Antimicrobial Resistance Genes in Porcine Pasteurella multocida Are Not Associated with Its Antimicrobial Susceptibility Pattern
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Resistance
2.2. Description of Resistance Genes
2.3. Analysis of the Association between the Presence of Resistance Genes and Antimicrobial Patterns
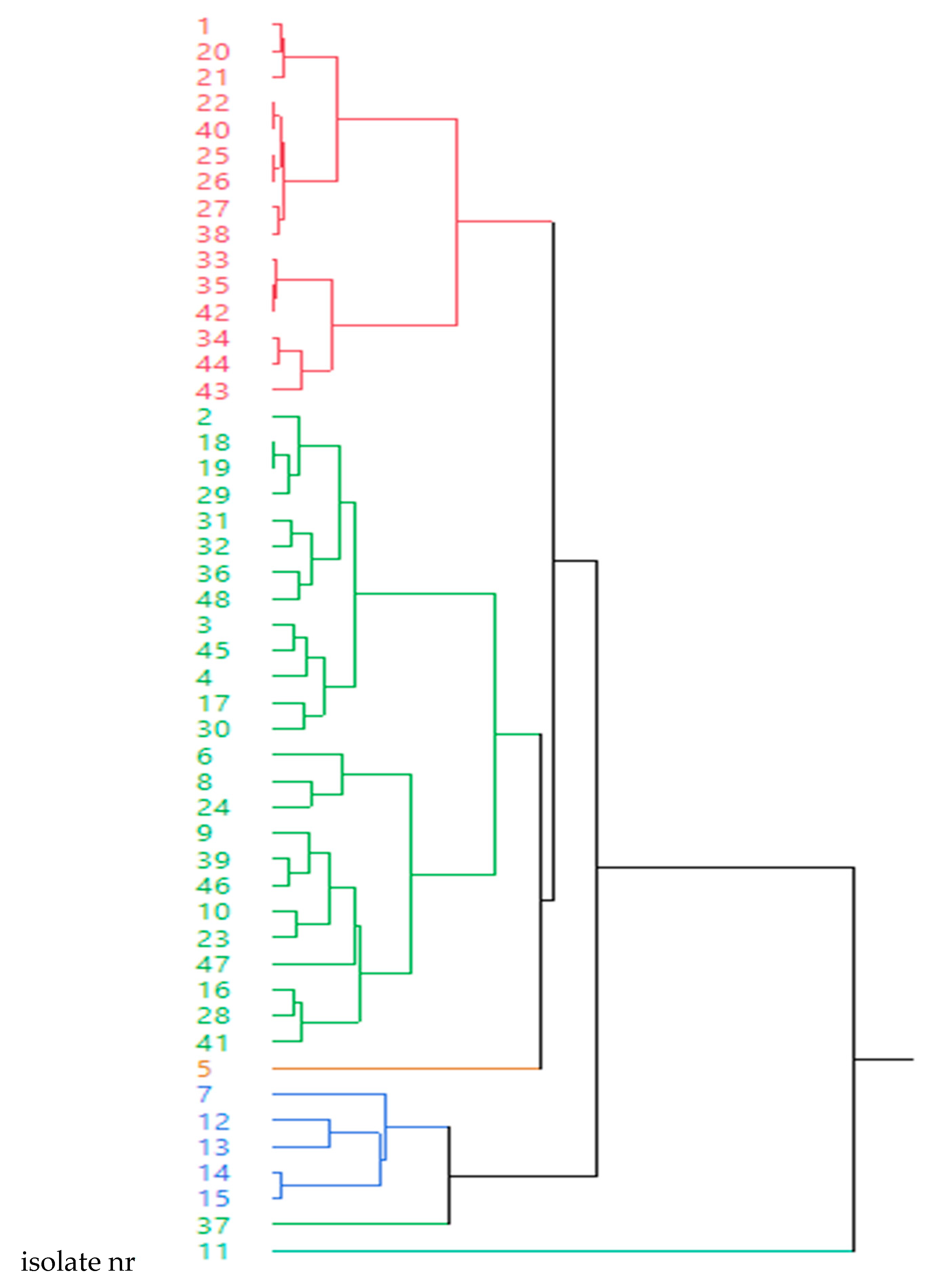
2.4. Relationship between the Presence of Resistance Genes and Clusters based on Antimicrobial Susceptibility Pattern of 12 Antimicrobials
3. Discussion
4. Material and Methods
4.1. Clinical Samples
4.2. Bacterial Isolation and Identification
4.3. Antimicrobial Sensitivity Testing
4.4. Determination of Antimicrobial Resistance Genes
4.5. Data Analysis
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carr, J.; Chen, S.P.; Connor, J.F.; Kirkwood, R.; Segalés, S. Respiratory disorders. In Pig Health; Carr, J., Chen, S.P., Connor, J.F., Kirkwood, R., Segalés, S., Eds.; CRC Press Taylor & Francis: Boca Ratón, FL, USA, 2018; pp. 103–152. [Google Scholar]
- Kachooel, A.; Ranjbar, M.M.; Kachooel, S. Evaluation of Pasteurella multocida serotype B:2 resistance to immune serum and complement system. Vet. Res. 2017, 8, 179–184. [Google Scholar]
- De Alvis, M.C. Haemorrhagic septicaemia—A general review. Br. Vet. J. 1992, 148, 99–112. [Google Scholar] [CrossRef]
- Tang, X.; Zhao, Z.; Hu, J.; Wu, B.; Cai, X.; He, Q.; Chen, H. Isolation, antimicrobial resistance, and virulence genes of Pasteurella multocida strains from swine in China. J. Clin. Microbiol. 2009, 47, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Fraile, L. Antimicrobial Therapy in Swine; Practical approach; Editorial Servet: Zaragoza, Spain, 2013. [Google Scholar]
- Li, Y.; Cunha de Silva, G.; Li, Y.; Rossi, C.C.; Fernández-Crespo, R.; Williamson, S.M.; Langford, P.R.; Soares Bazzolli, D.M.; Bossé, J.T. Evidence of illegitimate recombination between two Pasteurellaceae plasmids resulting in a novel multi-resistance replicon, pM3362MDR in Actinobacillus pleuropneumoniae. Front. Microbiol. 2018, 9, 2489. [Google Scholar] [CrossRef]
- Holmer, I.; Salomonsen, C.M.; Jorsal, S.E.; Astrup, L.B.; Jensen, V.F.; Høg, B.B.; Pedersen, K. Antibiotic resistance in porcine pathogenic bacteria and relation to antibiotic usage. BMC Vet. Res. 2019, 15, 449. [Google Scholar] [CrossRef]
