Role of the Circadian Gas-Responsive Hemeprotein NPAS2 in Physiology and Pathology
Abstract
Simple Summary
Abstract
1. Introduction
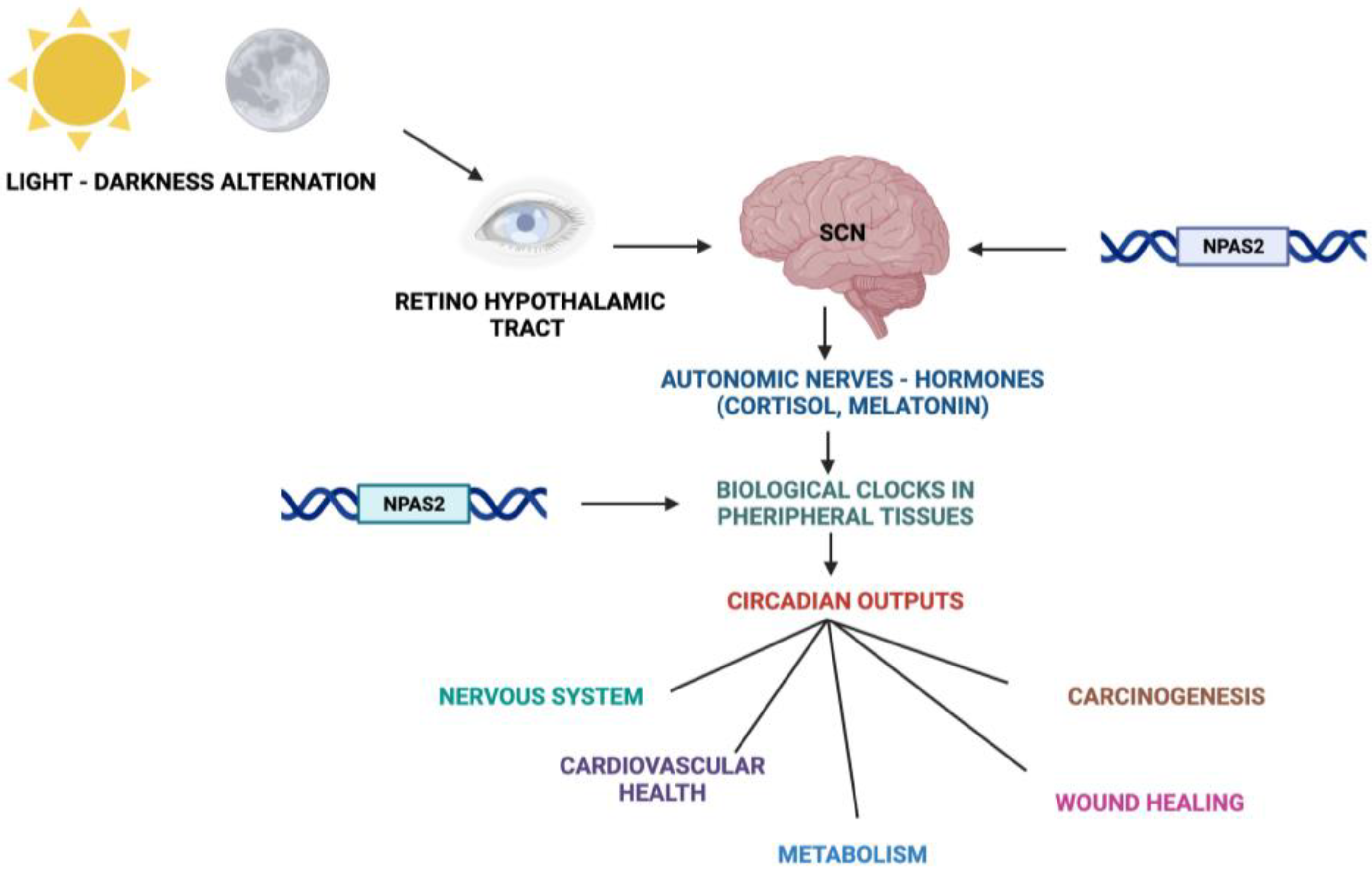
2. The Circadian Timing System
3. NPAS2 and the Molecular Clockwork
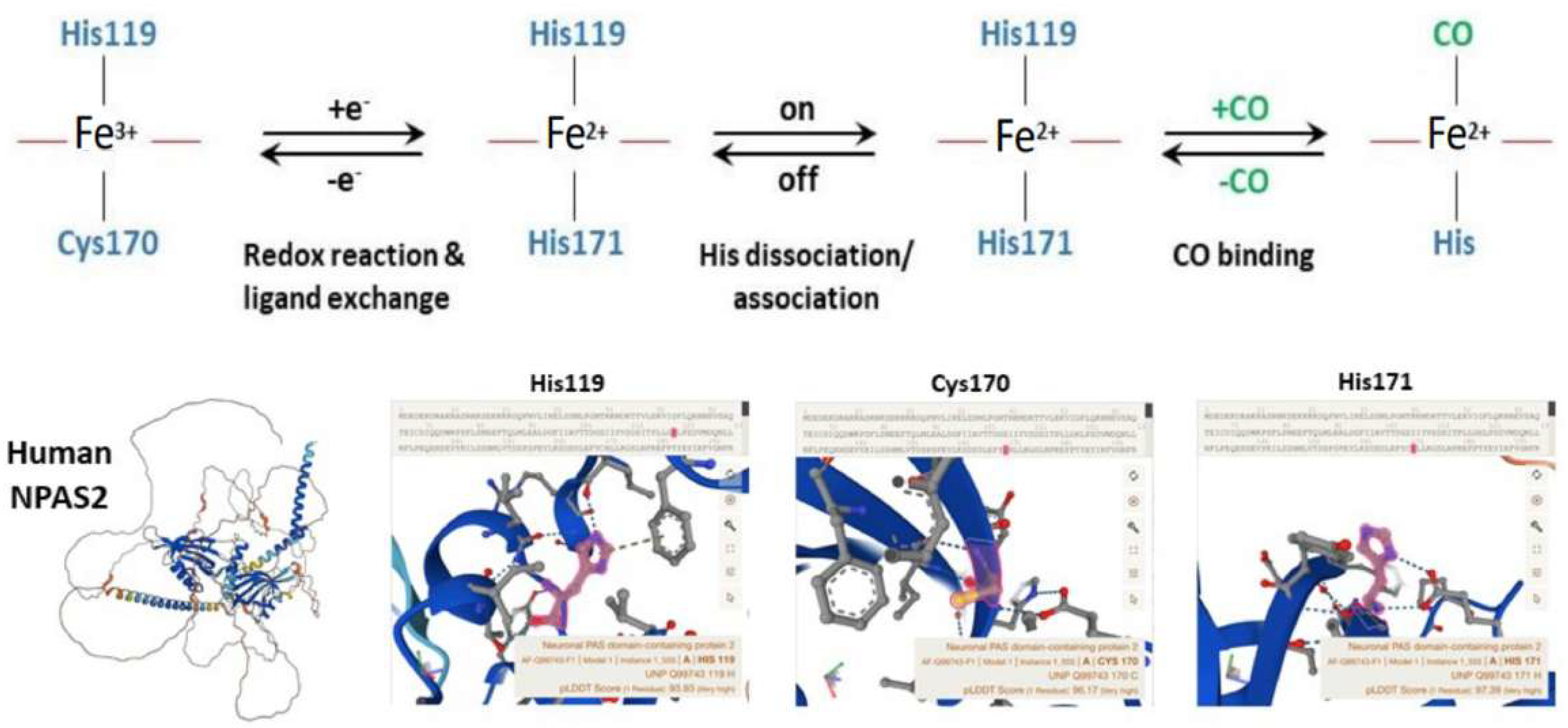
4. NPAS2, Hemeprotein and Gas-Responsive Transcription Factor
5. NPAS2 and the Metabolic Pathways
6. NPAS2 and the Central Nervous System in Mammals
7. NPAS2 in Cancer Onset and Progression
8. NPAS2 and the Cardiovascular System
9. NPAS2 and the Process of Wound Healing
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, Y.-D.; Barnard, M.; Tian, H.; Li, X.; Ring, H.Z.; Francke, U.; Shelton, J.; Richardson, J.; Russell, D.W.; McKnight, S.L. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc. Natl. Acad. Sci. USA 1997, 94, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.U.; Bai, G.; Pang, Y. Roles of NPAS2 in circadian rhythm and disease. Acta Biochim. Biophys. Sin. 2021, 53, 1257–1265. [Google Scholar] [CrossRef]
- Reick, M.; Garcia, J.A.; Dudley, C.; McKnight, S.L. NPAS2: An Analog of Clock Operative in the Mammalian Forebrain. Science 2001, 293, 506–509. [Google Scholar] [CrossRef]
- Chaix, A.; Zarrinpar, A.; Panda, S. The circadian coordination of cell biology. J. Cell Biol. 2016, 215, 15–25. [Google Scholar] [CrossRef]
- Hastings, M.H.; Reddy, A.B.; Maywood, E.S. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 2003, 4, 649–661. [Google Scholar] [CrossRef]
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2013, 24, 90–99. [Google Scholar] [CrossRef]
- Ray, S.; Reddy, A.B. Cross-talk between circadian clocks, sleep-wake cycles, and metabolic networks: Dispelling the darkness. BioEssays 2016, 38, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Bass, J.; Takahashi, J.S. Circadian Integration of Metabolism and Energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef]
- Patton, A.P.; Hastings, M.H. The Mammalian Circadian Time-Keeping System. J. Huntington’s Dis. 2023, 12, 91–104. [Google Scholar] [CrossRef]
- O’Neill, J.S.; Maywood, E.S.; Hastings, M.H. Cellular mechanisms of circadian pacemaking: Beyond transcriptional loops. Handb. Exp. Pharmacol. 2013, 217, 67–103. [Google Scholar] [CrossRef]
- Hastings, M.H.; Brancaccio, M.; Gonzalez-Aponte, M.F.; Herzog, E.D. Circadian Rhythms and Astrocytes: The Good, the Bad, and the Ugly. Annu. Rev. Neurosci. 2023, 46, 123–143. [Google Scholar] [CrossRef]
- Maywood, E.S.; O’Neill, J.S.; Reddy, A.B.; Chesham, J.E.; Prosser, H.M.; Kyriacou, C.P.; Godinho, S.I.H.; Nolan, P.M.; Hastings, M.H. Genetic and Molecular Analysis of the Central and Peripheral Circadian Clockwork of Mice. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Kim, J.Y. Mammalian circadian networks mediated by the suprachiasmatic nucleus. FEBS J. 2021, 289, 6589–6604. [Google Scholar] [CrossRef]
- Honma, S. The mammalian circadian system: A hierarchical multi-oscillator structure for generating circadian rhythm. J. Physiol. Sci. 2018, 68, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Astiz, M.; Heyde, I.; Oster, H. Mechanisms of Communication in the Mammalian Circadian Timing System. Int. J. Mol. Sci. 2019, 20, 343. [Google Scholar] [CrossRef]
- Rosenwasser, A.M.; Turek, F.W. Neurobiology of Circadian Rhythm Regulation. Sleep Med. Clin. 2015, 10, 403–412. [Google Scholar] [CrossRef]
- Patton, A.P.; Edwards, M.D.; Smyllie, N.J.; Hamnett, R.; Chesham, J.E.; Brancaccio, M.; Maywood, E.S.; Hastings, M.H. The VIP-VPAC2 neuropeptidergic axis is a cellular pacemaking hub of the suprachiasmatic nucleus circadian circuit. Nat. Commun. 2020, 11, 502–512. [Google Scholar] [CrossRef]
- Hamnett, R.; Chesham, J.E.; Maywood, E.S.; Hastings, M.H. The Cell-Autonomous Clock of VIP Receptor VPAC2 Cells Regulates Period and Coherence of Circadian Behavior. J. Neurosci. 2020, 41, 502–512. [Google Scholar] [CrossRef]
- Hastings, M.H.; Maywood, E.S.; Brancaccio, M. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 2018, 19, 453–469. [Google Scholar] [CrossRef] [PubMed]
- Varadarajan, S.; Tajiri, M.; Jain, R.; Holt, R.; Ahmed, Q.; LeSauter, J.; Silver, R. Connectome of the Suprachiasmatic Nucleus: New Evidence of the Core-Shell Relationship. Eneuro 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, M.; Patton, A.P.; Chesham, J.E.; Maywood, E.S.; Hastings, M.H. Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron 2017, 93, 1420–1435.e5. [Google Scholar] [CrossRef]
