Abstract
Staphylococcus aureus is one of the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens among which multidrug resistance has emerged. Resistance to methicillin has resulted in clinicians using the antibiotic of last resort, vancomycin, to treat infections caused by methicillin-resistant S. aureus (MRSA). However, excessive use and misuse of vancomycin are major causes of resistance among S. aureus strains. South Asia encompasses ~25% of the world’s population, and countries in South Asia are often characterized as low- and middle-income with poor healthcare infrastructure that may contribute to the emergence of antibiotic resistance. Here, we briefly highlight the mechanism of vancomycin resistance, its emergence in S. aureus, and the molecular epidemiology of non-susceptible S. aureus to vancomycin in the South Asian region.
1. Introduction
Staphylococcus aureus exists as a commensal of normal human microbiota, colonizing approximately 20–40% of healthy individuals [1]. However, S. aureus is also a widespread human pathogen that causes a range of community-acquired and nosocomial infections, ranging from minor skin and soft tissue infections to more serious and deadly conditions such as pneumonia, bacterial endocarditis, osteomyelitis, toxic mediated syndromes, and bacteremia [2,3,4]. Due to its metabolic flexibility, S. aureus adapts well to different environmental conditions and thus remains a major public health concern. With the advent of antibiotics such as penicillin and methicillin in the 20th century, S. aureus infections were effectively treated. However, resistance to both of these drugs emerged rapidly, resulting in difficulty in the treatment of infections [5]. During the last century, various reports have indicated a significant increase in healthcare-associated costs due to the emergence of antibiotic resistance in S. aureus strains [6]. Both hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) and community-associated MRSA (CA-MRSA) are major health concerns, and are on the rise globally [7]. The majority of HA-MRSA strains from various countries share the same genotype, as shown by molecular epidemiology research [8,9]. This suggests that a few healthcare-associated clones from Asia have spread internationally. However, a few relatively high-income countries in Asia, such as Taiwan, Japan, Korea, Hong Kong, and Singapore, have produced the majority of studies [9,10,11].
South Asia is the southern region of the continent Asia and consists of eight countries, including Pakistan, India, Bangladesh, Sri Lanka, Nepal, Maldives, Afghanistan, and Bhutan. South Asia covers about 2 million square miles, representing 3.5% of the world’s land area and 11.71% of the continent of Asia [12]. This region of South Asia has the highest population density on the entire planet, contributing to 24.89% of the world population, i.e., about 1.9 billion individuals [13]. The healthcare system of South Asia is amongst the world’s worst healthcare systems due to poor economic inequalities, and these inequalities compound the widespread gaps in access to healthcare systems [14]. Limited access to healthcare facilities and indiscriminate misuse of antibiotics are common practices in the region, contributing to the high burden of antimicrobial resistance to various antibiotics.
Vancomycin became the drug of choice for the effective treatment of MRSA in the late 1980s and is used to treat MRSA infections worldwide. However, S. aureus acquired resistance against vancomycin and became a great concern to global public health [15]. Several studies from South Asian countries, including India, Pakistan, Bangladesh, Sri Lanka, and Nepal, reported either reduced susceptibility or complete resistance to vancomycin, while no studies have been published from Afghanistan, Maldives, or Bhutan on the emergence of non-susceptible S. aureus strains against vancomycin [16,17,18,19].
Our understanding of the epidemiology of staphylococcal infections has been significantly constrained by the information being either incomplete or completely unavailable for the resource-constrained countries of Southeast Asia and South Asia. A recent systematic review reported a prevalence of 1.2% vancomycin-resistant S. aureus (VRSA) in Asia, 1.1% in Europe, 3.6% in America, and 2.5% in Africa [20]. Therefore, this review highlights the epidemiology of vancomycin-intermediate resistant S. aureus (VISA), VRSA, and heterogeneous vancomycin-intermediate resistant S. aureus (hVISA) in the South Asian region.
2. Antibiotic Resistance in S. aureus
Indiscriminate use of antibiotics played a significant role in the emergence of resistant bacteria, the increase in multi-drug resistant strains, and the spread of resistance among bacterial species [21]. Resistance to antibiotics in S. aureus is attributed to chromosomal mutations and extra-chromosomal elements such as plasmids. Mobile genetic elements such as staphylococcal cassette chromosomes, plasmids, bacteriophages, transposons, and pathogenicity islands are involved in horizontal gene transfer, which plays a significant role in the development and dissemination of antibiotic resistance in bacteria [22].
