Molecular Characterization and Comparative Genomics of IncQ-3 Plasmids Conferring Resistance to Various Antibiotics Isolated from a Wastewater Treatment Plant in Warsaw (Poland)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structural and Functional Genomics of Five Novel IncQ Plasmids
2.2. Comparative Genomics of Inc-Q3 Plasmids
2.3. IncQ-3 Plasmids as a Reservoir of Antibiotic Resistance Genes and Biogeography of IncQ-3 Plasmids
3. Materials and Methods
3.1. Isolation of IncQ-3 Plasmids
3.2. DNA Sequencing and Assembly
3.3. Gene Annotation and Bioinformatics
3.4. Plasmid Mobility Testing
3.5. Plasmid Sequences Accession Numbers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dröge, M.; Pühler, A.; Selbitschka, W. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol. Gen. Genet. 2000, 263, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yun, Z.; Ha, U.-H.; Lee, S.; Park, H.; Kwon, E.E.; Cho, Y.; Choung, S.; Oh, J.; Medriano, C.A.; et al. Transfer of antibiotic resistance plasmids in pure and activated sludge cultures in the presence of environmentally representative micro-contaminant concentrations. Sci. Total Environ. 2014, 468–469, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Marano, R.B.M.; Cytryn, E. The Mobile Resistome in Wastewater Treatment Facilities and Downstream Environments. In Antimicrobial Resistance in Wastewater Treatment Processes; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 129–155. ISBN 978-1-119-19242-8. [Google Scholar]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, J.A.; Wright, G.D. The antibiotic resistance “mobilome”: Searching for the link between environment and clinic. Front. Microbiol. 2013, 4, 138. [Google Scholar] [CrossRef] [Green Version]
- Krzemiński, P.; Markiewicz, Z.; Popowska, M. Entry Routes of Antibiotics and Antimicrobial Resistance in the Environment. In Antibiotics and Antimicrobial Resistance Genes: Environmental Occurrence and Treatment Technologies; Hashmi, M.Z., Ed.; Emerging Contaminants and Associated Treatment Technologies; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–26. ISBN 978-3-030-40422-2. [Google Scholar]
- Di Cesare, A.; Fontaneto, D.; Doppelbauer, J.; Corno, G. Fitness and Recovery of Bacterial Communities and Antibiotic Resistance Genes in Urban Wastewaters Exposed to Classical Disinfection Treatments. Environ. Sci. Technol. 2016, 50, 10153–10161. [Google Scholar] [CrossRef]
- Schlueter, A.; Szczepanowski, R.; Pühler, A.; Top, E. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 2007, 31, 449–477. [Google Scholar] [CrossRef]
- Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef]
- Boolchandani, M.; D’Souza, A.W.; Dantas, G. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019, 20, 356–370. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Ju, F.; Zhang, T. Exploring variation of antibiotic resistance genes in activated sludge over a four-year period through a metagenomic approach. Environ. Sci. Technol. 2013, 47, 10197–10205. [Google Scholar] [CrossRef]
- Schlueter, A.; Krause, L.; Szczepanowski, R.; Goesmann, A.; Pühler, A. Genetic diversity and composition of a plasmid metagenome from a wastewater treatment plant. J. Biotechnol. 2008, 136, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Szczepanowski, R.; Bekel, T.; Goesmann, A.; Krause, L.; Krömeke, H.; Kaiser, O.; Eichler, W.; Pühler, A.; Schlüter, A. Insight into the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to antimicrobial drugs analysed by the 454-pyrosequencing technology. J. Biotechnol. 2008, 136, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Agga, G.E.; Arthur, T.M.; Durso, L.M.; Harhay, D.M.; Schmidt, J.W. Antimicrobial-Resistant Bacterial Populations and Antimicrobial Resistance Genes Obtained from Environments Impacted by Livestock and Municipal Waste. PLoS ONE 2015, 10, e0132586. [Google Scholar] [CrossRef] [PubMed]
