Circadian Misalignment and Metabolic Disorders: A Story of Twisted Clocks
Abstract
Simple Summary
Abstract
1. Introduction
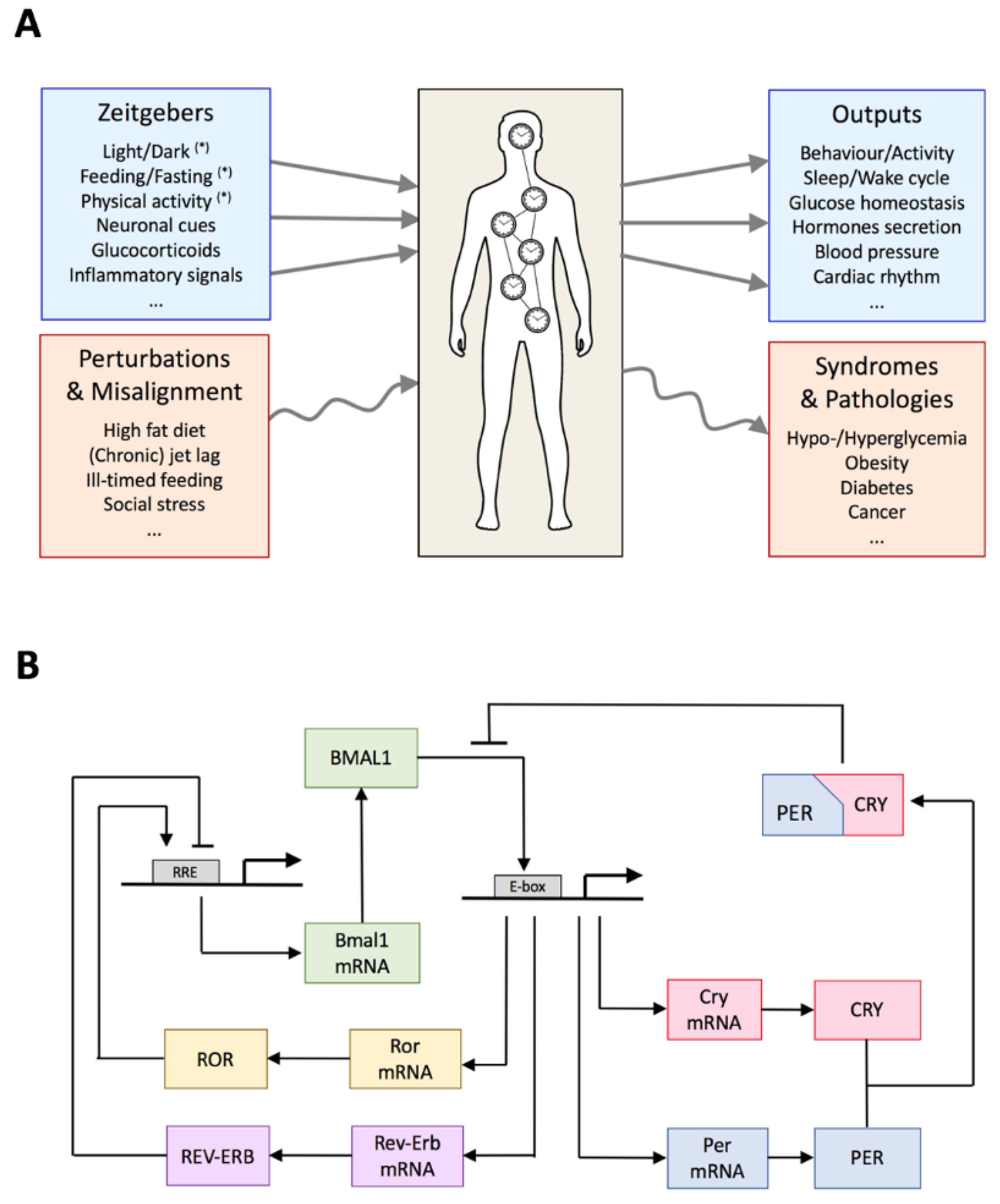
2. Global Organization of the Circadian System
3. Architecture of the Circadian Gene Regulatory Network
4. Circadian Timing of Clock Genes and Clock-Controlled Genes in Physiological Conditions
5. Ill-Timed Feeding Pattern and Twisted Clocks
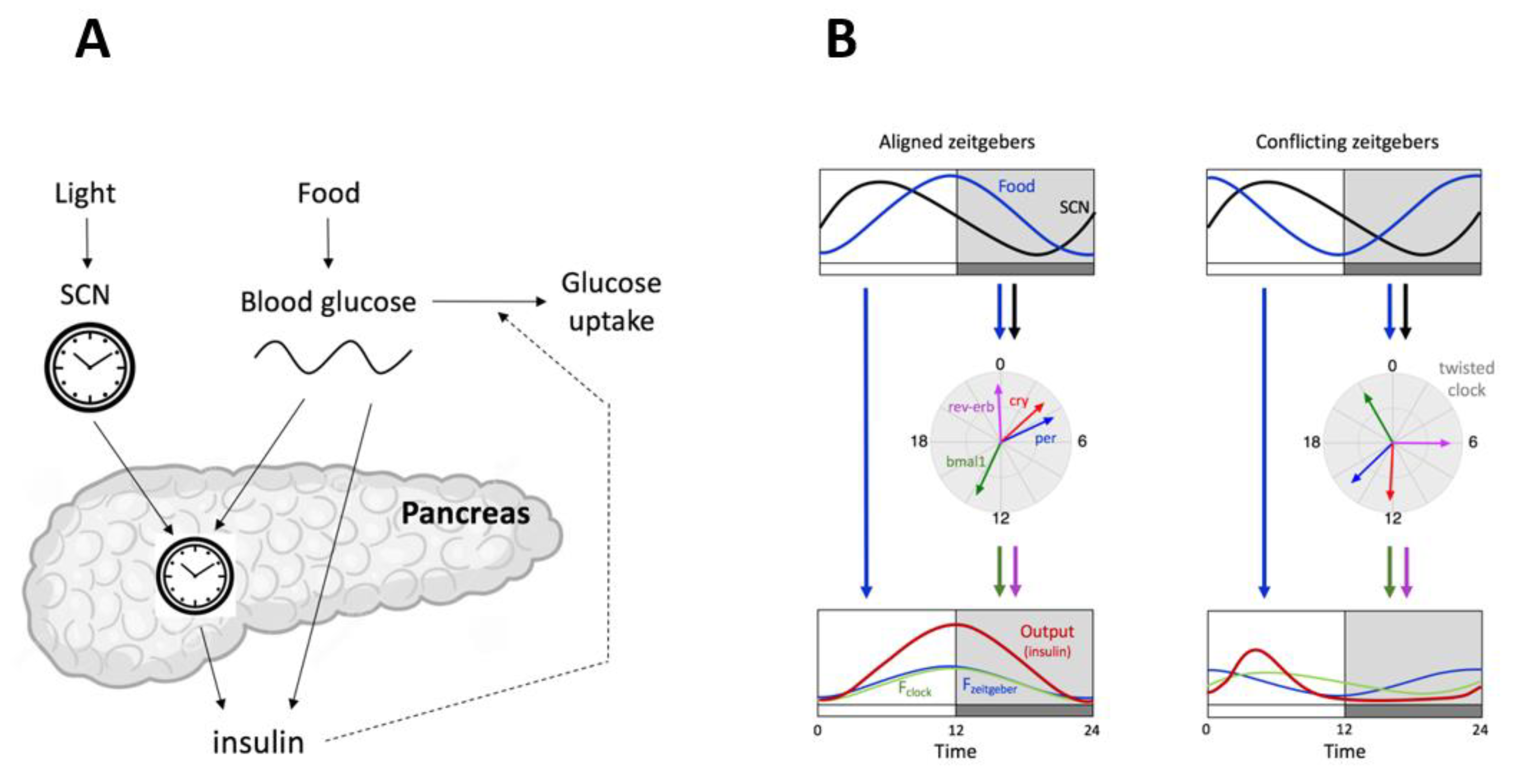
5.1. Internal Twist Explained by Mathematical Modeling
5.2. Another Conflicting Zeitgeber Paradigm
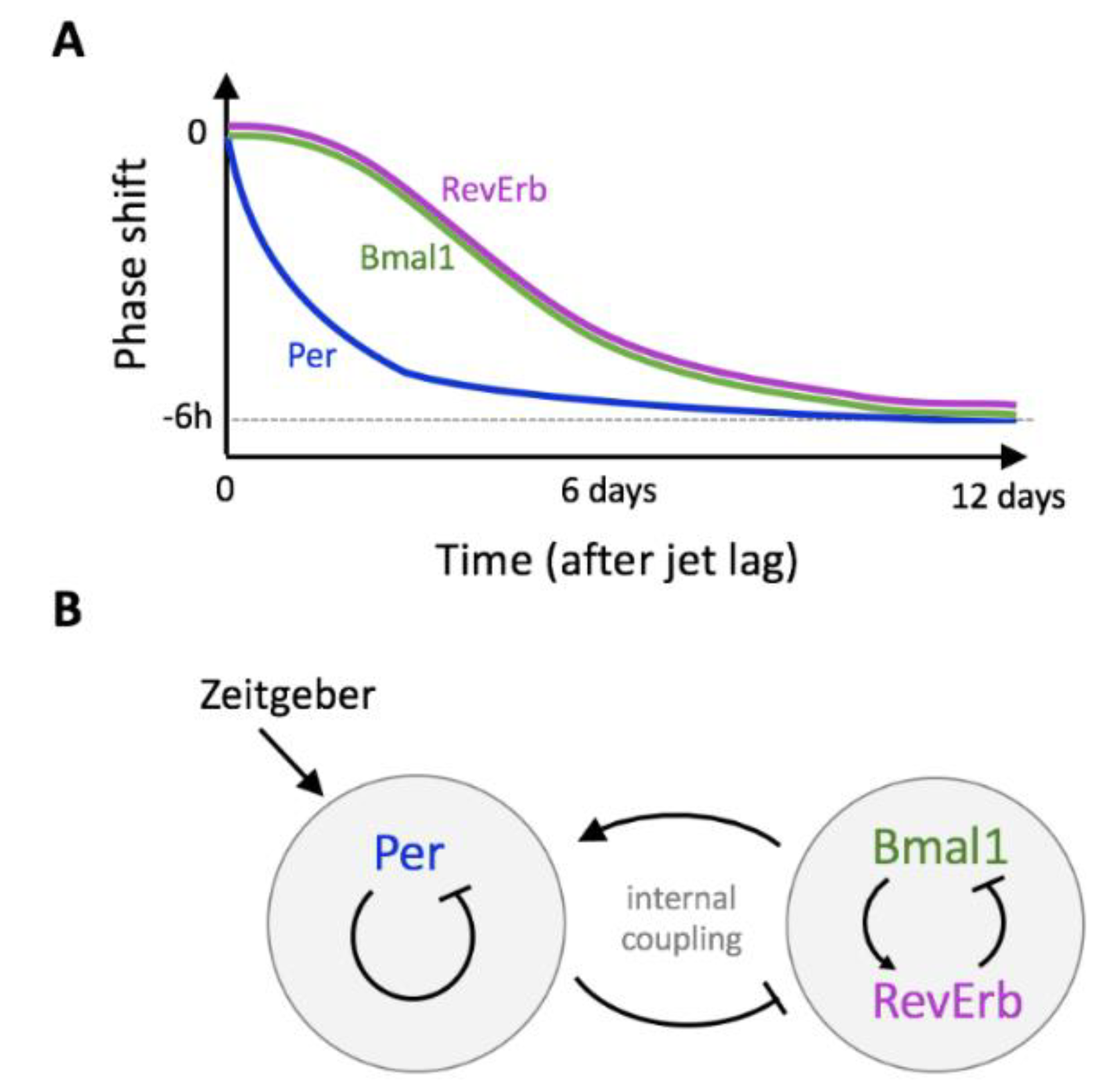
6. Jet Lag and Gene-Specific Resynchronization Time
Temporary Twist and Internal Decoupling Explained by Mathematical Modeling
7. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Chaix, A.; Zarrinpar, A.; Panda, S. The circadian coordination of cell biology. J. Cell Biol. 2016, 215, 15–25. [Google Scholar] [CrossRef]
- Finger, A.; Dibner, C.; Kramer, A. Coupled network of the circadian clocks: A driving force of rhythmic physiology. FEBS Lett. 2020, 594, 2734–2769. [Google Scholar] [CrossRef]
- Maury, E. Off the clock: From circadian disruption to metabolic disease. Int. J. Mol. Sci. 2019, 20, 1597. [Google Scholar] [CrossRef]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Hastings, M.H.; Maywood, E.S.; Brancaccio, M. The mammalian circadian timing system and the suprachias-matic nucleus as its pacemaker. Biology 2019, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Herzog, E.D. Neurons and networks in daily rhythms. Nat. Rev. Neurosci. 2007, 8, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Herzog, E.D.; Hermanstyne, T.; Smyllie, N.J.; Hastings, M.H. Regulating the suprachiasmatic nucleus (SCN) cir-cadian clockwork: Interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harb. Perspect. Biol. 2017, 9, a027706. [Google Scholar] [CrossRef]
- Saper, C.B.; Scammell, T.E.; Lu, J. Hypothalamic regulation of sleep and circadian rhythms. Nat. Cell Biol. 2005, 437, 1257–1263. [Google Scholar] [CrossRef]
- Challet, E. The circadian regulation of food intake. Nat. Rev. Endocrinol. 2019, 15, 393–405. [Google Scholar] [CrossRef]
- Panda, S. Circadian physiology of metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef]
- Buijs, F.N.; León-Mercado, L.; Guzmán-Ruiz, M.; Guerrero-Vargas, N.N.; Romo-Nava, F.; Buijs, R.M. The circadian system: A regulatory feedback network of periphery and brain. Physiology 2016, 31, 170–181. [Google Scholar] [CrossRef]
- Stenvers, D.J.; Scheer, F.A.J.L.; Schrauwen, P.; La Fleur, S.E.; Kalsbeek, A. Circadian clocks and insulin resistance. Nat. Rev. Endocrinol. 2019, 15, 75–89. [Google Scholar] [CrossRef]
- Mukherji, A.; Bailey, S.M.; Staels, B.; Baumert, T.F. The circadian clock and liver function in health and disease. J. Hepatol. 2019, 71, 200–211. [Google Scholar] [CrossRef]
