In Vitro Antibacterial Activity of Microbial Natural Products against Bacterial Pathogens of Veterinary and Zoonotic Relevance
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Natural Products
4.3. Determination of Minimal Inhibitory Concentrations (MICs) of the Natural Products
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patridge, E.; Gareiss, P.; Kinch, M.S.; Hoyer, D. An analysis of FDA-approved drugs: Natural products and their derivatives. Drug Discov. Today 2016, 21, 204–207. [Google Scholar] [CrossRef]
- Cui, C.Y.; Li, X.J.; Chen, C.; Wu, X.T.; He, Q.; Jia, Q.L.; Zhang, X.J.; Lin, Z.Y.; Li, C.; Fang, L.X.; et al. Comprehensive analysis of plasmid-mediated tet(X4)-positive Escherichia coli isolates from clinical settings revealed a high correlation with animals and environments-derived strains. Sci. Total Environ. 2022, 806, 150687. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, R.; Liu, D.J.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.B.; Ji, Q.J.; Wei, R.C.; Liu, Z.H.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- Li, Y.; Sun, X.R.; Xiao, X.; Wang, Z.Q.; Li, R.C. Global distribution and genomic characteristics of tet(X)-positive Escherichia coli among humans, animals, and the environment. Sci. Total Environ. 2023, 887, 164148. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef]
- Tiseo, K.; Huber, L.; Gilbert, M.; Robinson, T.P.; Van Boeckel, T.P. Global Trends in Antimicrobial Use in Food Animals from 2017 to 2030. Antibiotics 2020, 9, 918. [Google Scholar] [CrossRef]
- Allel, K.; Day, L.; Hamilton, A.; Lin, L.S.; Furuya-Kanamori, L.; Moore, C.E.; Van Boeckel, T.; Laxminarayan, R.; Yakob, L. Global antimicrobial-resistance drivers: An ecological country-level study at the human-animal interface. Lancet Planet. Health 2023, 7, E291–E303. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Kalemaki, D. Antimicrobial overuse and misuse in the community in Greece and link to antimicrobial resistance using methicillin-resistant S. aureus as an example. J. Infect. Public Health 2019, 12, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Pokharel, S.; Raut, S.; Adhikari, B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Glob. Health 2019, 4, e002104. [Google Scholar] [CrossRef] [PubMed]
- EU Health. EU Action on Antimicrobial Resistance. Available online: https://health.ec.europa.eu/antimicrobial-resistance/eu-action-antimicrobial-resistance_en (accessed on 7 October 2022).
- Pollock, J.; Low, A.S.; McHugh, R.E.; Muwonge, A.; Stevens, M.P.; Corbishley, A.; Gally, D.L. Alternatives to antibiotics in a One Health context and the role genomics can play in reducing antimicrobial use. Clin. Microbiol. Infect. 2020, 26, 1617–1621. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; International Natural Product Sciences Taskforce; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Bernal, F.A.; Hammann, P.; Kloss, F. Natural products in antibiotic development: Is the success story over? Curr. Opin. Biotechnol. 2022, 78, 102783. [Google Scholar] [CrossRef] [PubMed]
- WHO. Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis. 2020. Available online: https://www.who.int/publications/i/item/9789240021303 (accessed on 7 October 2022).