- Roberts, M.C. Tetracycline therapy: Update. Clin. Infect. Dis. 2003, 36, 462–467. [Google Scholar] [CrossRef]
- Blanco, M.; Gutiérrez-Martín, C.B.; Rodríguez-Ferri, E.F.; Roberts, M.C.; Navas, J. Distribution of tetracycline resistance genes in Actinobacillus pleuropneumoniae isolates from Spain. Antimicrob. Agents Chemother. 2006, 50, 702–708. [Google Scholar] [CrossRef]
- White, D.G.; Zhao, S.; Simjee, S.; Wagner, D.D.; McDermott, P.F. Antimicrobial resistance of foodborne pathogens. Microbes Infect. 2002, 4, 405–412. [Google Scholar] [CrossRef]
- Michael, G.B.; Bossé, J.T.; Schwarz, S. Antimicrobial resistance in Pasteurellaceae of veterinary origin. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Schwarz, S. Mechanisms of antimicrobial resistance in Pasteurellaceae. In Pasteurellaceae. Biology, Genomics and Molecular Aspects; Kuhnert, P., Christensen, H., Eds.; Caister Academic Press: Norfolk, UK, 2008; pp. 199–228. [Google Scholar]
- Zeineldin, M.M.; Megahed, A.; Blair, B.; Burton, B.; Aldridge, B.; Lowe, J. Negligible impact of perinatal tulathromycin metaphylaxis on the development dynamics of fecal microbiota and their accompanying antimicrobial resistome in piglets. Front. Microbiol. 2019, 10, 726. [Google Scholar] [CrossRef]
- González-Marín, C.; Spratt, D.A.; Millar, M.R.; Simmonds, M.; Kempley, S.T.; Allaker, R.P. Identification of bacteria and potential sources in neonates at risk of infection delivered by caesarean and vaginal birth. J. Med. Microbiol. 2012, 61, 31–41. [Google Scholar] [CrossRef] [PubMed]
- ECDC/EFSA/EMA. Second Joint Report on the Integrated Analysis of the Consumption of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Humans and Food-Producing Animals; John Wiley and Sons Ltd.: Solna, Sweden, 2017; p. 134. [Google Scholar]
- Petrocchi-Rilo, M.; Gutiérrez-Martín, C.B.; Méndez-Hernández, J.I.; Rodríguez-Ferri, E. Antimicrobial resistance of Pasteurella multocida isolates recovered from swine pneunomina in Spain throughout 2017 and 2018. Vet. Anim. Sci. 2019, 7, 100044. [Google Scholar] [CrossRef] [PubMed]
- San Millán, A.; Escudero, J.A.; Gutiérrez, B.; Hidalgo, L.; García, N.; Llagostera, M.; Domínguez, L.; González-Zorn, B. Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob. Agents Chemother. 2009, 53, 3399–3404. [Google Scholar] [CrossRef] [PubMed]