- Owino, S.; Buonfiglio, D.D.C.; Tchio, C.; Tosini, G. Melatonin signaling a key regulator of glucose homeostasis and energy metabolism. Front. Endocrinol. 2019, 10, 488. [Google Scholar] [CrossRef] [PubMed]
- Saini, C.; Suter, D.M.; Liani, A.; Gos, P.; Schibler, U. The mammalian circadian timing system: Synchronization of peripheral clocks. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Ralph, M.R.; Foster, R.G.; Davis, F.C.; Menaker, M. Transplanted suprachiasmatic nucleus determines circadian period. Science 1990, 247, 975–978. [Google Scholar] [CrossRef] [PubMed]
- Silver, R.; LeSauter, J.; Tresco, P.A.; Lehman, M.N. A diffusible coupling signal from the transplanted suprachi-asmatic nucleus controlling circadian locomotor rhythms. Nature 1996, 382, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Brown, S.A.; Marcacci, L.; Tronche, F.; Kellendonk, C.; Reichardt, H.M.; Schütz, G.; Schibler, U. Reset-ting of circadian time in peripheral tissues by glucocorticoid signaling. Science 2000, 289, 2344–2347. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Bernstein, E.L.; Jetton, A.E.; Matsumoto, S.I.; Markuns, J.F.; Lehman, M.N.; Bittman, E.L. Effects of suprachi-asmatic transplants on circadian rhythms of neuroendocrine function in golden hamsters. Endocrinology 1999, 140, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Damiola, F.; Le Minh, N.; Preitner, N.; Kornmann, B.; Fleury-Olela, F.; Schibler, U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000, 14, 2950–2961. [Google Scholar] [CrossRef]
- Stokkan, K.-A.; Yamazaki, S.; Tei, H.; Sakaki, Y.; Menaker, M. Entrainment of the circadian clock in the liver by feeding. Science 2001, 291, 490–493. [Google Scholar] [CrossRef]
- Li, A.J.; Wiater, M.F.; Oostrom, M.T.; Smith, B.R.; Wang, Q.; Dinh, T.T.; Roberts, B.L.; Jansen, H.T.; Ritter, S. Lep-tin-sensitive neurons in the arcuate nuclei contribute to endogenous feeding rhythms. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R1313–R1326. [Google Scholar] [CrossRef]
- Asher, G.; Gatfield, D.; Stratmann, M.; Reinke, H.; Dibner, C.; Kreppel, F.; Mostoslavsky, R.; Alt, F.W.; Schibler, U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008, 134, 317–328. [Google Scholar] [CrossRef]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D.; Guarente, L.P.; Sassone-Corsi, P. The NAD+-Dependent deacetylase SIRT1 modulates clock-mediated chromatin remodeling and circadian control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef]
- Lamia, K.A.; Sachdeva, U.M.; Di Tacchio, L.; Williams, E.C.; Alvarez, J.G.; Egan, D.F.; Vasquez, D.S.; Juguilon, H.; Panda, S.; Shaw, R.J.; et al. AMPK regulates the circadian clock by cryptochrome phosphoryla-tion and degradation. Science 2009, 326, 437–440. [Google Scholar] [CrossRef]