- Butler, M.S.; Gigante, V.; Sati, H.; Paulin, S.; Al-Sulaiman, L.; Rex, J.H.; Fernandes, P.; Arias, C.A.; Paul, M.; Thwaites, G.E.; et al. Analysis of the Clinical Pipeline of Treatments for Drug-Resistant Bacterial Infections: Despite Progress, More Action Is Needed. Antimicrob. Agents Chemother. 2022, 66, e0199121. [Google Scholar] [CrossRef] [PubMed]
- Hobson, C.; Chan, A.N.; Wright, G.D. The Antibiotic Resistome: A Guide for the Discovery of Natural Products as Antimicrobial Agents. Chem. Rev. 2021, 121, 3464–3494. [Google Scholar] [CrossRef]
- Privalsky, T.M.; Soohoo, A.M.; Wang, J.; Walsh, C.T.; Wright, G.D.; Gordon, E.M.; Gray, N.S.; Khosla, C. Prospects for Antibacterial Discovery and Development. J. Am. Chem. Soc. 2021, 143, 21127–21142. [Google Scholar] [CrossRef] [PubMed]
- Herold, K.; Gollmick, F.A.; Groth, I.; Roth, M.; Menzel, K.D.; Mollmann, U.; Grafe, U.; Hertweck, C. Cervimycin A-D: A polyketide glycoside complex from a cave bacterium can defeat vancomycin resistance. Chemistry 2005, 11, 5523–5530. [Google Scholar] [CrossRef]
- Herold, K.; Xu, Z.; Gollmick, F.A.; Grafe, U.; Hertweck, C. Biosynthesis of cervimycin C, an aromatic polyketide antibiotic bearing an unusual dimethylmalonyl moiety. Org. Biomol. Chem. 2004, 2, 2411–2414. [Google Scholar] [CrossRef]
- EFSA Panel on Animal Health and Welfare; Nielsen, S.S.; Bicout, D.J.; Calistri, P.; Canali, E.; Drewe, J.A.; Garin-Bastuji, B.; Gonzales Rojas, J.L.; Gortazar Schmidt, C.; Herskin, M.; et al. Scientific opinion on the assessment of animal diseases caused by bacteria resistant to antimicrobials: Cattle. EFSA J. 2021, 19, e06955. [Google Scholar] [CrossRef]
- Hampson, D.J.; Burrough, E.R. Swine Dysentery and Brachyspiral Colitis. In Diseases of Swine, 11th ed.; Zimmerman, J.J., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2019; pp. 951–970. [Google Scholar]
- Hampson, D.J.; Lugsomya, K.; La, T.; Phillips, N.D.; Trott, D.J.; Abraham, S. Antimicrobial resistance in Brachyspira-An increasing problem for disease control. Vet. Microbiol. 2019, 229, 59–71. [Google Scholar] [CrossRef]
- Herbst, W.; Schlez, K.; Heuser, J.; Baljer, G. Antimicrobial susceptibility of Brachyspira hyodysenteriae determined by a broth microdilution method. Vet. Rec. 2014, 174, 382. [Google Scholar] [CrossRef]
- La, T.; Phillips, N.D.; Dunlop, H.; Lugsomya, K.; Coiacetto, F.; Hampson, D.J. Testing the efficacy of kitasamycin for use in the control and treatment of swine dysentery in experimentally infected pigs. Aust. Vet. J. 2019, 97, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.G.; Andersen, A.A. Conjunctivitis caused by a swine Chlamydia trachomatis-like organism in gnotobiotic pigs. J. Vet. Diagn. Investig. 1999, 11, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, P.; Kirschvink, N.; Theegarten, D.; Berndt, A. An experimentally induced Chlamydia suis infection in pigs results in severe lung function disorders and pulmonary inflammation. Vet. Res. 2008, 39, 35. [Google Scholar] [CrossRef] [PubMed]
- Guscetti, F.; Schiller, I.; Sydler, T.; Heinen, E.; Pospischil, A. Experimental enteric infection of gnotobiotic piglets with Chlamydia suis strain S45. Vet. Microbiol. 2009, 135, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Bommana, S.; Polkinghorne, A. Mini Review: Antimicrobial Control of Chlamydial Infections in Animals: Current Practices and Issues. Front. Microbiol. 2019, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, P.; Liebler-Tenorio, E.; Sattler, S.; Sachse, K. Recurrence of Chlamydia suis infection in pigs after short-term antimicrobial treatment. Vet. J. 2011, 187, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S. Clostridium (Clostridioides) difficile in animals. J. Vet. Diagn. Investig. 2020, 32, 213–221. [Google Scholar] [CrossRef]