- Matter, D.; Rossano, A.; Limat, S.; Vorlet-Fawer, L.; Brodard, I.; Perreten, V. Antimicrobial resistance profile of Actinobacillus pleuropneumoniae and Actinobacillus porcitonsillarum. Vet. Microbiol. 2007, 122, 146–156. [Google Scholar] [CrossRef]
- San Millán, A.; Escudero, J.A.; Catalán, A.; Nieto, S.; Farelo, F.; Gibert, M.; Moreno, M.A.; Domínguez, L.; González-Zorn, L. β-lactam resistance in Haemophilus parasuis is mediated by plasmid pb1000 bearing blaROB1. Antimicrob. Agents Chemother. 2007, 51, 2260–2264. [Google Scholar] [CrossRef][Green Version]
- Dayao, D.A.E.; Gibson, J.S.; Blackall, P.J.; Turni, C. Antimicrobial resistance genes in Actinobacillus pleuropneumoniae, Haemophilus parasuis and Pasteurella multocida isolated from Australian pigs. Austr. Vet. J. 2016, 94, 227–231. [Google Scholar] [CrossRef]
- Vera-Lizarazo, Y.A.; Rodríguez-Ferri, E.F.; Martín de la Fuente, A.J.; Gutiérrez-Martín, C.B. Evaluation of changes in antimicrobial susceptibility patterns of Pasteurella multocida subsp. multocida isolates from pigs in Spain in 1987–1988 and 2003–2004. Am. J. Vet. Res. 2006, 67, 663–668. [Google Scholar]
- Chander, Y.; Oliveira, S.; Goyal, S.M. Characterization of ceftiofur resistance in swine bacterial pathogens. Vet. J. 2011, 187, 139–141. [Google Scholar] [CrossRef]
- Oh, Y.H.; Moon, D.C.; Lee, Y.J.; Hyun, B.H.; Lim, S.K. Genetic and phenotypic characterization of tetracycline-resistant Pasteurella multocida isolated from pigs. Vet. Microbiol. 2019, 233, 159–163. [Google Scholar] [CrossRef]
- Furian, T.Q.; Borges, K.A.; Laviniki, V.; da Silveira Rocha, S.L.; de Almeida, C.N.; do Nascimiento, V.P. Virulence genes and antimicrobial resistance of Pasteurella multocida isolated from poultry and swine. Brazil. J. Microbiol. 2016, 47, 210–216. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, S.J. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N.; Pfaller, M.A.; Rhomberg, P.R.; Walter, D.H. Tiamulin activity against fastidious and nonfastidious veterinary and human bacterial isolate: Initial development of in vitro susceptibility test methods. J. Clin. Microbiol. 2002, 40, 461–465. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 2018, 4th ed.; CLSI Supplement VET08; Wayne: Philadelphia, PA, USA.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 2018, 28th ed.; CLSI Supplement M100; Wayne: Philadelphia, PA, USA.