- Manaia, C.M.; Rocha, J.; Scaccia, N.; Marano, R.; Radu, E.; Biancullo, F.; Cerqueira, F.; Fortunato, G.; Iakovides, I.C.; Zammit, I.; et al. Antibiotic resistance in wastewater treatment plants: Tackling the black box. Environ. Int. 2018, 115, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Krzemiński, P.; Markiewicz, Z.; Popowska, M. Treatment Technologies for Removal of Antibiotics, Antibiotic Resistance Bacteria and Antibiotic-Resistant Genes. In Antibiotics and Antimicrobial Resistance Genes: Environmental Occurrence and Treatment Technologies; Hashmi, M.Z., Ed.; Emerging Contaminants and Associated Treatment Technologies; Springer International Publishing: Cham, Switzerland, 2020; pp. 415–434. ISBN 978-3-030-40422-2. [Google Scholar]
- LaPara, T.M.; Burch, T.R.; McNamara, P.J.; Tan, D.T.; Yan, M.; Eichmiller, J.J. Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor. Environ. Sci. Technol. 2011, 45, 9543–9549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manaia, C.; Novo, A.; Coelho, B.; Nunes, O. Ciprofloxacin Resistance in Domestic Wastewater Treatment Plants. Waterairand Soil Pollut. 2009, 208, 335–343. [Google Scholar] [CrossRef]
- Piotrowska, M.; Przygodzińska, D.; Matyjewicz, K.; Popowska, M. Occurrence and Variety of β-Lactamase Genes among Aeromonas spp. Isolated from Urban Wastewater Treatment Plant. Front. Microbiol. 2017, 8, 863. [Google Scholar] [CrossRef] [Green Version]
- Piotrowska, M.; Kowalska, S.; Popowska, M. Diversity of β-lactam resistance genes in gram-negative rods isolated from a municipal wastewater treatment plant. Ann. Microbiol. 2019, 69, 591–601. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo, P.; Adriana, A.; Jessica, S.; Carles, B.; Marinella, F.; Marta, L.; Luis, B.J.; Pierre, S. Antibiotic resistance in urban and hospital wastewaters and their impact on a receiving freshwater ecosystem. Chemosphere 2018, 206, 70–82. [Google Scholar] [CrossRef]
- Sakai, H.; Komano, T. DNA replication of IncQ broad-host-range plasmids in gram-negative bacteria. Biosci. Biotechnol. Biochem. 1996, 60, 377–382. [Google Scholar] [CrossRef]
- Guerry, P.; Van Embden, J.; Falkow, S. Molecular Nature of Two Nonconjugative Plasmids Carrying Drug Resistance Genes. J. Bacteriol. 1974, 117, 619–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardner, M.N.; Deane, S.M.; Rawlings, D.E. Isolation of a New Broad-Host-Range IncQ-Like Plasmid, pTC-F14, from the Acidophilic Bacterium Acidithiobacillus caldus and Analysis of the Plasmid Replicon. J. Bacteriol. 2001, 183, 3303–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loftie-Eaton, W.; Rawlings, D.E. Evolutionary Competitiveness of Two Natural Variants of the IncQ-Like Plasmids, pRAS3.1 and pRAS3.2. J. Bacteriol. 2010, 192, 6182–6190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loftie-Eaton, W.; Rawlings, D.E. Comparative Biology of Two Natural Variants of the IncQ-2 Family Plasmids, pRAS3.1 and pRAS3.2. J. Bacteriol. 2009, 191, 6436–6446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Y.; Pu, X.; Zheng, W.; Hu, G. High Prevalence of Plasmid-Mediated Quinolone Resistance and IncQ Plasmids Carrying qnrS2 Gene in Bacteria from Rivers near Hospitals and Aquaculture in China. PLoS ONE 2016, 11, e0159418. [Google Scholar] [CrossRef]
- Bönemann, G.; Stiens, M.; Pühler, A.; Schlüter, A. Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob. Agents Chemother. 2006, 50, 3075–3080. [Google Scholar] [CrossRef] [Green Version]
- Loftie-Eaton, W.; Rawlings, D.E. Diversity, biology and evolution of IncQ-family plasmids. Plasmid 2012, 67, 15–34. [Google Scholar] [CrossRef]