- Dureja, C.; Olaitan, A.O.; Hurdle, J.G. Mechanisms and impact of antimicrobial resistance in Clostridioides difficile. Curr. Opin. Microbiol. 2022, 66, 63–72. [Google Scholar] [CrossRef]
- Steele, J.; Feng, H.; Parry, N.; Tzipori, S. Piglet models of acute or chronic Clostridium difficile illness. J. Infect. Dis. 2010, 201, 428–434. [Google Scholar] [CrossRef]
- Michael, G.B.; Bossé, J.T.; Schwarz, S. Antimicrobial Resistance in Pasteurellaceae of Veterinary Origin. Microbiol. Spectr. 2018, 6, ARBA-0022-2017. [Google Scholar] [CrossRef]
- Schroedl, W.; Fuerll, B.; Reinhold, P.; Krueger, M.; Schuett, C. A novel acute phase marker in cattle: Lipopolysaccharide binding protein (LBP). J. Endotoxin Res. 2001, 7, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Fecteau, M.E. Paratuberculosis in Cattle. Vet. Clin. N. Am. Food Anim. Pract. 2018, 34, 209–222. [Google Scholar] [CrossRef]
- Saxena, S.; Spaink, H.P.; Forn-Cuni, G. Drug Resistance in Nontuberculous Mycobacteria: Mechanisms and Models. Biology 2021, 10, 96. [Google Scholar] [CrossRef]
- Köhler, H.; Soschinka, A.; Meyer, M.; Kather, A.; Reinhold, P.; Liebler-Tenorio, E. Characterization of a caprine model for the subclinical initial phase of Mycobacterium avium subsp. paratuberculosis infection. BMC Vet. Res. 2015, 11, 74. [Google Scholar] [CrossRef]
- Dudek, K.; Nicholas, R.A.J.; Szacawa, E.; Bednarek, D. Mycoplasma bovis Infections-Occurrence, Diagnosis and Control. Pathogens 2020, 9, 640. [Google Scholar] [CrossRef] [PubMed]
- Dudek, K.; Bednarek, D.; Ayling, R.D.; Kycko, A.; Reichert, M. Preliminary study on the effects of enrofloxacin, flunixin meglumine and pegbovigrastim on Mycoplasma bovis pneumonia. BMC Vet. Res. 2019, 15, 371. [Google Scholar] [CrossRef] [PubMed]
- Byrne, W.; Markey, B.; McCormack, R.; Egan, J.; Ball, H.; Sachse, K. Persistence of Mycoplasma bovis infection in the mammary glands of lactating cows inoculated experimentally. Vet. Rec. 2005, 156, 767–771. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, P.; Rabeling, B.; Günther, H.; Schimmel, D. Comparative evaluation of ultrasonography and lung function testing with the clinical signs and pathology of calves inoculated experimentally with Pasteurella multocida. Vet. Rec. 2002, 150, 109–114. [Google Scholar] [CrossRef]
- Coetzee, J.F.; Magstadt, D.R.; Sidhuid, P.K.; Follett, L.; Schuler, A.M.; Krull, A.C.; Cooper, V.L.; Engelken, T.J.; Kleinhenz, M.D.; O’Connor, A.M. Association between antimicrobial drug class for treatment and retreatment of bovine respiratory disease (BRD) and frequency of resistant BRD pathogen isolation from veterinary diagnostic laboratory samples. PLoS ONE 2019, 14, e0219104. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.S.; Pors, S.E.; Jensen, H.E.; Bille-Hansen, V.; Bisgaard, M.; Flachs, E.M.; Nielsen, O.L. An Investigation of the Pathology and Pathogens Associated with Porcine Respiratory Disease Complex in Denmark. J. Comp. Pathol. 2010, 143, 120–131. [Google Scholar] [CrossRef]