| Antimicrobial Agent | Range (μg/mL) | MIC50 (μg/mL) | MIC90 (μg/mL) | Breakpoint (μg/mL) * | Antimicrobial Resistance (%) |
|---|---|---|---|---|---|
| Amoxicillin | 1–8 | 0.25 | 8 | 0.5 $ | 2.1 |
| Ceftiofur | 0.06–0.25 | 0.06 | 0.12 | 2 | 0 |
| Doxycycline | 0.25–2 | 1 | >2 | 0.5 $$ | 52.1 |
| Enrofloxacin | 0.03–0.5 | 0.03 | 0.12 | 0.25 | 2.1 |
| Florfenicol | 0.5 | 0.5 | 0.5 | 2 | 0 |
| Marbofloxacin | 0.03–0.5 | 0.03 | 0.12 | 0.25 & | 4.2 |
| Oxytetracycline | 0.5–8 | 2 | >8 | 0.5 | 68.7 |
| Sulphamethoxazole/trimethoprim (19/1 ratio) § | 0.06–4 | 0.25 | >4 | 0.5 && 2 §§ | 43.7 31.2 |
| Tiamulin | 2–32 | 16 | >32 | 16 | 25 |
| Tildipirosin | 0.5–4 | 1 | 4 | 4 | 0 |
| Tilmicosin | 2–32 | 8 | 32 | 16 | 2.1 |
| Tulathromycin | 0.5–4 | 1 | 2 | 16 | 0 |
| Number of Isolate | Number of Antimicrobial Agents | Resistance to |
|---|---|---|
| 5 | 0 | No antimicrobial resistance |
| 2 | 1 | Oxytetracycline |
| 6 | 1 | Sulphamethoxazole/trimethoprim |
| 4 | 1 | Tiamulin |
| 12 | 2 | Doxycycline + oxytetracycline |
| 1 | 2 | Marbofloxacin + oxytetracycline |
| 3 | 2 | Oxytetracycline + sulphamethoxazole/trimethoprim |
| 1 | 2 | Oxytetracycline + tiamulin |
| 1 | 3 | Amoxicillin + doxycycline + oxytetracycline |
| 4 | 3 | Doxcycline + oxytetracycline + sulphamethoxazole/trimethoprim |
| 5 | 3 | Doxcycline + oxytetracycline + tiamulin |
| 1 | 3 | Oxytetracycline + tiamulin + tilmicosin |
| 1 | 4 | Doxycycline + enrofloxacin + oxytetracycline + tiamulin |
| 2 | 4 | Doxycycline + oxytetracycline + sulphamethoxazole/trimethoprim + tiamulin |
| Resistance Gene | Number of Isolates | Resistance or Sensitivity | Resistance or Sensitivity to |
|---|---|---|---|
| tetA | 3 | Resistance | Tetracyclines * |
| tetA | 1 | Oxytetracycline | |
| tetB | 11 | Tetracyclines * | |
| tetB | 3 | Oxytetracycline | |
| blaROB1 | 1 | Amoxicillin | |
| tetA | 2 | Sensitivity | Tetracyclines * |
| tetB | 5 | Tetracyclines * | |
| blaROB1 | 15 | Amoxicillin | |
| ermA | 8 | Macrolides $ | |
| ermC | 19 | Macrolides $ | |
| msrE | 12 | Macrolides $ | |
| mphE | 1 | Macrolides $ |
| Antimicrobial Resistance Genes * | β-lactams | Macrolides $ | Tetracyclines | ||
|---|---|---|---|---|---|
| Amoxicillin | Tilmicosin | Doxycycline | Oxitetracycline | ||
| β-lactam resistance genes | blaROB1 | 0.5536 | - | - | - |
| blaTEM | 0.8408 | - | - | - | |
| Macrolide resistance genes | ermA | - | 0.7764 | - | - |
| ermC | - | 0.6538 | - | - | |
| msrE | - | 0.7392 | - | - | |
| Tetracycline resistance genes | tetA | - | - | 0.9131 | 0.9063 |
| tetB | - | - | 0.5146 | 0.7255 | |
| Cluster | Isolate nr | MIC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Flor | Enrof | Amox | Marb | Ceft | Sulf | Tild | Dox | Oxitet | Tia | Tulat | Tilm | ||
| 1 | 38 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 4 | 0.5 | 0.25 | 0.5 | 16 | 1 | 2 |
| 27 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 4 | 0.5 | 0.5 | 0.5 | 16 | 1 | 2 | |
| 22 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 4 | 0.5 | 0.5 | 0.5 | 16 | 1 | 4 | |