- Majumdar, T.; Das, B.; Bhadra, R.K.; Dam, B.; Mazumder, S. Complete nucleotide sequence of a quinolone resistance gene (qnrS2) carrying plasmid of Aeromonas hydrophila isolated from fish. Plasmid 2011, 66, 79–84. [Google Scholar] [CrossRef]
- Poirel, L.; Carattoli, A.; Bernabeu, S.; Bruderer, T.; Frei, R.; Nordmann, P. A novel IncQ plasmid type harbouring a class 3 integron from Escherichia coli. J. Antimicrob. Chemother. 2010, 65, 1594–1598. [Google Scholar] [CrossRef] [Green Version]
- Yagi, J.M.; Sims, D.; Brettin, T.; Bruce, D.; Madsen, E.L. The genome of Polaromonas naphthalenivorans strain CJ2, isolated from coal tar-contaminated sediment, reveals physiological and metabolic versatility and evolution through extensive horizontal gene transfer. Environ. Microbiol. 2009, 11, 2253–2270. [Google Scholar] [CrossRef]
- Cyriaque, V.; Jacquiod, S.; Riber, L.; Abu Al-Soud, W.; Gillan, D.C.; Sørensen, S.J.; Wattiez, R. Selection and propagation of IncP conjugative plasmids following long-term anthropogenic metal pollution in river sediments. J. Hazard. Mater. 2020, 382, 121173. [Google Scholar] [CrossRef] [PubMed]
- Romaniuk, K.; Styczynski, M.; Decewicz, P.; Buraczewska, O.; Uhrynowski, W.; Fondi, M.; Wolosiewicz, M.; Szuplewska, M.; Dziewit, L. Diversity and Horizontal Transfer of Antarctic Pseudomonas spp. Plasmids. Genes 2019, 10, 850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, S.; Rohr, J.R.; Harwood, V.J. Vancomycin resistance plasmids affect persistence of Enterococcus faecium in water. Water Res. 2019, 166, 115069. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Jin, H.M.; Park, M.S.; Park, W.; Madsen, E.L.; Jeon, C.O. Recovery of plasmid pEMB1, whose toxin-antitoxin system stabilizes an ampicillin resistance-conferring β-lactamase gene in Escherichia coli, from natural environments. Appl. Environ. Microbiol. 2015, 81, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Barraud, O.; Casellas, M.; Dagot, C.; Ploy, M.-C. An antibiotic-resistant class 3 integron in an Enterobacter cloacae isolate from hospital effluent. Clin. Microbiol. Infect. 2013, 19, E306–E308. [Google Scholar] [CrossRef] [Green Version]
- Simo Tchuinte, P.L.; Stalder, T.; Venditti, S.; Ngandjio, A.; Dagot, C.; Ploy, M.-C.; Barraud, O. Characterisation of class 3 integrons with oxacillinase gene cassettes in hospital sewage and sludge samples from France and Luxembourg. Int. J. Antimicrob. Agents 2016, 48, 431–434. [Google Scholar] [CrossRef]
- Han, J.E.; Kim, J.H.; Choresca, C.H.; Shin, S.P.; Jun, J.W.; Chai, J.Y.; Park, S.C. A small IncQ-type plasmid carrying the quinolone resistance (qnrS2) gene from Aeromonas hydrophila. Lett. Appl. Microbiol. 2012, 54, 374–376. [Google Scholar] [CrossRef]
- Manageiro, V.; Romão, R.; Moura, I.B.; Sampaio, D.A.; Vieira, L.; Ferreira, E.; Caniça, M. Molecular Epidemiology and Risk Factors of Carbapenemase-Producing Enterobacteriaceae Isolates in Portuguese Hospitals: Results From European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE). Front. Microbiol. 2018, 9, 2834. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, E.; Sela, N.; Doron-Faigenboim, A.; Navon-Venezia, S.; Jurkevitch, E.; Cytryn, E. Genomic and Functional Characterization of qnr-Encoding Plasmids from Municipal Wastewater Biosolid Klebsiella pneumoniae Isolates. Front. Microbiol. 2015, 6, 1354. [Google Scholar] [CrossRef] [Green Version]
- Meyer, R. Replication and conjugative mobilization of Broad Host-range IncQ plasmids. Plasmid 2009, 62, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Scherzinger, E.; Haring, V.; Lurz, R.; Otto, S. Plasmid RSF1010 DNA Replication In Vitro Promoted by Purified RSF1010 RepA, RepB and RepC Proteins. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC333844/ (accessed on 9 February 2020).