- Crosby, P.; Hamnett, R.; Putker, M.; Hoyle, N.P.; Reed, M.; Karam, C.J.; Maywood, E.S.; Stangherlin, A.; Chesham, J.E.; Hayter, E.A.; et al. Insulin/IGF-1 drives PE-RIOD synthesis to entrain circadian rhythms with feeding time. Cell 2019, 177, 896–909. [Google Scholar] [CrossRef]
- Schiaffino, S.; Blaauw, B.; Dyar, K.A. The functional significance of the skeletal muscle clock: Lessons from Bmal1 knockout models. Skelet. Muscle 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Um, J.-H.; Pendergast, J.S.; Springer, D.A.; Foretz, M.; Viollet, B.; Brown, A.; Kim, M.K.; Yamazaki, S.; Chung, J.H. AMPK Regulates circadian rhythms in a tissue- and isoform-specific manner. PLoS ONE 2011, 6, e18450. [Google Scholar] [CrossRef]
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014, 24, 90–99. [Google Scholar] [CrossRef]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef]
- Hirano, A.; Fu, Y.-H.; Ptáček, L.J. The intricate dance of post-translational modifications in the rhythm of life. Nat. Struct. Mol. Biol. 2016, 23, 1053–1060. [Google Scholar] [CrossRef]
- St John, P.C.; Hirota, T.; Kay, S.A.; Doyle, F.J., III. Spatiotemporal separation of PER and CRY posttranslational regulation in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2014, 111, 2040–2045. [Google Scholar] [CrossRef] [PubMed]
- Toh, K.L.; Jones, C.R.; He, Y.; Eide, E.J.; Hinz, W.A.; Virshup, D.M.; Ptácek, L.J.; Fu, Y.H. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 2001, 291, 1040–1043. [Google Scholar] [CrossRef]
- Liu, Z.; Huang, M.; Wu, X.; Shi, G.; Xing, L.; Dong, Z.; Qu, Z.; Yan, J.; Yang, L.; Panda, S.; et al. PER1 phosphorylation specifies feeding rhythm in mice. Cell Rep. 2014, 7, 1509–1520. [Google Scholar] [CrossRef]
- Sato, T.K.; Panda, S.; Miraglia, L.J.; Reyes, T.M.; Rudic, R.D.; McNamara, P.; Naik, K.A.; FitzGerald, G.A.; Kay, S.A.; Hogenesch, J.B.; et al. A functional genomics strategy reveals rora as a component of the mammalian circadian clock. Neuron 2004, 43, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circa-dian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Bunger, M.K.; Wilsbacher, L.D.; Moran, S.M.; Clendenin, C.; Radcliffe, L.A.; Hogen Esch, J.B.; Simon, M.C.; Takahashi, J.S.; Bradfield, C.A. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 2020, 103, 1009–1017. [Google Scholar] [CrossRef]
- Liu, A.C.; Tran, H.G.; Zhang, E.E.; Priest, A.A.; Welsh, D.K.; Kay, S.A. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008, 4, e1000023. [Google Scholar] [CrossRef] [PubMed]
- Relógio, A.; Westermark, P.O.; Wallach, T.; Schellenberg, K.; Kramer, A.; Herzel, H. Tuning the mammalian circadian clock: Robust synergy of two loops. PLoS Comput. Biol. 2011, 7, e1002309. [Google Scholar] [CrossRef]