- Truswell, A.; Laird, T.J.; Jones, S.; O’Dea, M.; Blinco, J.; Abraham, R.; Morison, D.; Jordan, D.; Hampson, D.J.; Pang, S.; et al. Antimicrobial Resistance of and Genomic Insights into Pasteurella multocida Strains Isolated from Australian Pigs. Microbiol. Spectr. 2023, 11, e03784-22. [Google Scholar] [CrossRef] [PubMed]
- Brockmeier, S.L.; Halbur, P.G.; Thacker, E.L. Porcine Respiratory Disease Complex. In Polymicrobial Diseases, 1st ed.; Brogden, K.A., Guthmiller, J.M., Eds.; ASM Press: Washington, DC, USA; John Wiley & Sons: Hoboken, NJ, USA, 2002; pp. 231–258. [Google Scholar]
- Uchil, R.R.; Kohli, G.S.; Katekhaye, V.M.; Swami, O.C. Strategies to combat antimicrobial resistance. J. Clin. Diagn. Res. 2014, 8, ME01–ME04. [Google Scholar] [CrossRef]
- Chiriac, A.I.; Kloss, F.; Kramer, J.; Vuong, C.; Hertweck, C.; Sahl, H.G. Mode of action of closthioamide: The first member of the polythioamide class of bacterial DNA gyrase inhibitors. J. Antimicrob. Chemother. 2015, 70, 2576–2588. [Google Scholar] [CrossRef]
- Lachowicz-Wolak, A.; Klimowicz-Bodys, M.D.; Ploneczka-Janeczko, K.; Bykowy, M.; Siedlecka, M.; Cinciala, J.; Rypula, K. The Prevalence, Coexistence, and Correlations between Seven Pathogens Detected by a PCR Method from South-Western Poland Dairy Cattle Suffering from Bovine Respiratory Disease. Microorganisms 2022, 10, 1487. [Google Scholar] [CrossRef]
- Cid, D.; Pinto, C.; Dominguez, L.; Vela, A.I.; Fernandez-Garayzabal, J.F. Strength of association between isolation of Pasteurella multocida and consolidation lesions in ovine pneumonic pasteurellosis. Vet. Microbiol. 2020, 248, 108823. [Google Scholar] [CrossRef]
- Wilson, B.A.; Ho, M. Pasteurella multocida: From zoonosis to cellular microbiology. Clin. Microbiol. Rev. 2013, 26, 631–655. [Google Scholar] [CrossRef]
- Snowder, G.D.; Van Vleck, L.D.; Cundiff, L.V.; Bennett, G.L. Bovine respiratory disease in feedlot cattle: Environmental, genetic, and economic factors. J. Anim. Sci. 2006, 84, 1999–2008. [Google Scholar] [CrossRef]
- Dubrovsky, S.A.; Van Eenennaam, A.L.; Karle, B.M.; Rossitto, P.V.; Lehenbauer, T.W.; Aly, S.S. Bovine respiratory disease (BRD) cause-specific and overall mortality in preweaned calves on California dairies: The BRD 10K study. J. Dairy Sci. 2019, 102, 7320–7328. [Google Scholar] [CrossRef]
- Teixeira, A.G.V.; McArt, J.A.A.; Bicalho, R.C. Thoracic ultrasound assessment of lung consolidation at weaning in Holstein dairy heifers: Reproductive performance and survival. J. Dairy Sci. 2017, 100, 2985–2991. [Google Scholar] [CrossRef]
- Bach, A. Associations between several aspects of heifer development and dairy cow survivability to second lactation. J. Dairy Sci. 2011, 94, 1052–1057. [Google Scholar] [CrossRef]
- Dunn, T.R.; Ollivett, T.L.; Renaud, D.L.; Leslie, K.E.; LeBlanc, S.J.; Duffield, T.F.; Kelton, D.F. The effect of lung consolidation, as determined by ultrasonography, on first-lactation milk production in Holstein dairy calves. J. Dairy Sci. 2018, 101, 5404–5410. [Google Scholar] [CrossRef] [PubMed]
- Melchner, A.; van de Berg, S.; Scuda, N.; Feuerstein, A.; Hanczaruk, M.; Schumacher, M.; Straubinger, R.K.; Marosevic, D.; Riehm, J.M. Antimicrobial Resistance in Isolates from Cattle with Bovine Respiratory Disease in Bavaria, Germany. Antibiotics 2021, 10, 1538. [Google Scholar] [CrossRef]