- Martínez-Martínez, L.; Pascual, A.; Jacoby, G.A. Quinolone resistance from a transferable plasmid. Lancet 1998, 351, 797–799. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Strahilevitz, J.; Hooper, D.C. Plasmid-Mediated Quinolone Resistance. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elnekave, E.; Hong, S.L.; Lim, S.; Hayer, S.S.; Boxrud, D.; Taylor, A.J.; Lappi, V.; Noyes, N.; Johnson, T.J.; Rovira, A.; et al. Circulation of Plasmids Harboring Resistance Genes to Quinolones and/or Extended-Spectrum Cephalosporins in Multiple Salmonella enterica Serotypes from Swine in the United States. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Yu, X.; Shang, Y.; Xu, H.; Guo, L.; Liang, Y.; Kang, Y.; Song, L.; Sun, J.; Yue, F.; et al. Emergence and Characterization of a Novel IncP-6 Plasmid Harboring blaKPC-2 and qnrS2 Genes in Aeromonas taiwanensis Isolates. Front. Microbiol. 2019, 10, 2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-W.; Thawng, C.N.; Lee, S.H.; Cha, C.-J. Unique Features of Aeromonas Plasmid pAC3 and Expression of the Plasmid-Mediated Quinolone Resistance Genes. Msphere 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.B.; Park, C.H.; Kim, C.J.; Kim, E.-C.; Jacoby, G.A.; Hooper, D.C. Prevalence of Plasmid-Mediated Quinolone Resistance Determinants over a 9-Year Period. Antimicrob. Agents Chemother. 2009, 53, 639–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.-Y.; Zhao, S.-M.; Mu, X.-D.; Xiao, Z. Genetic characterization of plasmid-mediated quinolone resistance gene qnrS2 in Pseudoalteromonas and Shewanella isolates from seawater. FEMS Microbiol. Lett. 2017, 364, fnw295. [Google Scholar] [CrossRef] [Green Version]
- Gniadkowski, M. Evolution of extended-spectrum beta-lactamases by mutation. Clin. Microbiol. Infect. 2008, 14 (Suppl. 1), 11–32. [Google Scholar] [CrossRef] [Green Version]