- Brancaccio, M.; Wolfes, A.C.; Ness, N. Astrocyte Circadian Timekeeping in Brain Health and Neurodegeneration. Adv. Exp. Med. Biol. 2021, 1344, 87–110. [Google Scholar] [CrossRef]
- Bell-Pedersen, D.; Cassone, V.M.; Earnest, D.J.; Golden, S.S.; Hardin, P.E.; Thomas, T.L.; Zoran, M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005, 6, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.S.; Díaz, N.M.; D’souza, S.; Buhr, E.D. The molecular clockwork of mammalian cells. Semin. Cell Dev. Biol. 2022, 126, 87–96. [Google Scholar] [CrossRef]
- DeBruyne, J.P.; Weaver, D.R.; Reppert, S.M. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci. 2007, 10, 543–545. [Google Scholar] [CrossRef] [PubMed]
- Bertolucci, C.; Cavallari, N.; Colognesi, I.; Aguzzi, J.; Chen, Z.; Caruso, P.; Foá, A.; Tosini, G.; Bernardi, F.; Pinotti, M. Evidence for an Overlapping Role of CLOCK and NPAS2 Transcription Factors in Liver Circadian Oscillators. Mol. Cell. Biol. 2008, 28, 3070–3075. [Google Scholar] [CrossRef]
- Landgraf, D.; Wang, L.L.; Diemer, T.; Welsh, D.K. NPAS2 Compensates for Loss of CLOCK in Peripheral Circadian Oscillators. PLoS Genet. 2016, 12, e1005882. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2016, 18, 164–179. [Google Scholar] [CrossRef]
- Milev, N.B.; Rhee, S.-G.; Reddy, A.B. Cellular Timekeeping: It’s Redox o’Clock. Cold Spring Harb. Perspect. Biol. 2018, 10, a027698. [Google Scholar] [CrossRef]
- Robinson, I.; Reddy, A. Molecular mechanisms of the circadian clockwork in mammals. FEBS Lett. 2014, 588, 2477–2483. [Google Scholar] [CrossRef]
- Reddy, A.B. Genome-wide analyses of circadian systems. Handb. Exp. Pharmacol. 2013, 217, 379–388. [Google Scholar] [CrossRef]
- Cardone, L.; Hirayama, J.; Giordano, F.; Tamaru, T.; Palvimo, J.J.; Sassone-Corsi, P. Circadian Clock Control by SUMOylation of BMAL1. Science 2005, 309, 1390–1394. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, Y.; Lee, M.J.; Park, E.; Kang, S.H.; Chung, C.H.; Lee, K.H.; Kim, K. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol. Cell. Boil. 2008, 28, 6056–6065. [Google Scholar] [CrossRef]
- Sahar, S.; Zocchi, L.; Kinoshita, C.; Borrelli, E.; Sassone-Corsi, P. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS ONE 2010, 5, e8561. [Google Scholar] [CrossRef] [PubMed]
- Duez, H.; Staels, B.; Pierre, K.; Schlesinger, N.; Androulakis, I.P.; He, L.; Hamm, J.A.; Reddy, A.; Sams, D.; Peliciari-Garcia, R.A.; et al. Rev-erb-α: An integrator of circadian rhythms and metabolism. J. Appl. Physiol. 2009, 107, 1972–1980. [Google Scholar] [CrossRef]
- Crumbley, C.; Wang, Y.; Kojetin, D.J.; Burris, T.P. Characterization of the Core Mammalian Clock Component, NPAS2, as a REV-ERBα/RORα Target Gene. J. Biol. Chem. 2010, 285, 35386–35392. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, R.; Matsubara, C.; Node, K.; Takumi, T.; Akashi, M. Nuclear Receptor-mediated Cell-autonomous Oscillatory Expression of the Circadian Transcription Factor, Neuronal PAS Domain Protein 2 (NPAS2). J. Biol. Chem. 2013, 288, 36548–36553. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Kang, H.S.; Angers, M.; Jetten, A.M. Retinoic acid-related orphan receptor γ directly regulates neuronal PAS domain protein 2 transcription in vivo. Nucleic Acids Res. 2011, 39, 4769–4782. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hughes, M.E.; DiTacchio, L.; Hayes, K.R.; Vollmers, C.; Pulivarthy, S.; Baggs, J.E.; Panda, S.; Hogenesch, J.B. Harmonics of Circadian Gene Transcription in Mammals. PLoS Genet. 2009, 5, e1000442. [Google Scholar] [CrossRef] [PubMed]
- Bozek, K.; Relógio, A.; Kielbasa, S.M.; Heine, M.; Dame, C.; Kramer, A.; Herzel, H. Regulation of Clock-Controlled Genes in Mammals. PLoS ONE 2009, 4, e4882. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, S.; Yamaguchi, S.; Matsuo, T.; Ishida, Y.; Okamura, H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Minerva Anestesiol. 2001, 15, 995–1006. [Google Scholar] [CrossRef]
- Ueda, H.R.; Hayashi, S.; Chen, W.; Sano, M.; Machida, M.; Shigeyoshi, Y.; Iino, M.; Hashimoto, S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005, 37, 187–192. [Google Scholar] [CrossRef]