- Schink, A.K.; Hanke, D.; Semmler, T.; Brombach, J.; Bethe, A.; Lübke-Becker, A.; Teske, K.; Müller, K.E.; Schwarz, S. Novel multiresistance-mediating integrative and conjugative elements carrying unusual antimicrobial resistance genes in Mannheimia haemolytica and Pasteurella multocida. J. Antimicrob. Chemother. 2022, 77, 2033–2035. [Google Scholar] [CrossRef]
- Heinisch, L.; Roemer, E.; Jutten, P.; Haas, W.; Werner, W.; Mollmann, U. Semisynthetic derivatives of madurahydroxylactone and their antibacterial activities. J. Antibiot. 1999, 52, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, R.; Satoh, K.; Kikuchi, N.; Kikuchi, H.; Saigusa, D.; Al-Mamun, M.E.; Siddique, M.A.H.; Omura, J.; Satoh, T.; Sunamura, S.; et al. Identification of Celastramycin as a Novel Therapeutic Agent for Pulmonary Arterial Hypertension. Circ. Res. 2019, 125, 309–327. [Google Scholar] [CrossRef]
- Altenburg, J.; de Graaff, C.S.; van der Werf, T.S.; Boersma, W.G. Immunomodulatory effects of macrolide antibiotics-part 1: Biological mechanisms. Respiration 2011, 81, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Bode, C.; Diedrich, B.; Muenster, S.; Hentschel, V.; Weisheit, C.; Rommelsheim, K.; Hoeft, A.; Meyer, R.; Boehm, O.; Knuefermann, P.; et al. Antibiotics regulate the immune response in both presence and absence of lipopolysaccharide through modulation of Toll-like receptors, cytokine production and phagocytosis in vitro. Int. Immunopharmacol. 2014, 18, 27–34. [Google Scholar] [CrossRef]
- Yang, J.H.; Bhargava, P.; McCloskey, D.; Mao, N.; Palsson, B.O.; Collins, J.J. Antibiotic-Induced Changes to the Host Metabolic Environment Inhibit Drug Efficacy and Alter Immune Function. Cell Host Microbe 2017, 22, 757–765.e3. [Google Scholar] [CrossRef]
- Colclough, A.; Corander, J.; Sheppard, S.K.; Bayliss, S.C.; Vos, M. Patterns of cross-resistance and collateral sensitivity between clinical antibiotics and natural antimicrobials. Evol. Appl. 2019, 12, 878–887. [Google Scholar] [CrossRef]
- Michael, A.; Kelman, T.; Pitesky, M. Overview of Quantitative Methodologies to Understand Antimicrobial Resistance via Minimum Inhibitory Concentration. Animals 2020, 10, 1405. [Google Scholar] [CrossRef]
- Hannan, P.C. Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. International Research Programme on Comparative Mycoplasmology. Vet. Res. 2000, 31, 373–395. [Google Scholar] [CrossRef]
- Kikuchi, H.; Sekiya, M.; Katou, Y.; Ueda, K.; Kabeya, T.; Kurata, S.; Oshima, Y. Revised structure and synthesis of celastramycin a, a potent innate immune suppressor. Org. Lett. 2009, 11, 1693–1695. [Google Scholar] [CrossRef]
- Pullen, C.; Schmitz, P.; Meurer, K.; Bamberg, D.D.; Lohmann, S.; De Castro Franca, S.; Groth, I.; Schlegel, B.; Mollmann, U.; Gollmick, F.; et al. New and bioactive compounds from Streptomyces strains residing in the wood of Celastraceae. Planta 2002, 216, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Lincke, T.; Behnken, S.; Ishida, K.; Roth, M.; Hertweck, C. Closthioamide: An unprecedented polythioamide antibiotic from the strictly anaerobic bacterium Clostridium cellulolyticum. Angew. Chem. Int. Ed. 2010, 49, 2011–2013. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, A.; Steffens, U.; Gajdiss, M.; Boschert, A.L.; Droge, J.K.; Szekat, C.; Sass, P.; Malik, I.T.; Bornikoel, J.; Reinke, L.; et al. Cervimycin-Resistant Staphylococcus aureus Strains Display Vancomycin-Intermediate Resistant Phenotypes. Microbiol. Spectr. 2022, 10, e0256722. [Google Scholar] [CrossRef] [PubMed]