Several waves represent the emergence of antibiotic resistance in S. aureus including the emergence of penicillin-resistant S. aureus (PSRA) followed by MRSA, CA-MRSA, HA-MRSA, VISA, and VRSA, respectively [23]. PSRA was reported after a few years of treatment in the early 1940s, and its infections in humans spread across both community and hospital settings in the 1950s to early 1960s [5,24]. The blaZ gene, which encodes a β-lactamase, primarily conferred resistance in S. aureus by hydrolyzing the β-lactam ring of the antibiotic, thus inactivating penicillin [25]. After the emergence of PRSA, methicillin was introduced to treat the infections. Despite its efficacy in treating the PRSA infection, methicillin resistance soon emerged, and MRSA strains were reported consistently within two years of its use in clinical practice [26]. Currently, MRSA has spread worldwide, accounting for a large number of CA-MRSA and HA-MRSA infections and causing increased morbidity and mortality. Several surveys in different countries have reported the number of deaths caused by MRSA [27,28,29]. In America alone, ~19,000 hospitalized patients have died per year due to MRSA, while over 17,000 patients infected with S. aureus died every year due to antimicrobial resistance (AMR) against β-lactam antibiotics [30,31]. The emergence of CA-MRSA outside of healthcare settings in healthy individuals has led to complications in managing infections [32]. All MRSA strains have staphylococcal cassette chromosome (SCC), which carries the mecA gene. This gene encodes low-affinity penicillin-binding protein 2a, imparting resistance to β-lactam antibiotics [33]. Staphylococcal cassette chromosome mec (SCCmec) is the mobile genetic element of S. aureus, playing a significant role in staphylococci pathogenesis. SCCmec and mecA genes are incorporated into the chromosome of S. aureus, causing the evolution of resistance and resulting in the emergence of MRSA [34]. The two genes, namely, the cassette chromosome recombinase (ccr) gene and the mec gene complex, combine to form the SCCmec gene [35]. In addition, a new type of mec gene, mecC, with 63% residue identity to mecA, was discovered and found to be predominantly associated with a single lineage of MRSA in European countries [36]. About 615 MRSA strains carry this SCCmec gene and have been isolated in 11 different countries of Asia [37]. Eleven different SCCmec types have been reported. Of those, SCCmec types IV and V are typically carried by CA-MRSA, while SCCmec types I, II, and III typically belong to HA-MRSA [33,38,39]. Moreover, CA-MRSA strains carry genes such as lukS-PV and lukF-PV, which encode a bi-component pore-forming cytolytic toxin, Panton–Valentine leukocidin (PVL), that targets neutrophils, monocytes, and macrophages [38,40].
Since the 1980s, the preferred medication for the treatment of severe infections caused by MRSA has been vancomycin [41]. In 1997, the reduced susceptibility against vancomycin (VISA, minimum inhibitory concentration (MIC) = 8 µg/mL) was reported in Japan [42]. Shortly after the VISA report in Japan, two cases were reported in America, while the first VRSA case emerged in 2002 and was reported in a diabetic patient [43,44]. Since then, several reduced susceptible and resistant strains of S. aureus have been reported worldwide. Two methods for molecular characterization of S. aureus are widely used, including multilocus sequence typing (MLST) and spa typing. MLST is a molecular typing technique used to identify short internal fragments of nucleotide sequences (usually 400–500 bp) in multiple housekeeping genes, while spa typing is a rapid and affordable technique with highly discriminatory abilities to determine the variability in the polymorphic X region of staphylococcal protein A gene (spa gene), found in all strains of S. aureus [45,46]. Non-susceptible vancomycin S. aureus isolates were tested by Shekarabi et al., who found three different sequence types (ST) (ST5, ST8, and ST239) to be associated with VRSA [47]. Spa typing revealed that spa types t030 and t70 belonged to VISA, while t008, t37, and t002 were present in VRSA strains. Shekarabi et al. also showed the MIC values for specific types of VRSA strains and found that ST239-SCCmec III/t030 and ST22-SCCmec IV/t790 strains had MIC values ≥ 8 μg/mL, ST8-SCCmecIV/t008 strains had MIC values of 64 μg/mL, and ST239-SCCmec III/t037 and ST5-SCCmec II/t002 strains had MIC values of 512 μg/mL [47]. ST239 is endemic in the Asian region and is divided into three different clades: the European, Asian, and South American clades. Moreover, multi-resistant ST239 is more prevalent in Asia, the USA, and European countries [48]. Several studies have reported ST239 MRSA strains that are non-susceptible to vancomycin (VISA and VRSA) in this region [49,50].