- Yu, X.; Rollins, D.; Ruhn, K.A.; Stubblefield, J.J.; Green, C.B.; Kashiwada, M.; Rothman, P.B.; Takahashi, J.S.; Hooper, L.V. T H 17 Cell Differentiation Is Regulated by the Circadian Clock. Science 2013, 342, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Finger, A.; Kramer, A. Mammalian circadian systems: Organization and modern life challenges. Acta Physiol. 2020, 231, e13548. [Google Scholar] [CrossRef] [PubMed]
- Noshiro, M.; Furukawa, M.; Honma, S.; Kawamoto, T.; Hamada, T.; Honma, K.-I.; Kato, Y. Tissue-Specific Disruption of Rhythmic Expression of Dec1 and Dec2 in Clock Mutant Mice. J. Biol. Rhythm. 2005, 20, 404–418. [Google Scholar] [CrossRef] [PubMed]
- Sahar, S.; Sassone-Corsi, P. The epigenetic language of circadian clocks. Handb. Exp. Pharmacol. 2013, 217, 29–44. [Google Scholar] [CrossRef]
- Doi, M.; Hirayama, J.; Sassone-Corsi, P. Circadian Regulator CLOCK Is a Histone Acetyltransferase. Cell 2006, 125, 497–508. [Google Scholar] [CrossRef]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D.; Guarente, L.P.; Sassone-Corsi, P. The NAD+-Dependent Deacetylase SIRT1 Modulates CLOCK-Mediated Chromatin Remodeling and Circadian Control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef]
- Asher, G.; Gatfield, D.; Stratmann, M.; Reinke, H.; Dibner, C.; Kreppel, F.; Mostoslavsky, R.; Alt, F.W.; Schibler, U. SIRT1 Regulates Circadian Clock Gene Expression through PER2 Deacetylation. Cell 2008, 134, 317–328. [Google Scholar] [CrossRef]
- Nakahata, Y.; Sahar, S.; Astarita, G.; Kaluzova, M.; Sassone-Corsi, P. Circadian Control of the NAD + Salvage Pathway by CLOCK-SIRT1. Science 2009, 324, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, K.M.; Yoshino, J.; Brace, C.S.; Abrassart, D.; Kobayashi, Y.; Marcheva, B.; Hong, H.-K.; Chong, J.L.; Buhr, E.D.; Lee, C.; et al. Circadian Clock Feedback Cycle Through NAMPT-Mediated NAD + Biosynthesis. Science 2009, 324, 651–654. [Google Scholar] [CrossRef]
- Itoh, R.; Fujita, K.-I.; Mu, A.; Kim, D.H.T.; Tai, T.T.; Sagami, I.; Taketani, S. Imaging of heme/hemeproteins in nucleus of the living cells expressing heme-binding nuclear receptors. FEBS Lett. 2013, 587, 2131–2136. [Google Scholar] [CrossRef] [PubMed]
- Kaasik, K.; Lee, C.C. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 2004, 430, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Uchida, T.; Sato, E.; Sato, A.; Sagami, I.; Shimizu, T.; Kitagawa, T. CO-dependent Activity-controlling Mechanism of Heme-containing CO-sensor Protein, Neuronal PAS Domain Protein 2. J. Biol. Chem. 2005, 280, 21358–21368. [Google Scholar] [CrossRef]
- Uchida, T.; Sagami, I.; Shimizu, T.; Ishimori, K.; Kitagawa, T. Effects of the bHLH domain on axial coordination of heme in the PAS-A domain of neuronal PAS domain protein 2 (NPAS2): Conversion from His119/Cys170 coordination to His119/His171 coordination. J. Inorg. Biochem. 2012, 108, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Mukaiyama, Y.; Uchida, T.; Sato, E.; Sasaki, A.; Sato, Y.; Igarashi, J.; Kurokawa, H.; Sagami, I.; Kitagawa, T.; Shimizu, T. Spectroscopic and DNA-binding characterization of the isolated heme-bound basic helix-loop-helix-PAS-A domain of neuronal PAS protein 2 (NPAS2), a transcription activator protein associated with circadian rhythms. FEBS J. 2006, 273, 2528–2539. [Google Scholar] [CrossRef] [PubMed]
- Koudo, R.; Kurokawa, H.; Sato, E.; Igarashi, J.; Uchida, T.; Sagami, I.; Kitagawa, T.; Shimizu, T. Spectroscopic characterization of the isolated heme-bound PAS-B domain of neuronal PAS domain protein 2 associated with circadian rhythms. FEBS J. 2005, 272, 4153–4162. [Google Scholar] [CrossRef] [PubMed]
- Rutter, J.; Reick, M.; Wu, L.C.; McKnight, S.L. Regulation of Clock and NPAS2 DNA Binding by the Redox State of NAD Cofactors. Science 2001, 293, 510–514. [Google Scholar] [CrossRef]
- Dioum, E.M.; Rutter, J.; Tuckerman, J.R.; Gonzalez, G.; Gilles-Gonzalez, M.-A.; McKnight, S.L. NPAS2: A Gas-Responsive Transcription Factor. Science 2002, 298, 2385–2387. [Google Scholar] [CrossRef]