| 40 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 4 | 0.5 | 0.5 | 0.5 | 16 | 1 | 4 | |
| 25 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 4 | 0.5 | 0.5 | 1 | 16 | 1 | 4 | |
| 26 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 4 | 0.5 | 0.5 | 1 | 16 | 1 | 4 | |
| 34 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 4 | 0.5 | 1 | 8 | 16 | 1 | 2 | |
| 44 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 4 | 0.5 | 1 | 8 | 16 | 1 | 4 | |
| 21 | 0.5 | 0.03 | 0.25 | 0.03 | 0.12 | 4 | 0.5 | 0.25 | 0.5 | 16 | 1 | 4 | |
| 20 | 0.5 | 0.03 | 0.25 | 0.03 | 0.12 | 4 | 0.5 | 0.5 | 0.5 | 16 | 1 | 2 | |
| 35 | 0.5 | 0.03 | 0.25 | 0.03 | 0.12 | 4 | 0.5 | 1 | 8 | 16 | 1 | 2 | |
| 42 | 0.5 | 0.03 | 0.25 | 0.03 | 0.12 | 4 | 0.5 | 1 | 8 | 16 | 1 | 2 | |
| 33 | 0.5 | 0.03 | 0.25 | 0.03 | 0.12 | 4 | 1 | 1 | 8 | 16 | 1 | 2 | |
| 43 | 0.5 | 0.03 | 0.5 | 0.03 | 0.06 | 4 | 0.5 | 2 | 8 | 16 | 1 | 4 | |
| 1 | 0.5 | 0.03 | 0.5 | 0.03 | 0.12 | 4 | 0.5 | 0.5 | 1 | 16 | 1 | 2 | |
| 2 | 17 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 0.06 | 0.5 | 1 | 2 | 2 | 1 | 2 |
| 45 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 0.06 | 0.5 | 2 | 2 | 8 | 0.5 | 4 | |
| 3 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 0.06 | 1 | 2 | 4 | 8 | 1 | 8 | |
| 36 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 0.25 | 0.5 | 0.25 | 0.5 | 16 | 1 | 4 | |
| 29 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 0.06 | 1 | 2 | 2 | 16 | 1 | 8 | |
| 18 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 1 | 1 | 2 | 2 | 16 | 1 | 8 | |
| 19 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 1 | 1 | 2 | 2 | 16 | 1 | 8 | |
| 2 | 0.5 | 0.03 | 0.5 | 0.03 | 0.06 | 0.06 | 1 | 2 | 4 | 16 | 2 | 8 | |
| 31 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 0.25 | 2 | 1 | 2 | 16 | 2 | 16 | |
| 32 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 0.06 | 2 | 0.5 | 0.5 | 16 | 4 | 16 | |
| 46 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 1 | 1 | 0.25 | 0.5 | 32 | 1 | 8 | |
| 8 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 2 | 1 | 1 | 8 | 32 | 1 | 16 | |
| 41 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 0.06 | 2 | 2 | 2 | 32 | 2 | 16 | |
| 24 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 0.06 | 2 | 2 | 8 | 32 | 2 | 16 | |
| 28 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 0.25 | 2 | 2 | 2 | 32 | 2 | 8 | |
| 16 | 0.5 | 0.03 | 0.25 | 0.03 | 0.06 | 1 | 2 | 2 | 4 | 32 | 2 | 8 | |
| 48 | 0.5 | 0.03 | 0.25 | 0.06 | 0.06 | 0.06 | 1 | 1 | 2 | 16 | 2 | 8 | |
| 4 | 0.5 | 0.03 | 0.5 | 0.03 | 0.06 | 0.06 | 2 | 2 | 2 | 8 | 2 | 16 | |
| 6 | 0.5 | 0.03 | 0.5 | 0.03 | 0.06 | 0.06 | 2 | 1 | 8 | 16 | 2 | 8 | |
| 39 | 0.5 | 0.03 | 0,5 | 0.03 | 0.06 | 0.12 | 2 | 0.5 | 0.5 | 32 | 2 | 8 | |
| 47 | 0.5 | 0.03 | 0,5 | 0.03 | 0.06 | 0.06 | 4 | 0.5 | 1 | 32 | 4 | 32 | |
| 30 | 0.5 | 0.06 | 0.12 | 0.12 | 0.06 | 0.06 | 1 | 1 | 2 | 8 | 1 | 4 | |
| 10 | 0.5 | 0.06 | 0.25 | 0.06 | 0.12 | 0.5 | 2 | 0.5 | 0.5 | 32 | 4 | 8 | |
| 9 | 0.5 | 0.06 | 0.25 | 0.12 | 0.06 | 1 | 2 | 0.25 | 0.5 | 32 | 4 | 16 | |