2.1. Vancomycin’s Mechanism of Action against S. aureus and the Emergence of Resistance
Vancomycin exerts bactericidal activity by interrupting the cell wall synthesis of the bacteria, but many bacteria have overcome its effect and become resistant due to multiple mutations in chromosomal genes that affect cell wall synthesis [51]. The hydrophilic molecules of vancomycin interact with the terminal D-alanyl-D-alanine (d-Ala-d-Ala) moieties of the precursor lipid II through hydrogen bonding. Vancomycin binding causes a conformational change that prevents the inclusion of the precursor in the developing peptidoglycan chain and the subsequent trans-peptidation, resulting in cell wall lysis [15].
However, the increased usage of vancomycin has resulted in the establishment of two other forms of S. aureus that show reduced susceptibility or resistance to glycopeptides. The first one, known as VISA, has a thicker cell wall, along with insufficient cross-linking in the cell wall, which causes a buildup of acyl-d-alanyl-d-alanine (X-d-Ala-d-Ala) targets that entrap glycopeptides at the periphery. The second form is VRSA, which is due to transposon Tn1546-carrying the vanA operon acquired from Enterococcus species, resulting in the establishment of high-level resistance (Figure 1) [52,53,54]. The risk of transmission to S. aureus and other susceptible medically significant microbes due to the vancomycin-resistant enterococci is high [15].
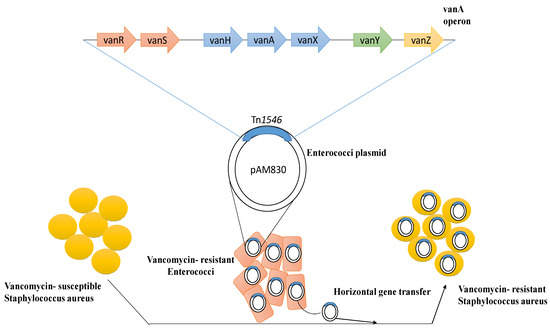
Figure 1.
Mechanism of emergence of vancomycin resistance among susceptible Staphylococcus aureus through horizontal gene transfer.
The first hVISA strain to be identified was Mu3, which was found in a patient with pneumonia in Japan in 1996 who had not responded to vancomycin [28]. The first instance of VISA (Mu50) was documented in a patient who had undergone heart surgery for a surgical wound infection in 1997 [42]. Vancomycin-intermediate resistant strains exhibit certain characteristics, such as reduced autolysis, weakened virulence, and thickened cell walls. The potential resistance is due to an increase in cell wall turnover, which generates non-cross-linked d-anyl-d-alanine side chains, resulting in less exposure of vancomycin to intracellular target molecules [55]. The reported VISA strains evolved from HA-MRSA are mainly from clonal complex 5 or 8 (CC5/CC8), in particular, from ST239 and ST5. However, the MSSA-derived CA-MRSA clone USA300-derived VISA was also reported [56,57]. With the high incidence of MRSA and increased reliability of the use of glycopeptides in the South Asian region, the emergence of non-susceptible strains of S. aureus was not surprising. However, non-susceptible strains have not disseminated widely in south Asia, but have been detected sporadically in patients who have previously used vancomycin or were on long-term vancomycin therapy for persistent S. aureus infections [58].
2.2. Interpretation Criteria for Vancomycin-Susceptible S. aureus
Vancomycin susceptibility testing can be conducted using different methods, including the E test, broth micro-dilution, agar dilution method, etc. The Clinical Laboratory Standards Institute (CLSI) decreased the clinical susceptible MIC breakpoint from 4 μg/mL to 2 μg/mL in 2006 [59]. VRSA has a MIC of >16 µg/mL; VISA a MIC range of 4–8 µg/mL; and hVISA a MIC range of ≤2 µg/mL, with a subpopulation of MIC = 4–8 µg/mL, which could all be classified as S. aureus with reduced susceptibility towards vancomycin [60]. The MIC to vancomycin of clinical S. aureus isolates has continuously increased with increasing vancomycin prescription, which may lead to more vancomycin treatment failures [59,61].