- Chudejova, K.; Rotova, V.; Skalova, A.; Medvecky, M.; Adamkova, V.; Papagiannitsis, C.C.; Hrabak, J. Emergence of sequence type 252 Enterobacter cloacae producing GES-5 carbapenemase in a Czech hospital. Diagn. Microbiol. Infect. Dis. 2018, 90, 148–150. [Google Scholar] [CrossRef]
- Japoni-Nejad, A.; Farshad, S.; van Belkum, A.; Ghaznavi-Rad, E. Novel cassette array in a class 1 integron in clinical isolates of Acinetobacter baumannii from central Iran. Int. J. Med. Microbiol. 2013, 303, 645–650. [Google Scholar] [CrossRef]
- Jones-Dias, D.; Manageiro, V.; Ferreira, E.; Barreiro, P.; Vieira, L.; Moura, I.B.; Caniça, M. Architecture of Class 1, 2, and 3 Integrons from Gram Negative Bacteria Recovered among Fruits and Vegetables. Front. Microbiol. 2016, 7, 1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, Q.; Cerdeira, L.; Fernandes, M.R.; Vianello, M.A.; Lincopan, N. Novel class 1 integron (In1390) harboring blaGES-5 in a Morganella morganii strain recovered from a remote community. Diagn. Microbiol. Infect. Dis. 2018, 91, 345–347. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Carrër, A.; Pitout, J.D.; Nordmann, P. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob. Agents Chemother. 2009, 53, 2492–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziurzyński, M. Drewniak & Dziewit Research Group. Available online: http://maisen.ddg.biol.uw.edu.pl/ (accessed on 10 December 2018).
- Carver, T.; Berriman, M.; Tivey, A.; Patel, C.; Böhme, U.; Barrell, B.G.; Parkhill, J.; Rajandream, M.-A. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 2008, 24, 2672–2676. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [Green Version]
- Bartosik, D.; Baj, J.; Sochacka, M.; Piechucka, E.; Wlodarczyk, M. Molecular characterization of functional modules of plasmid pWKS1 of Paracoccus pantotrophus DSM 11072The GenBank accession number for the sequence reported in this paper is AF482428. 2002, 148, 2847–2856. Microbiology 2002, 148, 2847–2856. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: New York, NY, USA, 2001. [Google Scholar]