| 23 | 0.5 | 0.12 | 0.25 | 0.12 | 0.12 | 0.06 | 1 | 0.5 | 1 | 32 | 2 | 8 | |
| 3 | 5 | 0.5 | 0.03 | 8 | 0.03 | 0.06 | 0.25 | 2 | 1 | 8 | 16 | 4 | 16 |
| 4 | 7 | 0.5 | 0,03 | 0.5 | 0.03 | 0.25 | 0.12 | 0.5 | 0.5 | 1 | 16 | 2 | 4 |
| 14 | 0.5 | 0.25 | 0.25 | 0.25 | 0.12 | 0.12 | 0.5 | 0.5 | 0.5 | 16 | 1 | 4 | |
| 12 | 0.5 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 1 | 0.5 | 0.5 | 16 | 2 | 8 | |
| 15 | 0.5 | 0.25 | 0.5 | 0.25 | 0.12 | 0.12 | 0.5 | 0.5 | 1 | 16 | 2 | 4 | |
| 13 | 0.5 | 0.25 | 0.5 | 0.5 | 0.25 | 0.12 | 0.5 | 0.5 | 1 | 16 | 2 | 4 | |
| 5 | 37 | 0.5 | 0.5 | 0.25 | 0.5 | 0.06 | 0.06 | 1 | 2 | 4 | 32 | 2 | 8 |
| 6 | 11 | 2 | 0.5 | 0.25 | 0.5 | 0.12 | 0.25 | 64 | 8 | 8 | 16 | 64 | 64 |
| Resistance Gene | Primer | Amplicon Size | Annealing Temperature | Reference |
|---|---|---|---|---|
| tetA | F: 5′-GTA ATT CTG AGC ACT GTC GC-3′ | 1057 pb | 62 °C | [20] |
| R: 5′-CTG CCT GGA CAA CAT TGT TT-3′ | ||||
| tetB | F: 5′CCT TAT CAT GCC AGT CTT GC-3′ | 774 pb | 50 °C | [20] |
| R: 5′ ACT GCC GTT TTT TTC GCC-3′ | ||||
| blaROB1 | F: 5′ CAT TAA CGG CTT GTT CGC-3′ | 852 pb | 55 °C | [20] |
| R: 5′-CTT GCT TTG CTG CAT CTT-3′ | ||||
| blaTEM | F: 5′GAG TAT TCA ACA TTT TCG T-3′ | 856 pb | 55 °C | [20] |
| R: 5′-ACC AAT GCT TAA TCA GTG A-3′ | ||||
| ermA | F: 5′-ACG ATA TTC ACG GTT TAC CCA CTT-A-3′ | 610 pb | 53 °C | [20] |
| R: 5-AAC CAG AAA AAC CCT AAA GAC ACG-3′ | ||||
| ermC | F: 5′-AAT-CGG CTC AGG AAA AGG-3′ | 562 pb | 55 °C | [20] |
| R: 5′-ATC GTC ATT TCC TGC ATG-3′ | ||||
| msrE | F: 5′-TAT AGC GAC TTT AGC GCC AA-3′ | 271 pb | 58 °C | [20] |
| R: 3′-GCC GTA GAA TAT GAG CTG AT-3′ | ||||
| mphE | F: 5′-ATG CCC AGC ATA TAA ATC GC-3′ | 295 pb | 58 °C | [20] |
| R: 5′-ATA TGG ACA AAG ATAGCC CG-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrocchi-Rilo, M.; Gutiérrez-Martín, C.-B.; Pérez-Fernández, E.; Vilaró, A.; Fraile, L.; Martínez-Martínez, S. Antimicrobial Resistance Genes in Porcine Pasteurella multocida Are Not Associated with Its Antimicrobial Susceptibility Pattern. Antibiotics 2020, 9, 614. https://doi.org/10.3390/antibiotics9090614
Petrocchi-Rilo M, Gutiérrez-Martín C-B, Pérez-Fernández E, Vilaró A, Fraile L, Martínez-Martínez S. Antimicrobial Resistance Genes in Porcine Pasteurella multocida Are Not Associated with Its Antimicrobial Susceptibility Pattern. Antibiotics. 2020; 9(9):614. https://doi.org/10.3390/antibiotics9090614
Chicago/Turabian StylePetrocchi-Rilo, Máximo, César-B. Gutiérrez-Martín, Esther Pérez-Fernández, Anna Vilaró, Lorenzo Fraile, and Sonia Martínez-Martínez. 2020. "Antimicrobial Resistance Genes in Porcine Pasteurella multocida Are Not Associated with Its Antimicrobial Susceptibility Pattern" Antibiotics 9, no. 9: 614. https://doi.org/10.3390/antibiotics9090614
APA StylePetrocchi-Rilo, M., Gutiérrez-Martín, C.-B., Pérez-Fernández, E., Vilaró, A., Fraile, L., & Martínez-Martínez, S. (2020). Antimicrobial Resistance Genes in Porcine Pasteurella multocida Are Not Associated with Its Antimicrobial Susceptibility Pattern. Antibiotics, 9(9), 614. https://doi.org/10.3390/antibiotics9090614