Due to the presence of hVISA and the inadequacy of standard clinical antimicrobial susceptibility testing to precisely identify VISA, there were still troubling issues even after these interpretation breakpoints were lowered to assure concordance with clinical treatment feedback. Since 2009, it has been impossible to evaluate the vancomycin susceptibility of S. aureus using the disc diffusion method, since it is unable to distinguish between vancomycin-susceptible S. aureus (VSSA), VISA, and VRSA strains [62]. Clinical laboratories in Taiwan use automated platforms such as the BD PhoenixTM automated testing system, the VITEK® 2 automated instrument, and the MicroScan system because the time-consuming and labor-intensive standard dilution methods (micro- or macro-dilution and agar dilution) are not appropriate for routine mass clinical use. One of the techniques suggested by CLSI to identify vancomycin resistance in S. aureus is vancomycin screening agar, which includes vancomycin at 6 µg/mL and is being widely used in the region to determine vancomycin-resistant enterococci (VRE). However, there is the possibility that this screening may overlook VISA strains with vancomycin MICs of 4–6 μg/mL [63].
2.3. Selection Criteria
A comprehensive literature search was performed on the PubMed, Google Scholar, and Web of Science databases for original research publications from 1997 to March 2023. The terms used in the search strategy included vancomycin-resistant Staphylococcus aureus, vancomycin intermediate Staphylococcus aureus, heterogeneous vancomycin Staphylococcus aureus, VRSA, VISA, and hVISA in each of the South Asian countries. The bibliographies of relevant articles were also searched to identify other related studies. The number of publications found to be relevant to the epidemiology of non-susceptible vancomycin S. aureus (VISA, VRSA, hVISA) in South Asia was <40, showing the lack of reporting in the region, especially in India and Pakistan, where the AMR burden is alarmingly high.
3. Vancomycin Non-Susceptible S. aureus in South Asia
3.1. Pakistan
Since the emergence of VRSA in the USA, many other countries have also reported vancomycin resistance in clinical isolates of MRSA. Azhar et al. reported 7.3% VRSA in the clinical isolates, and the vanA operon was detected in 74% of those VRSA isolates [64]. A study reported the persistence of VRSA in the blood of postoperative cardiac surgery patients, and isolates were found to carry vanA on a plasmid and icaA on the chromosome [65]. Under conditions of sub-inhibitory vancomycin concentrations, the isolated strain also formed some extracellular polymeric matrix substance and showed resistance to triton-X100-caused autolysis [65]. A study conducted by Taj et al. in Pakistan identified one strain as VRSA (MIC = 32 µg/mL) and four strains as VISA (two strains with MIC = 8 µg/mL and two strains with MIC = 16 µg/mL) [66]. A retrospective study conducted in Pakistan reported a prevalence of 8.33% VRSA (n = 565) and 11.68% VISA (n = 792), which was the highest reported frequency found so far in Pakistan [67]. A prevalence of 13% VISA in the clinical isolates of MRSA strains was reported in a study conducted in Karachi, Pakistan [68]. In 2019, Saeed et al. [69] selected 30 isolates that were phenotypically resistant to methicillin and vancomycin to find resistance genes. Out of 30 MRSA isolates, 10 (33.33%) were successfully amplified for the mecA partial region of the antibiotic resistance gene, while vanA was amplified from 14 (46.6%) isolates. Similarly, another study was conducted in which a total of 86 VRSA isolates were subjected to the amplification of vancomycin-resistance encoding genes. It was found that 40 isolates showed the presence of vanA, whereas 51 isolates showed the presence of vanB. Most of the isolates that were vanB-positive also contained vanA [16]. The resistant strains of S. aureus against methicillin and vancomycin were reported in a study conducted in a tertiary care hospital of Lahore, Pakistan. The prevalences of MRSA (25.5%), VISA (11%), and VRSA (2.5%) were reported in patients with skin injuries [70]. A similar study evaluated the antimicrobial resistance patterns of S. aureus isolated from different clinical samples and found a presence of 12% VRSA [71]. Table 1 presents the epidemiology S. aureus strains that exhibit non-susceptibility to vancomycin in South Asian countries, including Pakistan.