- Rey, G.; Valekunja, U.K.; Feeney, K.A.; Wulund, L.; Milev, N.B.; Stangherlin, A.; Ansel-Bollepalli, L.; Velagapudi, V.; O’neill, J.S.; Reddy, A.B. The Pentose Phosphate Pathway Regulates the Circadian Clock. Cell Metab. 2016, 24, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, K.; Tajima, F.; Ishijima, S.; Sagami, I. Changes in pH and NADPH Regulate the DNA Binding Activity of Neuronal PAS Domain Protein 2, a Mammalian Circadian Transcription Factor. Biochemistry 2015, 54, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Boehning, D.; Snyder, S.H. Carbon Monoxide and Clocks. Science 2002, 298, 2339–2340. [Google Scholar] [CrossRef] [PubMed]
- Klemz, R.; Reischl, S.; Wallach, T.; Witte, N.; Jürchott, K.; Klemz, S.; Lang, V.; Lorenzen, S.; Knauer, M.; Heidenreich, S.; et al. Reciprocal regulation of carbon monoxide metabolism and the circadian clock. Nat. Struct. Mol. Biol. 2016, 24, 15–22. [Google Scholar] [CrossRef]
- Ascenzi, P.; Bocedi, A.; Leoni, L.; Visca, P.; Zennaro, E.; Milani, M.; Bolognesi, M. CO Sniffing through Heme-based Sensor Proteins. IUBMB Life 2004, 56, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Gilun, P.; Stefanczyk-Krzymowska, S.; Romerowicz-Misielak, M.; Tabecka-Lonczynska, A.; Przekop, F.; Koziorowski, M. Carbon monoxide-mediated humoral pathway for the transmission of light signal to the hypothalamus. J. Physiol. Pharmacol. 2013, 64, 761–772. [Google Scholar]
- Minegishi, S.; Sagami, I.; Negi, S.; Kano, K.; Kitagishi, H. Circadian clock disruption by selective removal of endogenous carbon monoxide. Sci. Rep. 2018, 8, 11996. [Google Scholar] [CrossRef]
- Mazzoccoli, G.; De Cosmo, S.; Mazza, T. The Biological Clock: A Pivotal Hub in Non-alcoholic Fatty Liver Disease Pathogenesis. Front. Physiol. 2018, 9, 193. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Zhang, Y.; Tsuchiya, H.; Smalling, R.; Jetten, A.M.; Wang, L. Small heterodimer partner/neuronal PAS domain protein 2 axis regulates the oscillation of liver lipid metabolism. Hepatology 2015, 61, 497–505. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Cen, H.; Guo, L.; Zhang, T.; Yang, Y.; Dong, D.; Wu, B. Circadian oscillator NPAS2 regulates diurnal expression and activity of CYP1A2 in mouse liver. Biochem. Pharmacol. 2022, 206, 115345. [Google Scholar] [CrossRef]
- Zhou, Z.; Lin, Y.; Gao, L.; Yang, Z.; Wang, S.; Wu, B. Cyp3a11 metabolism-based chronotoxicity of brucine in mice. Toxicol. Lett. 2019, 313, 188–195. [Google Scholar] [CrossRef]
- Musiek, E.S.; Lim, M.M.; Yang, G.; Bauer, A.Q.; Qi, L.; Lee, Y.; Roh, J.H.; Ortiz-Gonzalez, X.; Dearborn, J.T.; Culver, J.P.; et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J. Clin. Investig. 2013, 123, 5389–5400. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.A.; Zhang, D.; Estill, S.J.; Michnoff, C.; Rutter, J.; Reick, M.; Scott, K.; Diaz-Arrastia, R.; McKnight, S.L. Impaired Cued and Contextual Memory in NPAS2-Deficient Mice. Science 2000, 288, 2226–2230. [Google Scholar] [CrossRef]
- Ozburn, A.R.; Kern, J.; Parekh, P.K.; Logan, R.W.; Liu, Z.; Falcon, E.; Becker-Krail, D.; Purohit, K.; Edgar, N.M.; Huang, Y.; et al. NPAS2 Regulation of Anxiety-Like Behavior and GABAA Receptors. Front. Mol. Neurosci. 2017, 10, 360. [Google Scholar] [CrossRef] [PubMed]
- Ozburn, A.R.; Falcon, E.; Twaddle, A.; Nugent, A.L.; Gillman, A.G.; Spencer, S.M.; Arey, R.N.; Mukherjee, S.; Lyons-Weiler, J.; Self, D.W.; et al. Direct Regulation of Diurnal Drd3 Expression and Cocaine Reward by NPAS2. Biol. Psychiatry 2014, 77, 425–433. [Google Scholar] [CrossRef]
- DePoy, L.M.; Becker-Krail, D.D.; Zong, W.; Petersen, K.; Shah, N.M.; Brandon, J.H.; Miguelino, A.M.; Tseng, G.C.; Logan, R.W.; McClung, C.A. Circadian-Dependent and Sex-Dependent Increases in Intravenous Cocaine Self-Administration in Npas2 Mutant Mice. J. Neurosci. 2021, 41, 1046–1058. [Google Scholar] [CrossRef]
- Puig, S.; Shelton, M.A.; Barko, K.; Seney, M.L.; Logan, R.W. Sex-specific role of the circadian transcription factor NPAS2 in opioid tolerance, withdrawal and analgesia. Genes, Brain Behav. 2022, 21, e12829. [Google Scholar] [CrossRef]