- Gräfe, U.; Schade, W.; Roth, M.; Radics, L.; Incze, M.; Ujszaszy, K. Griseochelin, a novel carboxylic acid antibiotic from Streptomyces griseus. J. Antibiot. 1984, 37, 836–846. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Walther, E.; Boldt, S.; Kage, H.; Lauterbach, T.; Martin, K.; Roth, M.; Hertweck, C.; Sauerbrei, A.; Schmidtke, M.; Nett, M. Zincophorin-biosynthesis in Streptomyces griseus and antibiotic properties. GMS Infect. Dis. 2016, 4, Doc08. [Google Scholar] [CrossRef]
- Ino, A.; Hasegawa, Y.; Murabayashi, A. Total synthesis of the antimycoplasma antibiotic micacocidin. Tetrahedron Lett. 1998, 39, 3509–3512. [Google Scholar] [CrossRef]
- Kage, H.; Kreutzer, M.F.; Wackler, B.; Hoffmeister, D.; Nett, M. An iterative type I polyketide synthase initiates the biosynthesis of the antimycoplasma agent micacocidin. Chem. Biol. 2013, 20, 764–771. [Google Scholar] [CrossRef]
- Fleck, W.F.; Strauss, D.G.; Meyer, J.; Porstendorfer, G. Fermentation, isolation, and biological activity of maduramycin: A new antibiotic from Actinomadura rubra. Z. Allg. Mikrobiol. 1978, 18, 389–398. [Google Scholar] [CrossRef]
- Kim, B.M.; Choi, H.Y.; Kim, G.W.; Zheng, C.J.; Kim, Y.H.; Kim, W.G. Madurahydroxylactone, an Inhibitor of Staphylococcus aureus FtsZ from Nonomuraea sp. AN100570. J. Microbiol. Biotechnol. 2017, 27, 1994–1998. [Google Scholar] [CrossRef]
- Feßler, A.T.; Wang, Y.; Burbick, C.R.; Diaz-Campos, D.; Fajt, V.R.; Lawhon, S.D.; Li, X.-Z.; Lubbers, B.V.; Maddock, K.; Miller, R.A.; et al. Antimicrobial susceptibility testing in veterinary medicine: Performance, interpretation of results, best practices and pitfalls. One Health Adv. 2023, 1, 26. [Google Scholar] [CrossRef]
- CLSI Document Vet01-A4; Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard-Fourth Edition. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2013; Volume 33.
- CLSI Document M24-A2; Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes; Approved Standard-Second Edition. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2011.
- Rodwell, A.W.; Whitcomb, R.F. Methods for direct and indirect measurement of mycoplasma growth. In Methods in Mycoplasmology; Razin, S., Tully, J.G., Eds.; Academic Press: NewYork, NY, USA, 1983; pp. 185–196. [Google Scholar]
- CLSI Document M11-A8; Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria; Approved Standard-Eighth Edition. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012; Volume 32.
- Kehrenberg, C.; Catry, B.; Haesebrouck, F.; de Kruif, A.; Schwarz, S. tet(L)-mediated tetracycline resistance in bovine Mannheimia and Pasteurella isolates. J. Antimicrob. Chemother. 2005, 56, 403–406. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Salmon, S.A.; Watts, J.L.; Schwarz, S. Tetracycline resistance genes in isolates of Pasteurella multocida, Mannheimia haemolytica, Mannheimia glucosida and Mannheimia varigena from bovine and swine respiratory disease: Intergeneric spread of the tet(H) plasmid pMHT1. J. Antimicrob. Chemother. 2001, 48, 631–640. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Schwarz, S. Identification of a truncated, but functionally active tet(H) tetracycline resistance gene in Pasteurella aerogenes and Pasteurella multocida. FEMS Microbiol. Lett. 2000, 188, 191–195. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Werckenthin, C.; Schwarz, S. Tn5706, a transposon-like element from Pasteurella multocida mediating tetracycline resistance. Antimicrob. Agents. Chemother. 1998, 42, 2116–2118. [Google Scholar] [CrossRef]