3.2. India
A routine nasal carriage survey in the intensive care unit of a hospital in northern India identified two VISA isolates (MIC = 8 mg/L) carrying the vanA gene [72]. The study was considered as the first study from India to report the presence of the vanA gene among VISA isolates. The asymptomatic colonization of vanA-positive VRSA raised apprehension about the spread of resistant strains among hospital staff and the local community. The study by Goud et al. in India reported a 1.4% VRSA prevalence confirmed by the presence of vanA in the swab samples collected from the anterior nares, forearms, and hands of 1000 healthy individuals [73]. Vancomycin resistance in MRSA isolates from the ICUs of tertiary care hospitals in Hyderabad, India was tested, and the presence of 16 VISA (MIC = 4–8 mg/L) and 7 VRSA (MIC = 16–64 mg/L) strains among the clinical isolates were reported; 6 of these VRSA strains were vanA-positive. A recent study from India reported the presence of 7 (6.08%) VRSA strains and 53 (46.08%) VISA strains among clinical isolates [74]. A reduced susceptibility of 11.6% (n = 33) of the isolates to vancomycin was also observed in Eastern India. Among the 33 strains with reduced susceptibility, 18 were VISA and 13 were hVISA [75]. In 2004, the Asian Network for Surveillance of Resistant Pathogens (ANSORP) reported the presence of hVISA among 1357 MRSA isolates in Asian countries, which showed a prevalence of 2.1% in Thailand, Singapore (2.3%), Vietnam (2.4%), the Philippines (3.6%), South Korea (6.1%), India (6.3%) and Japan (8.2%) [76]. The spa type t601, which belongs to CC5, showed a frequency of 33.3% for hVISA, and t002 of CC5 accounted for a 6.9% prevalence among hVISA strains in a study conducted by ANSORP (Asian Network for Surveillance of Resistant Pathogens) in 2004–2006 [77]. A study in Puducherry, India determined the prevalence of 12.4% hVISA among clinical isolates. Moreover, the study also determined the distribution of SCCmec types among hVISA isolates and found SCCmec type III among 8%, SCCmec type IV in 17.7%, and SCCmec type V in 50% of the strains [78]. A similar study carried out in India found a prevalence of 7% (n = 33) hVISA/VISA strains among the clinical isolates of MRSA and MSSA. Out of these 33 hVISA/VISA strains, 42.4% (n = 14) belonged to agr I, 24.2% (n = 8) belonged to agr II, 30% (n = 10) belonged to agr III, and 3% (n = 1) belonged to agr IV [79]. A study from a tertiary care hospital in Coastal Karnataka, India, reported a prevalence of 6.4% hVISA among MRSA isolates [80].
3.3. Bangladesh
The study conducted in Bangladesh by Islam et al. reported the presence of 2 (13.3%) VRSA strains among the samples collected from wounds of hospitalized patients, and found the presence of vanB in the isolated strains [81]. Bangladesh also reported the emergence of VRSA among MRSA clinical isolates and detected the presence of vancomycin resistance in 3 (7.89%) of the MRSA isolates. A similar study reported a prevalence of 28% (n = 8) VRSA, with 6 strains having MIC values of 32 μg/mL and 2 strains having MIC values of 64 μg/mL, and 55% (n = 16) were VISA [19]. The study conducted in 2010–2011 in Bangladesh reported a prevalence of 93.44% VISA among clinical isolates, indicating the growing antimicrobial-resistant strains in the region [82].
S. aureus strains isolated from patients in the North Central region of Bangladesh were analyzed for their molecular features and genetic backgrounds. In a study of 430 S. aureus clinical isolates, 31% were MRSA, with 73% having SCCmec type IV and 14% having type V, and most belonging to coagulase (coa) genotypes IIa, IIIa, IVb, and XIa. The most common coa type in MSSA was IIIa, followed by Va, IIa, and VIa. The study also reported that CC88 (ST88, ST2884) and ST772 were the potential prevailing lineages of PVL-positive MRSA/MSSA. Additionally, the newly discovered CC80 clade was among the primary PVL-negative MRSA lineages distributed in an endemic manner throughout Bangladesh [83]. A research investigation was conducted to ascertain the prevalence of MRSA and VRSA nasal colonization among healthcare providers. The findings of the study revealed that there was a complete absence of resistance to vancomycin, whereas the prevalence of MRSA was 7.2% [84]. A cross-sectional observational study was conducted at the Department of Microbiology, Mymensingh Medical College, Mymensingh, Bangladesh, to identify the presence of MRSA and its susceptibility to various antibiotics. The research revealed a prevalence rate of 26.4% among clinical isolates, and the isolates exhibited a sensitivity rate of 100% towards vancomycin and gentamicin [85]. In a related investigation carried out by Pervaz et al., it was discovered that the frequency of MRSA was 58.4% (n = 38) among clinical samples collected from Dhaka City in Bangladesh. This included 22 cases of CA-MRSA and 16 cases of HA-MRSA. However, all MRSA isolates were observed to be receptive to vancomycin [86].