- Becker-Krail, D.D.; Parekh, P.K.; Ketchesin, K.D.; Yamaguchi, S.; Yoshino, J.; Hildebrand, M.A.; Dunham, B.; Ganapathiraju, M.K.; Logan, R.W.; McClung, C.A. Circadian transcription factor NPAS2 and the NAD+-dependent deacetylase SIRT1 interact in the mouse nucleus accumbens and regulate reward. Eur. J. Neurosci. 2022, 55, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Gamble, M.C.; Chuan, B.; Gallego-Martin, T.; Shelton, M.A.; Puig, S.; O’donnell, C.P.; Logan, R.W. A role for the circadian transcription factor NPAS2 in the progressive loss of non-rapid eye movement sleep and increased arousal during fentanyl withdrawal in male mice. Psychopharmacology 2022, 239, 3185–3200. [Google Scholar] [CrossRef] [PubMed]
- Parekh, P.K.; Logan, R.W.; Ketchesin, K.D.; Becker-Krail, D.; Shelton, M.A.; Hildebrand, M.A.; Barko, K.; Huang, Y.H.; McClung, C.A. Cell-Type-Specific Regulation of Nucleus Accumbens Synaptic Plasticity and Cocaine Reward Sensitivity by the Circadian Protein, NPAS2. J. Neurosci. 2019, 39, 4657–4667. [Google Scholar] [CrossRef]
- Herichová, I.; Hasáková, K.; Lukáčová, D.; Mravec, B.; Horváthová, Ľ.; Kavická, D. Prefrontal Cortex and Dorsomedial Hypothalamus Mediate Food Reward-Induced Effects via npas2 and egr1 Expression in Rat. Physiol. Res. 2017, 66, S501–S510. [Google Scholar] [CrossRef] [PubMed]
- Franken, P.; Dudley, C.A.; Estill, S.J.; Barakat, M.; Thomason, R.; O’Hara, B.F.; McKnight, S.L. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: Genotype and sex interactions. Proc. Natl. Acad. Sci. USA 2006, 103, 7118–7123. [Google Scholar] [CrossRef]
- Evans, D.S.; Parimi, N.; Nievergelt, C.M.; Blackwell, T.; Redline, S.; Ancoli-Israel, S.; Orwoll, E.S.; Cummings, S.R.; Stone, K.L.; Tranah, G.J.; et al. Common Genetic Variants in ARNTL and NPAS2 and at Chromosome 12p13 are Associated with Objectively Measured Sleep Traits in the Elderly. Sleep 2013, 36, 431–446. [Google Scholar] [CrossRef]
- Dall’Ara, I.; Ghirotto, S.; Ingusci, S.; Bagarolo, G.; Bertolucci, C.; Barbujani, G. Demographic history and adaptation account for clock gene diversity in humans. Heredity 2016, 117, 165–172. [Google Scholar] [CrossRef]
- Kovanen, L.; Saarikoski, S.T.; Aromaa, A.; Lönnqvist, J.; Partonen, T. ARNTL (BMAL1) and NPAS2 Gene Variants Contribute to Fertility and Seasonality. PLoS ONE 2010, 5, e10007. [Google Scholar] [CrossRef]
- Partonen, T.; Treutlein, J.; Alpman, A.; Frank, J.; Johansson, C.; Depner, M.; Aron, L.; Rietschel, M.; Wellek, S.; Soronen, P.; et al. Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann. Med. 2007, 39, 229–238. [Google Scholar] [CrossRef]
- Geoffroy, P.A.; Lajnef, M.; Bellivier, F.; Jamain, S.; Gard, S.; Kahn, J.-P.; Henry, C.; Leboyer, M.; Etain, B. Genetic association study of circadian genes with seasonal pattern in bipolar disorders. Sci. Rep. 2015, 5, 10232. [Google Scholar] [CrossRef] [PubMed]
- Yeim, S.; Boudebesse, C.; Etain, B.; Belliviera, F. Biomarqueurs et gènes circadiens dans le trouble bipolaire. L’Encéphale 2015, 41, S38–S44. [Google Scholar] [CrossRef]
- Kim, H.-I.; Lee, H.-J.; Cho, C.-H.; Kang, S.-G.; Yoon, H.-K.; Park, Y.-M.; Lee, S.-H.; Moon, J.-H.; Song, H.-M.; Lee, E.; et al. Association of CLOCK, ARNTL, and NPAS2 gene polymorphisms and seasonal variations in mood and behavior. Chrono. Int. 2015, 32, 785–791. [Google Scholar] [CrossRef]
- Etain, B.; Milhiet, V.; Boudebesse, C.; Bellivier, F.; Drouot, X.; Henry, C.; Leboyer, M. Circadian abnormalities as markers of susceptibility in bipolar disorders. Front. Biosci. 2014, 6, 120–137. [Google Scholar] [CrossRef] [PubMed]
- Partonen, T. Clock gene variants in mood and anxiety disorders. J. Neural Transm. 2012, 119, 1133–1145. [Google Scholar] [CrossRef] [PubMed]
- Soria, V.; Martínez-Amorós, E.; Escaramís, G.; Valero, J.; Pérez-Egea, R.; García, C.; Gutiérrez-Zotes, A.; Puigdemont, D.; Bayés, M.; Crespo, J.M.; et al. Differential Association of Circadian Genes with Mood Disorders: CRY1 and NPAS2 are Associated with Unipolar Major Depression and CLOCK and VIP with Bipolar Disorder. Neuropsychopharmacology 2010, 35, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Johansson, C.; Willeit, M.; Smedh, C.; Ekholm, J.; Paunio, T.; Kieseppä, T.; Lichtermann, D.; Praschak-Rieder, N.; Neumeister, A.; Nilsson, L.-G.; et al. Circadian Clock-Related Polymorphisms in Seasonal Affective Disorder and their Relevance to Diurnal Preference. Neuropsychopharmacology 2002, 28, 734–739. [Google Scholar] [CrossRef]
- Yang, Y.; Lindsey-Boltz, L.A.; Vaughn, C.M.; Selby, C.P.; Cao, X.; Liu, Z.; Hsu, D.S.; Sancar, A. Circadian clock, carcinogenesis, chronochemotherapy connections. J. Biol. Chem. 2021, 297, 101068. [Google Scholar] [CrossRef]