- Hanahan, D.; Jessee, J.; Bloom, F.R. [4] Plasmid transformation of Escherichia coli and other bacteria. In Methods in Enzymology; Bacterial Genetic Systems; Elsevier Inc.: Amsterdam, The Netherlands, 1991; Volume 204, pp. 63–113. [Google Scholar]
- Irani, V.R.; Rowe, J.J. Enhancement of transformation in Pseudomonas aeruginosa PAO1 by Mg2+ and heat. BioTechniques 1997, 22, 54–56. [Google Scholar] [CrossRef]
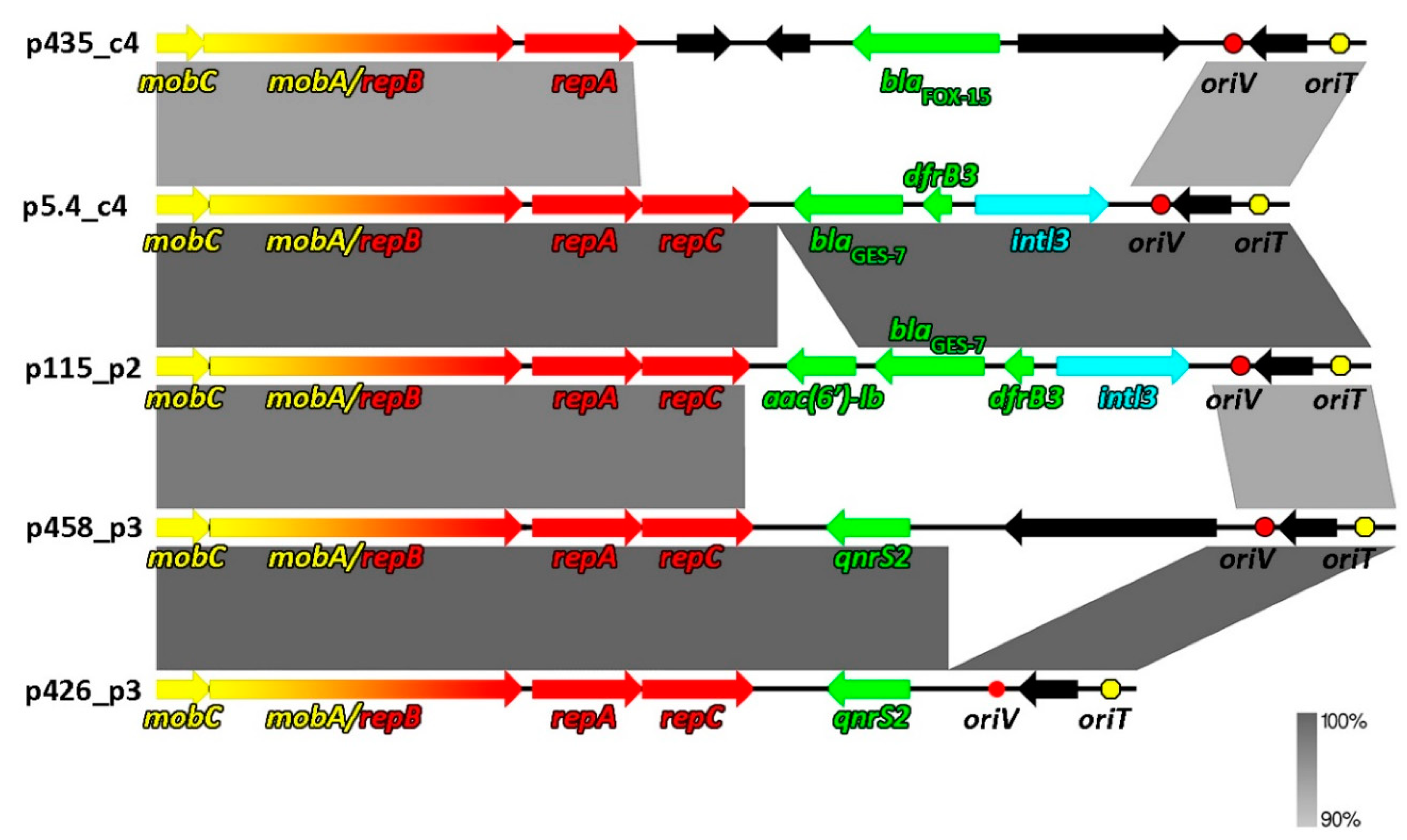
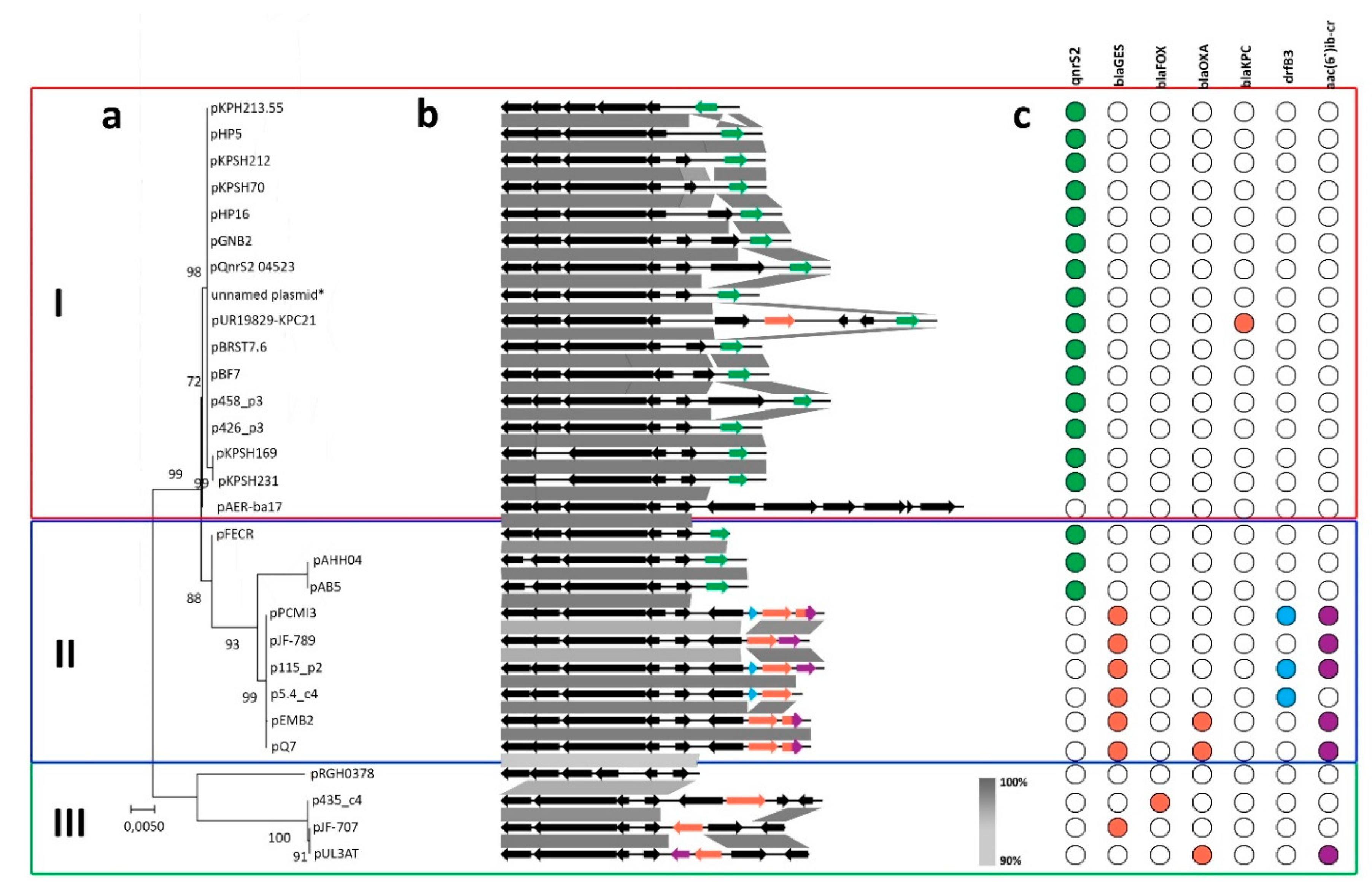
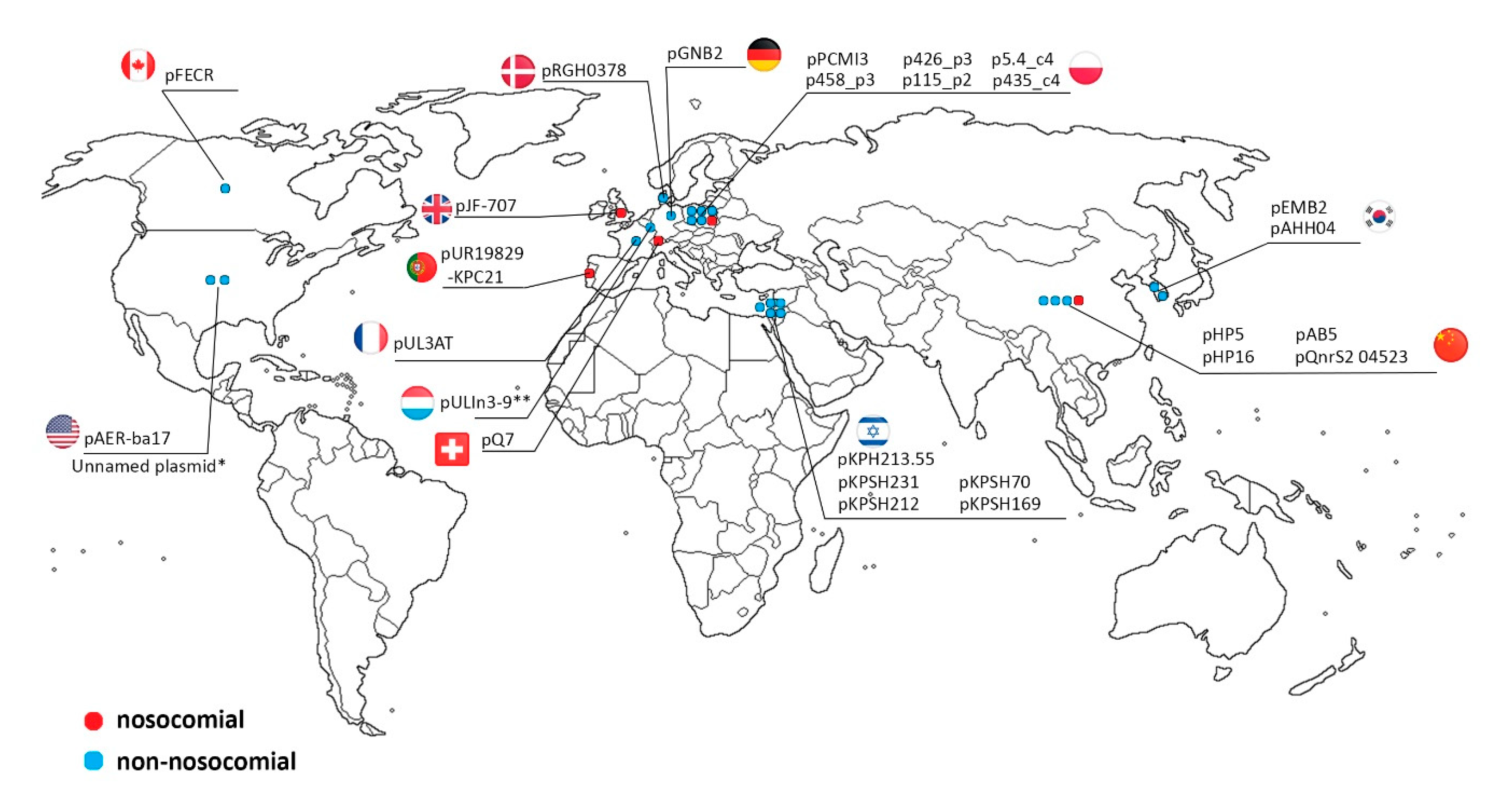
| Plasmid Name | Plasmid Size (bp) | GC Content (%) | Host Strain (16S rDNA GenBank acc. no.) | Type of Wastewater | Antibiotic Resistance Genes | Integron Integrase Genes | GenBank Accession Number |
|---|---|---|---|---|---|---|---|
| p5.4_c4 | 8814 | 62 | Aeromonas sp. (MF461069, MF461079) | RW | blaGES-7, dfrB3 | intI3 | MT231818 |
| p115_p2 | 9448 | 61.5 | Aeromonas sp. (MF461085); Raoultella sp. (MF457856- MF457866) | RW | aac(6′)ib, blaGES-7, dfrB3 | intI3 | MT231822 |
| p458_p3 | 9639 | 57 | Aeromonas sp. (MF461156) | TW | qnrS2 | - | MT231819 |
| p426_p3 | 7621 | 59.7 | Aeromonas sp. (MF461154) | TW | qnrS2 | - | MT231821 |
| p435_c4 | 9407 | 61.2 | Kluyvera sp. (MF457880) | AS | blaFOX-15 | - | MT231820 |
| Plasmid | Size (bp) | Host Strain | Sample Origin | Antibiotic Resistance Genes | Integron Integrase Genes | GenBank Accession Number | Reference |
|---|---|---|---|---|---|---|---|
| pPCMI3 | 9448 | Serratia marcescens subsp. marcescens S89 | clinical sample, urine (Poland) | blaOXA/aac(6’)-Ib-cr, blaGES-7, dfrB3 | intI3 | MH569711 | - |
| pJF-789 | 9016 | Klebsiella oxytoca H140960789 | clinical isolate | blaGES-5, aac(6′)-lb | intI1 | KX912254 | - |
| pEMB2 | 9042 | uncultured bacterium | WWTP (South Korea) | blaGES-1, blaOXA/aac(6′)-lb | intI3 | KJ631731 | [37] |