- Uriu, K.; Tei, H. Feedback loops interlocked at competitive binding sites amplify and facilitate genetic oscilla-tions. J. Theor. Biol. 2017, 428, 56–64. [Google Scholar] [CrossRef]
- Ukai-Tadenuma, M.; Yamada, R.G.; Xu, H.; Ripperger, J.A.; Liu, A.C.; Ueda, H.R. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 2011, 144, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.R.; Hayashi, S.; Chen, W.; Sano, M.; Machida, M.; Shigeyoshi, Y.; Iino, M.; Hashimoto, S. System-level identi-fication of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005, 37, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Male, V.; Nisoli, I.; Gascoyne, D.M.; Brady, H.J. E4BP4: An unexpected player in the immune response. Trends Immunol. 2012, 33, 98–102. [Google Scholar] [CrossRef]
- Zhao, Z.; Yin, L.; Wu, F.; Tong, X. Hepatic metabolic regulation by nuclear factor E4bpj. Mol. Endocrinol. 2021, 66, R15–R21. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.H.; Yamazaki, S.; Lowrey, P.L.; Shimomura, K.; Ko, C.H.; Buhr, E.D.; Siepka, S.M.; Hong, H.K.; Oh, W.J.; Yoo, O.J.; et al. PERIOD2: Luciferase real-time reporting of circadian dynamics reveals per-sistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 5339–5346. [Google Scholar] [CrossRef] [PubMed]
- Aton, S.J.; Colwell, C.S.; Harmar, A.J.; Waschek, J.; Herzog, E.D. Vasoactive intestinal polypeptide mediates circa-dian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 2005, 8, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Bernard, S.; Gonze, D.; Cajavec, B.; Herzel, H.; Kramer, A. Synchronization-induced rhythmicity of circadian os-cillators in the suprachiasmatic nucleus. PLoS Comput. Biol. 2007, 3, e68. [Google Scholar] [CrossRef] [PubMed]
- Webb, A.B.; Taylor, S.R.; Thoroughman, K.A.; Doyle, F.J., III.; Herzog, E.D. Weakly circadian cells improve resyn-chrony. PLoS Comput. Biol. 2012, 8, e1002787. [Google Scholar] [CrossRef]
- Webb, A.B.; Angelo, N.; Huettner, J.E.; Herzog, E.D. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc. Natl. Acad. Sci. USA 2009, 106, 16493–16498. [Google Scholar] [CrossRef]
- Shigeyoshi, Y.; Taguchi, K.; Yamamoto, S.; Takekida, S.; Yan, L.; Tei, H.; Moriya, T.; Shibata, S.; Loros, J.J.; Dunlap, J.C.; et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 1997, 91, 1043–1053. [Google Scholar] [CrossRef]
- Albrecht, U.; Sun, Z.S.; Eichele, G.; Lee, C.C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 1997, 91, 1055–1064. [Google Scholar] [CrossRef]
- Griffin, E.A., Jr.; Staknis, D.; Weitz, C.J. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 1999, 286, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Gau, D.; Lemberger, T.; Von Gall, C.; Kretz, O.; Le Minh, N.; Gass, P.; Schmid, W.; Schibler, U.; Korf, H.W.; Schütz, G.; et al. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron 2002, 34, 245–253. [Google Scholar] [CrossRef]