- Sancar, A.; Van Gelder, R.N. Clocks, cancer, and chronochemotherapy. Science 2021, 371, 6524. [Google Scholar] [CrossRef]
- Mocellin, S.; Tropea, S.; Benna, C.; Rossi, C.R. Circadian pathway genetic variation and cancer risk: Evidence from genome-wide association studies. BMC Med. 2018, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Lin, L. Circadian clock as a possible control point in colorectal cancer progression (Review). Int. J. Oncol. 2022, 61, 149. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Liu, F.; Han, Y.; Li, P.; Yuan, B.; Wang, X.; Chen, Y.; Kuang, Y.; Zhi, Q.; Zhao, H. Silencing NPAS2 promotes cell growth and invasion in DLD-1 cells and correlated with poor prognosis of colorectal cancer. Biochem. Biophys. Res. Commun. 2014, 450, 1058–1062. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Arnal, L.; Hakim, O.; Patel, V.R.; Baldi, P.; Hager, G.L.; Sassone-Corsi, P. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat. Struct. Mol. Biol. 2013, 20, 1206–1213. [Google Scholar] [CrossRef]
- Shu, X.-S.; Li, L.; Tao, Q. Chromatin regulators with tumor suppressor properties and their alterations in human cancers. Epigenomics 2012, 4, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Han, S.; Liu, X.; Wang, S.; Ma, H. Integrated single-cell and transcriptome sequencing analyses determines a chromatin regulator-based signature for evaluating prognosis in lung adenocarcinoma. Front. Oncol. 2022, 12, 1031728. [Google Scholar] [CrossRef]
- Tang, X.; Qi, C.; Zhou, H.; Liu, Y. A novel metabolic-immune related signature predicts prognosis and immunotherapy response in lung adenocarcinoma. Heliyon 2022, 8, e10164. [Google Scholar] [CrossRef] [PubMed]
- Chai, R.; Liao, M.; Ou, L.; Tang, Q.; Liang, Y.; Li, N.; Huang, W.; Wang, X.; Zheng, K.; Wang, S. Circadian Clock Genes Act as Diagnostic and Prognostic Biomarkers of Glioma: Clinic Implications for Chronotherapy. BioMed Res. Int. 2022, 2022, 9774879. [Google Scholar] [CrossRef]
- Zhang, Z.; Liang, Z.; Gao, W.; Yu, S.; Hou, Z.; Li, K.; Zeng, P. Identification of circadian clock genes as regulators of immune infiltration in Hepatocellular Carcinoma. J. Cancer 2022, 13, 3199–3208. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, Y.; Hu, S.; Fan, Z.; Wang, X.; Li, S. Bioinformatics Analysis of Differentially Expressed Rhythm Genes in Liver Hepatocellular Carcinoma. Front. Genet. 2021, 12, 680528. [Google Scholar] [CrossRef]
- Yuan, P.; Yang, T.; Mu, J.; Zhao, J.; Yang, Y.; Yan, Z.; Hou, Y.; Chen, C.; Xing, J.; Zhang, H.; et al. Circadian clock gene NPAS2 promotes reprogramming of glucose metabolism in hepatocellular carcinoma cells. Cancer Lett. 2020, 469, 498–509. [Google Scholar] [CrossRef]
- Yang, T.; Yuan, P.; Yang, Y.; Liang, N.; Wang, Q.; Li, J.; Lu, R.; Zhang, H.; Mu, J.; Yan, Z.; et al. NPAS2 Contributes to Liver Fibrosis by Direct Transcriptional Activation of Hes1 in Hepatic Stellate Cells. Mol. Ther. Nucleic Acids 2019, 18, 1009–1022. [Google Scholar] [CrossRef]
- Xu, T.; Jin, T.; Lu, X.; Pan, Z.; Tan, Z.; Zheng, C.; Liu, Y.; Hu, X.; Ba, L.; Ren, H.; et al. A signature of circadian rhythm genes in driving anaplastic thyroid carcinoma malignant progression. Cell. Signal. 2022, 95, 110332. [Google Scholar] [CrossRef]
- Lesicka, M.; Jabłońska, E.; Wieczorek, E.; Seroczyńska, B.; Siekierzycka, A.; Skokowski, J.; Kalinowski, L.; Wąsowicz, W.; Reszka, E. Altered circadian genes expression in breast cancer tissue according to the clinical characteristics. PLoS ONE 2018, 13, e0199622. [Google Scholar] [CrossRef]
- Yi, C.; Mu, L.; de la Longrais, I.A.R.; Sochirca, O.; Arisio, R.; Yu, H.; Hoffman, A.E.; Zhu, Y.; Katsaro, D. The circadian gene NPAS2 is a novel prognostic biomarker for breast cancer. Breast Cancer Res. Treat. 2010, 120, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Monsees, G.M.; Kraft, P.; Hankinson, S.E.; Hunter, D.J.; Schernhammer, E.S. Circadian genes and breast cancer susceptibility in rotating shift workers. Int. J. Cancer 2012, 131, 2547–2552. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Dai, Z.-M.; Zhao, Y.; Wang, X.-J.; Kang, H.-F.; Ma, X.-B.; Lin, S.; Wang, M.; Yang, P.-T.; Dai, Z.-J. Current evidence on the relationship between two common polymorphisms in NPAS2 gene and cancer risk. Int. J. Clin. Exp. Med. 2015, 8, 7176–7183. [Google Scholar] [PubMed]
