biology and Taxonomy. Biochem. Syst. Ecol. 2003, 31, 345–357. [Google Scholar] [CrossRef]
- Wessinger, C.A. From Pollen Dispersal to Plant Diversification: Genetic Consequences of Pollination Mode. New Phytol. 2021, 229, 3125–3132. [Google Scholar] [CrossRef]
- Jürgens, A. Comparative Floral Morphometrics in Day-Flowering, Night-Flowering and Self-Pollinated Caryophylloideae (Agrostemma, Dianthus, Saponaria, Silene, and Vaccaria). Plant Syst. Evol. 2006, 257, 233–250. [Google Scholar] [CrossRef]
- Collin, C.; Pennings, P.; Rueffler, C.; Widmer, A.; Shykoff, J. Natural Enemies and Sex: How Seed Predators and Pathogens Contribute to Sex-Differential Reproductive Success in a Gynodioecious Plant. Oecologia 2002, 131, 94–102. [Google Scholar] [CrossRef]
- Bakhtiari, M.; Formenti, L.; Caggìa, V.; Glauser, G.; Rasmann, S. Variable Effects on Growth and Defense Traits for Plant Ecotypic Differentiation and Phenotypic Plasticity along Elevation Gradients. Ecol. Evol. 2019, 9, 3740–3755. [Google Scholar] [CrossRef]
- Hardion, L.; Perrier, A.; Martinez, M.; Navrot, N.; Gaquerel, E.; Tournay, F.; Nguefack, J.; Combroux, I. Integrative Revision of Dianthus superbus Subspecies Reveals Different Degrees of Differentiation, from Plasticity to Species Distinction. Syst. Biodivers. 2020, 18, 255–268. [Google Scholar] [CrossRef]
- D’Antraccoli, M.; Carta, A.; Astuti, G.; Franzoni, J.; Giacò, A.; Tiburtini, M.; Pinzani, L.; Peruzzi, L. A Comprehensive Approach to Improving Endemic Plant Species Research, Conservation, and Popularization. JZBG 2023, 4, 490–506. [Google Scholar] [CrossRef]