| pQ7 | 9042 | Escherichia coli 7 | clinical sample (Switzerland) | blaGES-1, blaOXA/aac(6′)-lb | intI3 | NC_014356 | [32] |
| pJF-707 | 8300 | Klebsiella oxytoca H143640707 | clinical sample (UK) | blaGES-5 | intI3 | KX946994 | - |
| pUL3AT | 9005 | Enterobacter cloacae LIM73 | hospital effluent (France) | blaOXA-10, aac(6’)-Ib | intI3 | HE616889 | [38] |
| pULIn3-9 | Citrobacter freundii LIM86 | hospital effluent (Luxembourg) | blaOXA-2, blaGES-1 | - | - | [39] | |
| pAHH04 | 7191 | Aeromonas hydrophila SNUFPC-A10 | fish (South Korea) | qnrS2 | - | JN315883 | [40] |
| pAB5 | 7212 | Aeromonas caviae AB5 | river water (China) | qnrS2 | - | KU644674 | [28] |
| pFECR | 6671 | uncultured bacterium clone AA-102 | WWTP (Canada) | qnrS2 | - | MF554639 | - |
| pUR19829-KPC21 | 12748 | Escherichia coli INSRA19829 | clinical sample (Portugal) | qnrS2 | - | MH133192 | [41] |
| pHP5 | 7637 | Aeromonas allosaccharophila HP5 | aquatic environment near hospital (China) | qnrS2 | - | KU644676 | [28] |
| pHP16 | 8213 | Aeromonas caviae HP16 | aquatic environment near hospital (China) | qnrS2 | - | KU644675 | [28] |
| pKPSH212 | 7742 | Klebsiella pneumoniae I212 | WWTP/ reclamation project site (Israel) | qnrS2 | - | KT896501 | [42] |
| pKPSH70 | 7748 | Klebsiella pneumoniae I70 | WWTP/ reclamation project site (Israel) | qnrS2 | - | KT896500 | [42] |
| pBRST7.6 | 7621 | Aeromonas hydrophila AO1 | infected fish | qnrS2 | - | NC_011207 | [31] |
| pGNB2 | 8469 | - | activated sludge (Germany) | qnrS2 | - | NC_013773 | [29] |
| pKPSH213.55 | 6981 | Klebsiella pneumoniae I213 | WWTP/ reclamation project site (Israel) | qnrS2 | - | KT896502 | [42] |
| pKPSH231 | 7748 | Klebsiella pneumoniae I231 | WWTP/ reclamation project site (Israel) | qnrS2 | - | KT896503 | [42] |
| pKPSH169 | 7747 | Klebsiella pneumoniae I169 | WWTP/ reclamation project site (Israel) | qnrS2 | - | KT896499 | [42] |