- Zhu, Y.; Leaderer, D.; Guss, C.; Brown, H.N.; Zhang, Y.; Boyle, P.; Stevens, R.G.; Hoffman, A.; Qin, Q.; Han, X.; et al. Ala394Thr polymorphism in the clock geneNPAS2: A circadian modifier for the risk of non-Hodgkin’s lymphoma. Int. J. Cancer 2007, 120, 432–435. [Google Scholar] [CrossRef]
- Zheng, X.; Xiuyi, L.; Zhu, L.; Xu, K.; Shi, C.; Cui, L.; Ding, H. The Circadian Gene NPAS2 Act as a Putative Tumor Stimulative Factor for Uterine Corpus Endometrial Carcinoma. Cancer Manag. Res. 2021, 13, 9329–9343. [Google Scholar] [CrossRef]
- Hermyt, E.; Zmarzly, N.; Kruszniewska-Rajs, C.; Gola, J.; Jeda-Golonka, A.; Szczepanek, K.; Mazurek, U.; Witek, A. Expression pattern of circadian rhythm-related genes and its potential relationship with miRNAs activity in endometrial cancer. Ginekol. Polska 2023, 94, 33–40. [Google Scholar] [CrossRef]
- Feng, D.; Xiong, Q.; Zhang, F.; Shi, X.; Xu, H.; Wei, W.; Ai, J.; Yang, L. Identification of a Novel Nomogram to Predict Progression Based on the Circadian Clock and Insights Into the Tumor Immune Microenvironment in Prostate Cancer. Front. Immunol. 2022, 13, 777724. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chen, Y.; Quan, P.; Zhang, J.; Han, S.; Wang, G.; Qi, R.; Zhang, X.; Wang, F.; Yuan, J.; et al. NPAS2 promotes aerobic glycolysis and tumor growth in prostate cancer through HIF-1A signaling. BMC Cancer 2023, 23, 280. [Google Scholar] [CrossRef]
- Wendeu-Foyet, M.G.; Koudou, Y.; Cénée, S.; Trétarre, B.; Rébillard, X.; Cancel-Tassin, G.; Cussenot, O.; Boland, A.; Bacq, D.; Deleuze, J.; et al. Circadian genes and risk of prostate cancer: Findings from the EPICAP study. Int. J. Cancer 2019, 145, 1745–1753. [Google Scholar] [CrossRef]
- Zhu, Y.; Stevens, R.G.; Hoffman, A.E.; FitzGerald, L.M.; Kwon, E.M.; Ostrander, E.A.; Davis, S.; Zheng, T.; Stanford, J.L. Testing the Circadian Gene Hypothesis in Prostate Cancer: A Population-Based Case-Control Study. Cancer Res 2009, 69, 9315–9322. [Google Scholar] [CrossRef]
- Yu, C.-C.; Chen, L.-C.; Chiou, C.-Y.; Chang, Y.-J.; Lin, V.C.; Huang, C.-Y.; Lin, I.-L.; Chang, T.-Y.; Lu, T.-L.; Lee, C.-H.; et al. Genetic variants in the circadian rhythm pathway as indicators of prostate cancer progression. Cancer Cell Int. 2019, 19, 87. [Google Scholar] [CrossRef] [PubMed]
- Benna, C.; Rajendran, S.; Spiro, G.; Tropea, S.; Del Fiore, P.; Rossi, C.R.; Mocellin, S. Associations of clock genes polymorphisms with soft tissue sarcoma susceptibility and prognosis. J. Transl. Med. 2018, 16, 338. [Google Scholar] [CrossRef] [PubMed]
- Iyyanki, T.; Zhang, B.; Wang, Q.; Hou, Y.; Jin, Q.; Xu, J.; Yang, H.; Liu, T.; Wang, X.; Song, F.; et al. Subtype-associated epigenomic landscape and 3D genome structure in bladder cancer. Genome Biol. 2021, 22, 1–20. [Google Scholar] [CrossRef]
- Song, B.; Chen, Y.; Liu, Y.; Wan, C.; Zhang, L.; Zhang, W. NPAS2 regulates proliferation of acute myeloid leukemia cells via CDC25A -mediated cell cycle progression and apoptosis. J. Cell. Biochem. 2018, 120, 8731–8741. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Li, J.; Zhou, F.; Huang, Q.; Zhang, J.; Guo, X.; Lyu, Z.; Zhang, H.; Xing, J. NPAS2 promotes cell survival of hepatocellular carcinoma by transactivating CDC25A. Cell Death Dis. 2017, 8, e2704. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; D’alessandro, M.; Lee, C. miRNAs Are Required for Generating a Time Delay Critical for the Circadian Oscillator. Curr. Biol. 2013, 23, 1959–1968. [Google Scholar] [CrossRef]
- Zhao, F.; Pu, Y.; Qian, L.; Zang, C.; Tao, Z.; Gao, J. MiR-20a-5p promotes radio-resistance by targeting NPAS2 in nasopharyngeal cancer cells. Oncotarget 2017, 8, 105873–105881. [Google Scholar] [CrossRef] [PubMed]
- Franzoni, A.; Markova-Car, E.; Dević-Pavlić, S.; Jurišić, D.; Puppin, C.; Mio, C.; De Luca, M.; Petruz, G.; Damante, G.; Pavelić, S.K. A polymorphic GGC repeat in the NPAS2 gene and its association with melanoma. Exp. Biol. Med. 2017, 242, 1553–1558. [Google Scholar] [CrossRef]