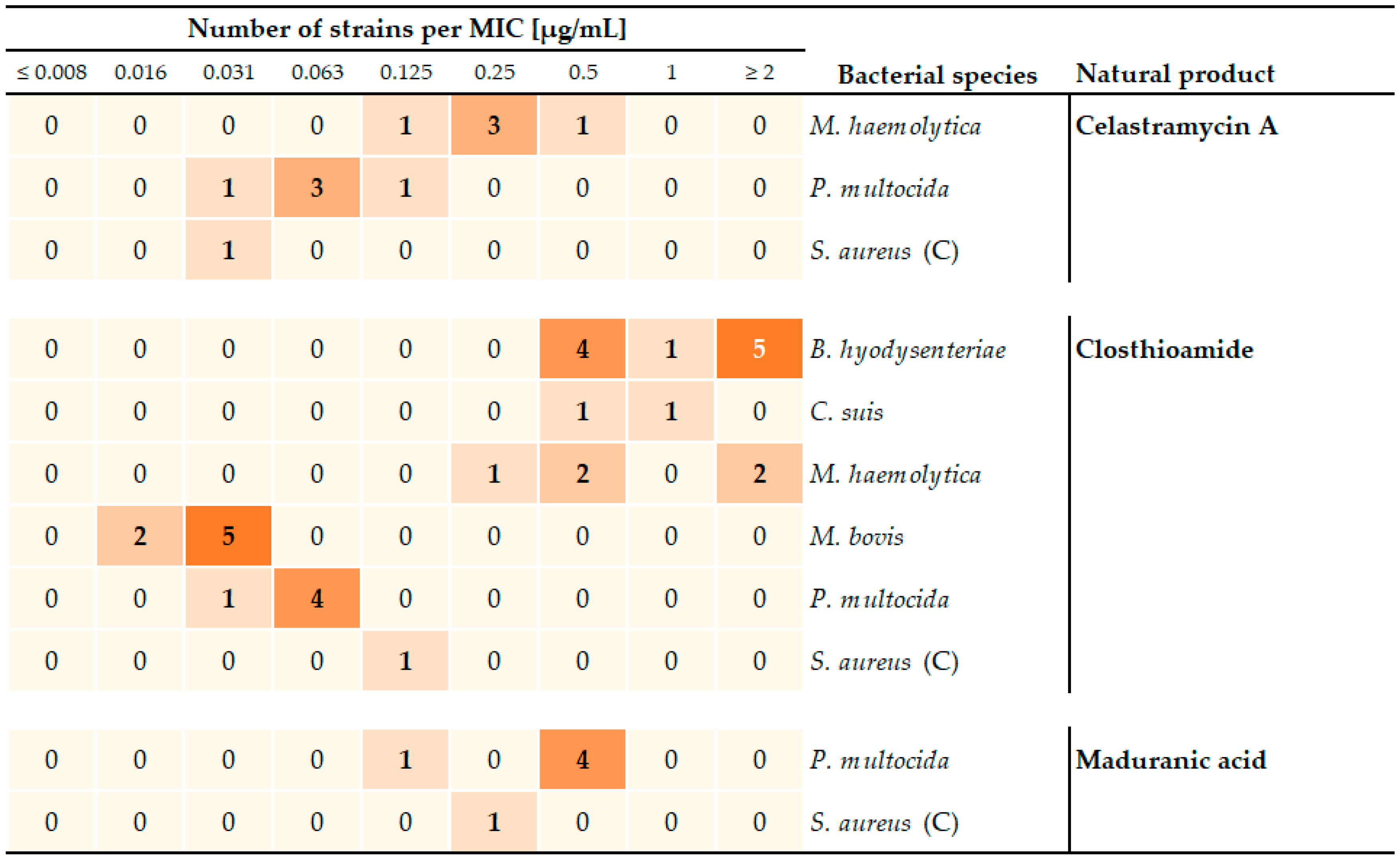
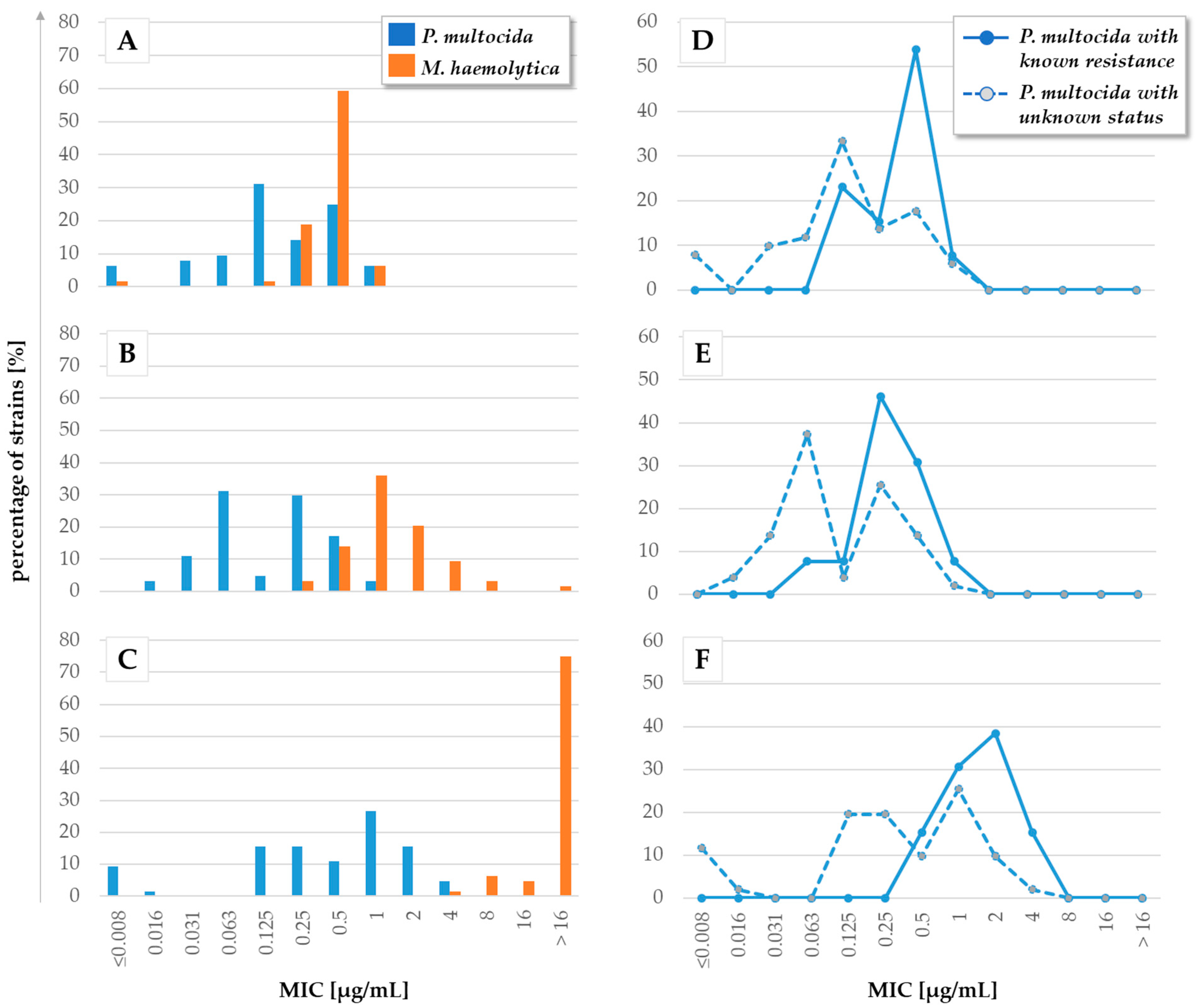
| Bacterial Species | Infectious Lifestyle | Disease in Animals (Most Relevant Hosts) | Reference | Antimicrobial Resistance (AMR) | Animal Model (Host, Organ System) | ||
|---|---|---|---|---|---|---|---|
| Antibiotic Class Affected | Reference | Species | Reference | ||||
| Brachyspira hyodysenteriae | Extracellular | Dysentery (Swine) | Hampson et al., 2019 [25] | Macrolides, Pleuromutilins | Hampson et al., 2019 [26] Herbst et al., 2014 [27] | Swine, Intestinal tract | La et al., 2019 [28] |
| Chlamydia suis | Intracellular | Respiratory disease, Diarrhea, Conjunctivitis (Swine) | Rogers et al., 1999 [29] Reinhold et al., 2008 [30] Guscetti et al., 2009 [31] | Tetracyclines | Bommana et al., 2019 [32] | Swine, Respiratory tract | Reinhold et al., 2011 [33] |
| Clostridioides difficile | Extracellular | Diarrhea (Piglets, Horses) | Weese, 2020 [34] | Fluoroquinolones, Macrolides, Ansamycins | Dureja et al., 2022 [35] | Swine, Intestinal tract | Steele et al., 2010 [36] |
| Mannheimia haemolytica | Extracellular and facultative intracellular | Respiratory disease (Cattle, Goats, Sheep) | Michael et al., 2018 [37] | β-Lactams, Macrolides, Tetracyclines | Michael et al., 2018 [37] | Cattle, Respiratory tract | Schroedl et al., 2001 [38] |
| Mycobacterium avium ssp. | Intracellular | Respiratory disease, Johne’s disease (Cattle) | Fecteau, 2018 [39] | Macrolides, Ansamycins | Saxena et al., 2021 [40] | Goat, Intestinal tract | Köhler et al., 2015 [41] |
| Mycoplasma bovis | Extracellular | Respiratory disease, Mastitis, Arthritis (Cattle) | Dudek et al., 2020 [42] | Macrolides, Phenicols, Tetracyclines | EFSA Panel on Animal Health and Welfare et al., 2021 [24] | Cattle, Respiratory tract | Dudek et al., 2019 [43] |
| Cattle, Mammary gland | Byrne et al., 2005 [44] | ||||||
| Pasteurella multocida | Extracellular and facultative intracellular | Respiratory disease (Cattle, Sheep, Swine), Mastitis (Sheep), Septicaemia (Cattle) | Michael et al., 2018 [37] | β-Lactams, Macrolides, Tetracyclines | Michael et al., 2018 [37] | Cattle, Respiratory tract | Reinhold et al., 2002 [45] |
| Bacterial Species | Family | Phylum | Cellular Properties | Test System | Reference | No. of Type (T) or Control (C) Strains 1 | No. of Field Isolates |
|---|---|---|---|---|---|---|---|
| Brachyspira hyodysenteriae | Brachyspiraceae | Spirochaetota | Gram negative | BMD | Herbst et al., 2014 [27] | - | 10 |
| Chlamydia suis | Chlamydiaceae | Chlamydiota | Gram negative | BMD | In-house method (cell culture with BGM cells, see Section 4.3) | 1 (T) | 1 |
| Clostridioides difficile | Peptostreptococcaceae | Bacillota | Gram positive, spore forming | AD | CLSI M11-A08 | - | 4 |
| Escherichia coli | Enterobacteriaceae | Pseudomonadota | Gram negative | BMD | CLSI Vet01 A4 | 1 (C) | - |
| Mannheimia haemolytica | Pasteurellaceae | Pseudomonadota | Gram negative | BMD | CLSI Vet01 A4 | - | 5 |
| Mycobacterium avium ssp. | Mycobacteriaceae | Actinomycetota | Acid-fast bacteria | BMD | CLSI M24-A02 | 1 (T), 1 (C) | 4 |
| Mycoplasma bovis | Mycoplasmataceae | Mycoplasmatota | Cell-wall free | BMD | Hannan, 2000 [69] | 1 (T) | 6 |