| pBF7.8 | 7844 | Aeromonas hydrophila IB101 | infected fish | qnrS2 | - | KM245123 | - |
| unnamed | 7555 | Salmonella enterica subsp. enterica serovar Typhimurium var. 5- strain 63 | swine (USA) | qnrS2 | - | MK191840 | - |
| pQnrS2_045523 | 9639 | Enterobacter kobei WCHEK045523 | clinical sample (China) | qnrS2 | - | CP032896 | - |
| pAER-ba17 | 13512 | Aeromonas sp. ASNIH2 | USA | - | - | CP026407 | - |
| pRGRH0378 | 5790 | uncultured prokaryote | from rat gut metagenome metamobilome, hospital sewer (Denmark) | - | - | LN853034 | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piotrowska, M.; Dziewit, L.; Ostrowski, R.; Chmielowska, C.; Popowska, M. Molecular Characterization and Comparative Genomics of IncQ-3 Plasmids Conferring Resistance to Various Antibiotics Isolated from a Wastewater Treatment Plant in Warsaw (Poland). Antibiotics 2020, 9, 613. https://doi.org/10.3390/antibiotics9090613
Piotrowska M, Dziewit L, Ostrowski R, Chmielowska C, Popowska M. Molecular Characterization and Comparative Genomics of IncQ-3 Plasmids Conferring Resistance to Various Antibiotics Isolated from a Wastewater Treatment Plant in Warsaw (Poland). Antibiotics. 2020; 9(9):613. https://doi.org/10.3390/antibiotics9090613
Chicago/Turabian StylePiotrowska, Marta, Lukasz Dziewit, Rafał Ostrowski, Cora Chmielowska, and Magdalena Popowska. 2020. "Molecular Characterization and Comparative Genomics of IncQ-3 Plasmids Conferring Resistance to Various Antibiotics Isolated from a Wastewater Treatment Plant in Warsaw (Poland)" Antibiotics 9, no. 9: 613. https://doi.org/10.3390/antibiotics9090613
APA StylePiotrowska, M., Dziewit, L., Ostrowski, R., Chmielowska, C., & Popowska, M. (2020). Molecular Characterization and Comparative Genomics of IncQ-3 Plasmids Conferring Resistance to Various Antibiotics Isolated from a Wastewater Treatment Plant in Warsaw (Poland). Antibiotics, 9(9), 613. https://doi.org/10.3390/antibiotics9090613