- Tischkau, S.A.; Mitchell, J.W.; Tyan, S.H.; Buchanan, G.F.; Gillette, M.U. Ca2+/cAMP response element-binding pro-tein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nu-cleus circadian clock. J. Biol. Chem. 2003, 278, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Travnickova-Bendova, Z.; Cermakian, N.; Reppert, S.M.; Sassone-Corsi, P. Bimodal regulation of mPeriod pro-moters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl. Acad. Sci. USA 2002, 99, 7728–7733. [Google Scholar] [CrossRef] [PubMed]
- Torra, I.P.; Tsibulsky, V.; Delaunay, F.; Saladin, R.; Laudet, V.; Fruchart, J.C.; Kosykh, V.; Staels, B. Circadian and glu-cocorticoid regulation of Rev-erba expression in liver. Endocrinology 2000, 141, 3799–3806. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lahens, N.F.; Balance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [PubMed]
- Etchegaray, J.-P.; Lee, C.; Wade, P.A.; Reppert, S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nat. Cell Biol. 2002, 421, 177–182. [Google Scholar] [CrossRef]
- Koike, N.; Yoo, S.-H.; Huang, H.-C.; Kumar, V.; Lee, C.; Kim, T.-K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef]
- Rey, G.; Cesbron, F.; Rougemont, J.; Reinke, H.; Brunner, M.; Naef, F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011, 9, e1000595. [Google Scholar] [CrossRef] [PubMed]
- Fang, B.; Everett, L.J.; Jager, J.; Briggs, E.; Armour, S.M.; Feng, D.; Roy, A.; Gerhart-Hines, Z.; Sun, Z.; Lazar, M.A. Circa-dian enhancers coordinate multiple phases of rhythmic gene transcription In Vivo. Cell 2014, 159, 1140–1152. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, F.; Lin, Y.; Wu, B. Targeting REV-ERBα for therapeutic purposes: Promises and challenges. Theranostics 2020, 10, 4168–4182. [Google Scholar] [CrossRef] [PubMed]
- Leloup, J.-C.; Goldbeter, A. Toward a detailed computational model for the mammalian circadian clock. Proc. Natl. Acad. Sci USA 2003, 100, 7051–7056. [Google Scholar] [CrossRef]
- Forger, D.B.; Peskin, C.S. A detailed predictive model of the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2003, 100, 14806–14811. [Google Scholar] [CrossRef]
- Mirsky, H.P.; Liu, A.C.; Welsh, D.K.; Kay, S.A.; Doyle, F.J. A model of the cell-autonomous mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2009, 106, 11107–11112. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nakahata, Y.; Soma, H.; Akashi, M.; Mamine, T.; Takumi, T. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol. Biol. 2004, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cai, Y.; Sothern, R.B.; Guan, Y.; Chan, P. Chronobiological Analysis of circadian patterns in transcription of seven key clock genes in six peripheral tissues in mice. Chronobiol. Int. 2007, 24, 793–820. [Google Scholar] [CrossRef]