- Cao, X.-M.; Kang, W.-D.; Xia, T.-H.; Yuan, S.-B.; Guo, C.-A.; Wang, W.-J.; Liu, H.-B. High expression of the circadian clock gene NPAS2 is associated with progression and poor prognosis of gastric cancer: A single-center study. World J. Gastroenterol. 2023, 29, 3645–3657. [Google Scholar] [CrossRef]
- Anea, C.B.; Merloiu, A.M.; Fulton, D.J.R.; Patel, V.; Rudic, R.D. Immunohistochemistry of the circadian clock in mouse and human vascular tissues. Vessel. Plus 2018, 2, 16. [Google Scholar] [CrossRef]
- Curtis, A.M.; Cheng, Y.; Kapoor, S.; Reilly, D.; Price, T.S.; FitzGerald, G.A. Circadian variation of blood pressure and the vascular response to asynchronous stress. Proc. Natl. Acad. Sci. USA 2007, 104, 3450–3455. [Google Scholar] [CrossRef]
- Leu, H.-B.; Chung, C.-M.; Lin, S.-J.; Chiang, K.-M.; Yang, H.-C.; Ho, H.-Y.; Ting, C.-T.; Lin, T.-H.; Sheu, S.-H.; Tsai, W.-C.; et al. Association of circadian genes with diurnal blood pressure changes and non-dipper essential hypertension: A genetic association with young-onset hypertension. Hypertens. Res. 2014, 38, 155–162. [Google Scholar] [CrossRef]
- Bray, M.S.; Shaw, C.A.; Moore, M.W.S.; Garcia, R.A.P.; Zanquetta, M.M.; Durgan, D.J.; Jeong, W.J.; Tsai, J.-Y.; Bugger, H.; Zhang, D.; et al. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am. J. Physiol. Circ. Physiol. 2008, 294, H1036–H1047. [Google Scholar] [CrossRef]
- Westgate, E.J.; Cheng, Y.; Reilly, D.F.; Price, T.S.; Walisser, J.A.; Bradfield, C.A.; FitzGerald, G.A. Genetic Components of the Circadian Clock Regulate Thrombogenesis In Vivo. Circulation 2008, 117, 2087–2095. [Google Scholar] [CrossRef]
- Takeda, N.; Maemura, K. The role of clock genes and circadian rhythm in the development of cardiovascular diseases. Cell. Mol. Life Sci. 2015, 72, 3225–3234. [Google Scholar] [CrossRef]
- Sun, Q.; Zhao, J.; Liu, L.; Wang, X.; Gu, X. Identification of the potential biomarkers associated with circadian rhythms in heart failure. PeerJ 2023, 11, e14734. [Google Scholar] [CrossRef] [PubMed]
- Chavva, H.; Brazeau, D.A.; Denvir, J.; Primerano, D.A.; Fan, J.; Seeley, S.L.; Rorabaugh, B.R. Methamphetamine-induced changes in myocardial gene transcription are sex-dependent. BMC Genom. 2021, 22, 259. [Google Scholar] [CrossRef] [PubMed]
- Cantone, L.; Tobaldini, E.; Favero, C.; Albetti, B.; Sacco, R.M.; Torgano, G.; Ferrari, L.; Montano, N.; Bollati, V. Particulate Air Pollution, Clock Gene Methylation, and Stroke: Effects on Stroke Severity and Disability. Int. J. Mol. Sci. 2020, 21, 3090. [Google Scholar] [CrossRef] [PubMed]
- Schallner, N.; Lieberum, J.-L.; Gallo, D.; LeBlanc, R.H.; Fuller, P.M.; Hanafy, K.A.; Otterbein, L.E.; Maury, E.; Ramsey, K.M.; Bass, J.; et al. Carbon Monoxide Preserves Circadian Rhythm to Reduce the Severity of Subarachnoid Hemorrhage in Mice. Stroke 2017, 48, 2565–2573. [Google Scholar] [CrossRef] [PubMed]
- Clements, A.; Shibuya, Y.; Hokugo, A.; Brooks, Z.; Roca, Y.; Kondo, T.; Nishimura, I.; Jarrahy, R. In vitro assessment of Neuronal PAS domain 2 mitigating compounds for scarless wound healing. Front. Med. 2023, 9, 1014763. [Google Scholar] [CrossRef]
- Shibuya, Y.; Hokugo, A.; Okawa, H.; Kondo, T.; Khalil, D.; Wang, L.; Roca, Y.; Clements, A.; Sasaki, H.; Berry, E.; et al. Therapeutic downregulation of neuronal PAS domain 2 (Npas2) promotes surgical skin wound healing. eLife 2022, 11, e71074. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murgo, E.; Colangelo, T.; Bellet, M.M.; Malatesta, F.; Mazzoccoli, G. Role of the Circadian Gas-Responsive Hemeprotein NPAS2 in Physiology and Pathology. Biology 2023, 12, 1354. https://doi.org/10.3390/biology12101354
Murgo E, Colangelo T, Bellet MM, Malatesta F, Mazzoccoli G. Role of the Circadian Gas-Responsive Hemeprotein NPAS2 in Physiology and Pathology. Biology. 2023; 12(10):1354. https://doi.org/10.3390/biology12101354
Chicago/Turabian StyleMurgo, Emanuele, Tommaso Colangelo, Maria Marina Bellet, Francesco Malatesta, and Gianluigi Mazzoccoli. 2023. "Role of the Circadian Gas-Responsive Hemeprotein NPAS2 in Physiology and Pathology" Biology 12, no. 10: 1354. https://doi.org/10.3390/biology12101354
APA StyleMurgo, E., Colangelo, T., Bellet, M. M., Malatesta, F., & Mazzoccoli, G. (2023). Role of the Circadian Gas-Responsive Hemeprotein NPAS2 in Physiology and Pathology. Biology, 12(10), 1354. https://doi.org/10.3390/biology12101354