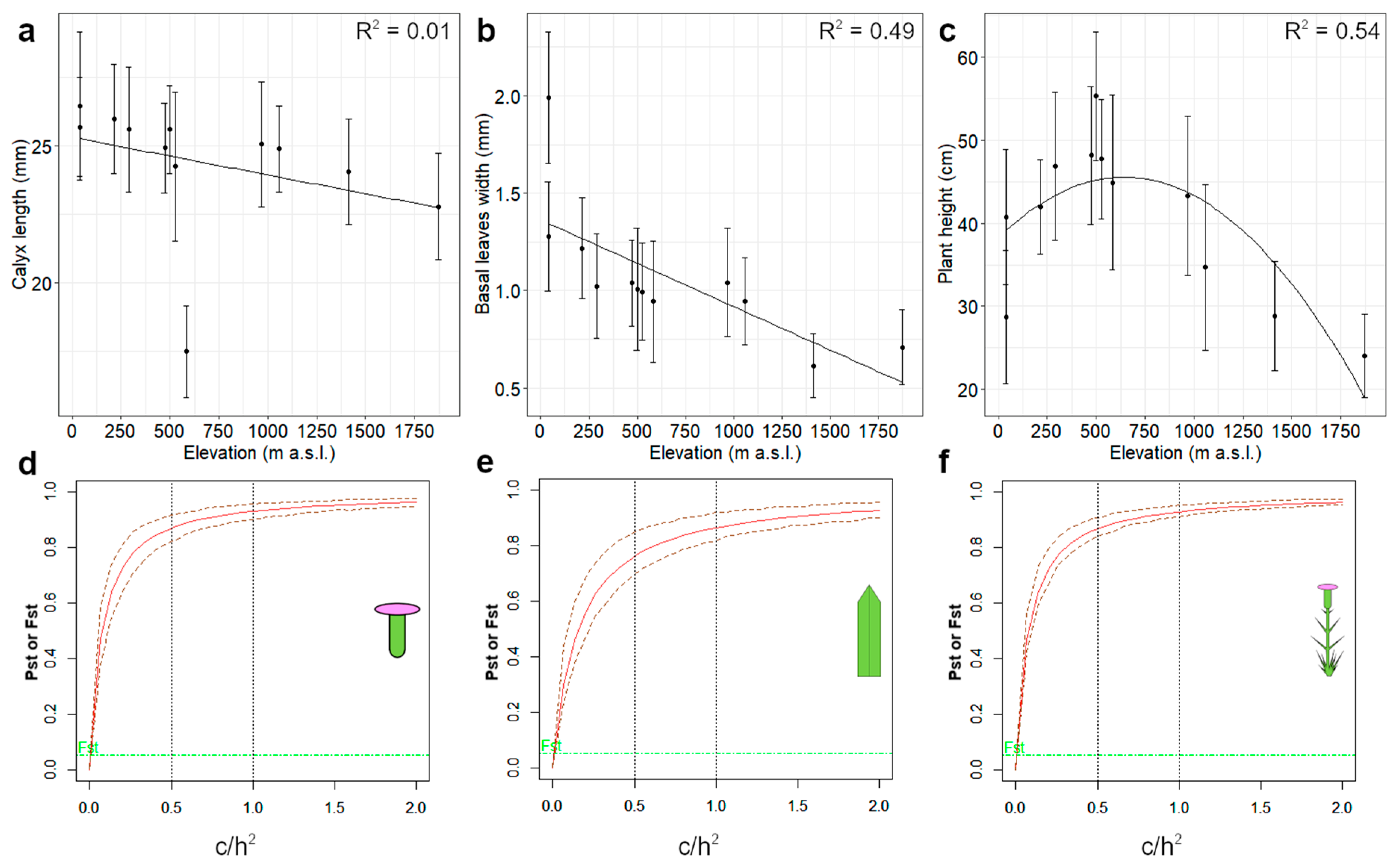
| Character | ID | c/h2 | Pst/Fst | 99% Lower CI | 99% Upper CI |
|---|---|---|---|---|---|
| Plant height | 1 | 1 | 0.929 | 0.903 | 0.957 |
| 0.1 | 0.566 | 0.484 | 0.684 | ||
| Basal leaf width | 6 | 1 | 0.864 | 0.800 | 0.925 |
| 0.1 | 0.388 | 0.285 | 0.555 | ||
| Calyx length | 16 | 1 | 0.930 | 0.887 | 0.961 |
| 0.1 | 0.571 | 0.449 | 0.718 | ||
| Genetic divergence at neutral loci | 0.049 | 0.043 | 0.053 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franzoni, J.; Astuti, G.; Peruzzi, L. Weak Genetic Isolation and Putative Phenotypic Selection in the Wild Carnation Dianthus virgineus (Caryophyllaceae). Biology 2023, 12, 1355. https://doi.org/10.3390/biology12101355
Franzoni J, Astuti G, Peruzzi L. Weak Genetic Isolation and Putative Phenotypic Selection in the Wild Carnation Dianthus virgineus (Caryophyllaceae). Biology. 2023; 12(10):1355. https://doi.org/10.3390/biology12101355
Chicago/Turabian StyleFranzoni, Jacopo, Giovanni Astuti, and Lorenzo Peruzzi. 2023. "Weak Genetic Isolation and Putative Phenotypic Selection in the Wild Carnation Dianthus virgineus (Caryophyllaceae)" Biology 12, no. 10: 1355. https://doi.org/10.3390/biology12101355
APA StyleFranzoni, J., Astuti, G., & Peruzzi, L. (2023). Weak Genetic Isolation and Putative Phenotypic Selection in the Wild Carnation Dianthus virgineus (Caryophyllaceae). Biology, 12(10), 1355. https://doi.org/10.3390/biology12101355