- Storch, K.F.; Lipan, O.; Leykin, I.; Viswanathan, N.; Davis, F.C.; Wong, W.H.; Weitz, C.J. Extensive and divergent cir-cadian gene expression in liver and heart. Nature 2002, 417, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Korenčič, A.; Bordyugov, G.; Košir, R.; Rozman, D.; Goličnik, M.; Herzel, H. The interplay of cis-regulatory ele-ments rules circadian rhythms in mouse liver. PLoS ONE 2012, 7, e46835. [Google Scholar] [CrossRef]
- Korenčič, A.; Košir, R.; Bordyugov, G.; Lehmann, R.; Rozman, D.; Herzel, H. Timing of circadian genes in mamma-lian tissues. Sci. Rep. 2014, 4, 5782. [Google Scholar] [CrossRef]
- Perelis, M.; Marcheva, B.; Ramsey, K.M.; Schipma, M.J.; Hutchison, A.L.; Taguchi, A.; Peek, C.B.; Hong, H.; Huang, W.; Omura, C.; et al. Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 2015, 350, 4250. [Google Scholar] [CrossRef] [PubMed]
- Kalsbeek, A.; La Fleur, S.; Fliers, E. Circadian control of glucose metabolism. Mol. Metab. 2014, 3, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Kettner, N.M.; Mayo, S.A.; Hua, J.; Lee, C.; Moore, D.D.; Fu, L. Circadian dysfunction induces leptin resistance in mice. Cell Metab. 2015, 22, 448–459. [Google Scholar] [CrossRef]
- Dibner, C.; Schibler, U. Circadian timing of metabolism in animal models and humans. J. Internal. Med. 2015, 277, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Oosterman, J.E.; Kalsbeek, A.; La Fleur, S.E.; Belsham, D.D. Impact of nutrients on circadian rhythmicity. Am. J. Physiol. Integr. Comp. Physiol. 2015, 308, R337–R350. [Google Scholar] [CrossRef]
- Qian, J.; Scheer, F.A. Circadian system and glucose metabolism: Implications for physiology and disease. Trends Endocrinol. Metab. 2016, 27, 282–293. [Google Scholar] [CrossRef]
- Challet, E. Keeping circadian time with hormones. Diabetes Obes. Metab. 2015, 17, 76–83. [Google Scholar] [CrossRef]
- Allada, R.; Bass, J. Circadian mechanisms in medicine. N. Engl. J. Med. 2021, 384, 550–561. [Google Scholar] [CrossRef]
- Kohsaka, A.; Laposky, A.D.; Ramsey, K.M.; Estrada, C.; Joshu, C.; Kobayashi, Y.; Turek, F.W.; Bass, J. High-fat diet dis-rupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007, 6, 414–421. [Google Scholar] [CrossRef]
- Bray, M.S.; Ratcliffe, W.F.; Grenett, M.H.; Brewer, R.A.; Gamble, K.L.; Young, M.E. Quantitative analysis of light-phase restricted feeding reveals metabolic dyssynchrony in mice. Int. J. Obes. 2013, 37, 843–852. [Google Scholar] [CrossRef]