| Pasteurella multocida | Pasteurellaceae | Pseudomonadota | Gram negative | BMD | CLSI Vet01 A4 | - | 5 |
| Staphylococcus aureus | Staphylococcaceae | Bacillota | Gram positive | BMD | CLSI Vet01 A4 | 1 (C) | - |
| Natural Product | Origin | Structure | MW [g/mol] | Production | Mode of Action | Toxicity CC50 [mg/L] LD50 [mg/kg] | Reference |
|---|---|---|---|---|---|---|---|
| Celastramycin A | Streptomyces MaB-QuH-8 | 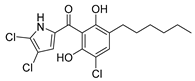 | 390.69 | Synthetic | Not known | CC50 16.9 a | Kikuchi et al., 2009 [70]; Pullen et al., 2002 [71] |
| Closthioamide | Ruminiclostridium cellulolyticum |  | 695.04 | Synthetic | Inhibition of topoisomerase II | CC50 5.2 ± 0.5 a | Chiriac et al., 2015 [51]; Lincke et al., 2010 [72] |
| Cervimycin C | Streptomyces tendae HKI-179 | 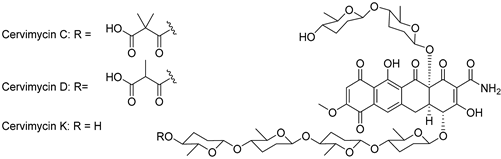 | 1228.29 | Microbial | Not known | CC50 >50 a | Herold et al., 2004 [23]; Herold et al., 2005 [22] |
| Cervimycin D | Streptomyces tendae HKI-179 | 1214.26 | Microbial | Not known | CC50 >50 a | Herold et al., 2004 [23]; Herold et al., 2005 [22] | |
| Cervimycin K1 | Streptomyces tendae HKI-179 | 1114.19 | Microbial | Not known | CC50 11.0 a | Dietrich et al., 2022 [73] | |
| Griseochelin | Streptomyces griseus HKI 0741 | 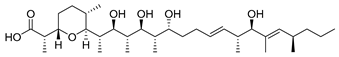 | 568.83 | Microbial | Not known, but ionophore (Zn2+) activity | CC50 0.6 ± 0.3 b; 13.7 ± 15.4 c | Gräfe et al., 1984 [74]; Walther et al., 2016 [75] |
| Micacocidin | Ralstonia solanacearum GMI 1000 and Pseudomonas sp. No. 57-250 | 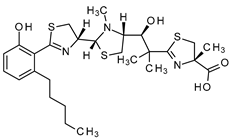 | 565.81 | Microbial | Not known | CC50 >50 a | Ino et al., 1998 [76]; Kage et al., 2013 [77] |
| Maduranic acid/madurahdroxylacton | Nonomuria rubra DSM12263 | 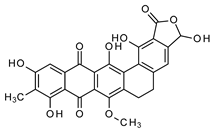 | 490.42 | Microbial | Suggested FtsZ inhibitor | LD50 >250 d | Fleck et al., 1978 [78]; Kim et al., 2017 [79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barth, S.A.; Preussger, D.; Pietschmann, J.; Feßler, A.T.; Heller, M.; Herbst, W.; Schnee, C.; Schwarz, S.; Kloss, F.; Berens, C.; et al. In Vitro Antibacterial Activity of Microbial Natural Products against Bacterial Pathogens of Veterinary and Zoonotic Relevance. Antibiotics 2024, 13, 135. https://doi.org/10.3390/antibiotics13020135
Barth SA, Preussger D, Pietschmann J, Feßler AT, Heller M, Herbst W, Schnee C, Schwarz S, Kloss F, Berens C, et al. In Vitro Antibacterial Activity of Microbial Natural Products against Bacterial Pathogens of Veterinary and Zoonotic Relevance. Antibiotics. 2024; 13(2):135. https://doi.org/10.3390/antibiotics13020135
Chicago/Turabian StyleBarth, Stefanie A., Daniel Preussger, Jana Pietschmann, Andrea T. Feßler, Martin Heller, Werner Herbst, Christiane Schnee, Stefan Schwarz, Florian Kloss, Christian Berens, and et al. 2024. "In Vitro Antibacterial Activity of Microbial Natural Products against Bacterial Pathogens of Veterinary and Zoonotic Relevance" Antibiotics 13, no. 2: 135. https://doi.org/10.3390/antibiotics13020135
APA StyleBarth, S. A., Preussger, D., Pietschmann, J., Feßler, A. T., Heller, M., Herbst, W., Schnee, C., Schwarz, S., Kloss, F., Berens, C., & Menge, C. (2024). In Vitro Antibacterial Activity of Microbial Natural Products against Bacterial Pathogens of Veterinary and Zoonotic Relevance. Antibiotics, 13(2), 135. https://doi.org/10.3390/antibiotics13020135