- Mukherji, A.; Kobiita, A.; Chambon, P. Shifting the feeding of mice to the rest phase creates metabolic altera-tions, which, on their own, shift the peripheral circadian clocks by 12 hour. Proc. Natl. Acad. Sci. USA 2015, 112, E6683–E6690. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, A.; Kobiita, A.; Damara, M.; Misra, N.; Meziane, H.; Champy, M.-F.; Chambon, P. Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc. Natl. Acad. Sci. USA 2015, 112, E6691–E6698. [Google Scholar] [CrossRef]
- Opperhuizen, A.-L.; Wang, D.; Foppen, E.; Jansen, R.; Boudzovitch-Surovtseva, O.; De Vries, J.; Fliers, E.; Kalsbeek, A. Feeding during the resting phase causes profound changes in physiology and desynchronization between liver and muscle rhythms of rats. Eur. J. Neurosci. 2016, 44, 2795–2806. [Google Scholar] [CrossRef] [PubMed]
- De Goede, P.; Sen, S.; Oosterman, J.E.; Foppen, E.; Jansen, R.; la Fleur, S.E.; Challet, E.; Kalsbeek, A. Differential effects of diet composition and timing of feeding behavior on rat brown adipose tissue and skeletal muscle periph-eral clocks. Neurobiol. Sleep Circadian Rhythms 2018, 4, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Bur, I.M.; Zouaoui, S.; Fontanaud, P.; Coutry, N.; Molino, F.; Martin, A.O.; Mollard, P.; Bonnefont, X. The comparison between circadian oscillators in mouse liver and pituitary gland reveals different integration of feeding and light schedules. PLoS ONE 2010, 5, e15316. [Google Scholar] [CrossRef] [PubMed]
- Woller, A.; Gonze, D. Modeling clock-related metabolic syndrome due to conflicting light and food cues. Sci. Rep. 2018, 8, 13641. [Google Scholar] [CrossRef] [PubMed]
- Nakao, R.; Yamamoto, S.; Horikawa, K.; Yasumoto, Y.; Nikawa, T.; Mukai, C.; Oishi, K. Atypical expression of cir-cadian clock genes in denervated mouse skeletal muscle. Chronobiol. Int. 2015, 32, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Heyde, I.; Oster, H. Differentiating external zeitgeber impact on peripheral circadian clock resetting. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Yamazaki, S.; Numano, R.; Abe, M.; Hida, A.; Takahashi, R.; Ueda, M.; Block, G.D.; Sakaki, Y.; Menaker, M.; Tei, H. Re-setting central and peripheral circadian oscillators in transgenic rats. Science 2000, 288, 682–685. [Google Scholar] [CrossRef]
- Davidson, A.J.; Castanon-Cervantes, O.; Leise, T.L.; Molyneux, P.C.; Harrington, M.E. Visualizing jet lag in the mouse suprachiasmatic nucleus and peripheral circadian timing system. Eur. J. Neurosci. 2009, 29, 171–180. [Google Scholar] [CrossRef]
- Reddy, A.B.; Field, M.D.; Maywood, E.S.; Hastings, M.H. Differential resynchronisation of circadian clock gene expression within the suprachiasmatic nuclei of mice subjected to experimental jet lag. J. Neurosci. 2002, 22, 7326–7330. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, S.; Eichele, G.; Oster, H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J. Clin. Investig. 2010, 120, 2600–2609. [Google Scholar] [CrossRef]
- Oster, H.; Damerow, S.; Kiessling, S.; Jakubcakova, V.; Abraham, D.; Tian, J.; Hoffmann, M.W.; Eichele, G. The circa-dian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006, 4, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Ono, D.; Honma, S.; Nakajima, Y.; Kuroda, S.; Enoki, R.; Honma, K.-I. Dissociation of Per1 and Bmal1 circadian rhythms in the suprachiasmatic nucleus in parallel with behavioral outputs. Proc. Natl. Acad. Sci. USA 2017, 114, E3699–E3708. [Google Scholar] [CrossRef] [PubMed]
- Schmal, C.; Ono, D.; Myung, J.; Pett, J.P.; Honma, S.; Honma, K.I.; Herzel, H.; Tokuda, I.T. Weak coupling between in-tracellular feedback loops explains dissociation of clock gene dynamics. PLoS Comput. Biol. 2019, 15, e1007330. [Google Scholar] [CrossRef] [PubMed]
- Forger, D.; Gonze, D.; Virshup, D.M.; Welsh, D.K. Beyond intuitive modeling: Combining biophysical models with innovative experiments to move the circadian clock field forward. J. Biol. Rhythm. 2007, 22, 200–210. [Google Scholar] [CrossRef]
- Almeida, S.; Chaves, M.; Delaunay, F. Transcription-based circadian mechanism controls the duration of mo-lecular clock states in response to signaling inputs. J. Theor. Biol. 2020, 484, 110015. [Google Scholar] [CrossRef] [PubMed]
- Woller, A.; Duez, H.; Staels, B.; Lefranc, M. A mathematical model of the liver circadian clock linking feeding and fasting cycles to clock function. Cell Rep. 2016, 17, 1087–1097. [Google Scholar] [CrossRef]
- Bae, S.-A.; Androulakis, I.P. The synergistic role of light-feeding phase relations on entraining robust circadian rhythms in the periphery. Gene Regul. Syst. Biol. 2017, 11. [Google Scholar] [CrossRef]
- Bae, S.-A.; Androulakis, I.P. Mathematical analysis of circadian disruption and metabolic re-entrainment of hepatic gluconeogenesis: The intertwining entraining roles of light and feeding. Am. J. Physiol. Metab. 2018, 314, E531–E542. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woller, A.; Gonze, D. Circadian Misalignment and Metabolic Disorders: A Story of Twisted Clocks. Biology 2021, 10, 207. https://doi.org/10.3390/biology10030207
Woller A, Gonze D. Circadian Misalignment and Metabolic Disorders: A Story of Twisted Clocks. Biology. 2021; 10(3):207. https://doi.org/10.3390/biology10030207
Chicago/Turabian StyleWoller, Aurore, and Didier Gonze. 2021. "Circadian Misalignment and Metabolic Disorders: A Story of Twisted Clocks" Biology 10, no. 3: 207. https://doi.org/10.3390/biology10030207
APA StyleWoller, A., & Gonze, D. (2021). Circadian Misalignment and Metabolic Disorders: A Story of Twisted Clocks. Biology, 10(3), 207. https://doi.org/10.3390/biology10030207