3.4. Sri Lanka
According to a 2004–2006 ANSOPR study, the prevalence of nosocomial S. aureus isolates in Sri Lanka was 86.5% [87]. According to a study conducted in 2017 by Jayaweera et al., MRSA had become a severe hazard to Sri Lanka’s public health, and its isolation rate was high (40.2%). All of the MRSA isolates had MICs of less than 2 μg/mL [88]. Out of 100 isolates, 21 (21%) were CA-MRSA and 79 (79%) were HA-MRSA, according to a report from the National Hospital of Sri Lanka [89]. Among the HA S. aureus infections, the rates of MRSA were quite high in Sri Lanka (86.5%), while among CA S. aureus infections, the proportion of MRSA was 38.8% in Sri Lanka [87]. In 2019, an observational study revealed the presence of PVL-positive ST5-MRSA-IVc belonging to CC5 isolates from Sri Lanka, which is one of the major clones spread in different countries such as Australia, the United Kingdom, and Argentina, and might be a threat to neighboring countries [90]. The first case of reduced susceptibility against vancomycin emerged in 2002 in a 60-year-old man with a surgical site infection [91]. None of the MRSA isolates in any other study reported the emergence of resistance against vancomycin.
3.5. Nepal
A cross-sectional study undertaken for the antimicrobial susceptibility profiling of the isolates from chronic dacryocystitis in 2010 in Nepal showed the emergence of vancomycin resistance in 81.48% (n = 22) of the isolated strains of S. aureus [92]. Other similar studies were conducted in Nepal, but no other studies reported the emergence of complete valid resistance to vancomycin in S. aureus strains. An increase in the MIC values of vancomycin among MRSA strains also emerged in Nepal, and 4 (9.52%) strains of MRSA were reported as VISA in a study conducted in Nepal [93].
The prevalence of MRSA in Nepal has shown an upward trajectory, ranging from 29.1% to 68%, as reported in previous studies [94,95,96]. However, there have been few research studies conducted on the identification of VISA, VRSA, and their associated genetic markers. The prevalence of VRSA among MRSA isolates was reported to be 11.11% (n = 5) in a hospital-based cross-sectional study conducted at the Annapurna Neurological Institute and Allied Sciences, Kathmandu, Nepal. The VRSA isolates underwent screening to determine the presence of vanA and vanB. The results indicated that vanA was present in two of the isolates, whereas vanB was not detected in any of the isolates [97]. A research investigation was carried out to ascertain the MICs of vancomycin in 47 strains of MRSA that were obtained from clinical specimens gathered from a medical facility located in Nepal. The results of the study revealed that the MIC values ranged from 0.125 μg/mL to 1 μg/mL, and no VISA or VRSA was detected [96].
3.6. Afghanistan
A study was conducted to assess the antimicrobial susceptibility patterns of S. aureus strains obtained from patients at two major healthcare centers situated in Kabul, Afghanistan. The study revealed that a significant proportion of the isolates (56.2%) were MRSA, while vancomycin exhibited no resistance against the pathogen [98].
The multidrug-resistant S. aureus including MRSA has also been reported in a few studies from war-torn Afghanistan [99,100]. The most commonly detected clones were the Bengal Bay clone (ST772-MRSA-V PVL+), the Southwest Pacific clone (CC30-MRSA-IV PVL+), and the CC22-MRSA-IV TSST-1+ clone [99]. No specific study from Afghanistan has reported the emergence of vancomycin non-susceptible S. aureus.

Table 1.
Characteristics and molecular epidemiology of VRSA, VISA, and hVISA isolates in South Asian countries.
Table 1.
Characteristics and molecular epidemiology of VRSA, VISA, and hVISA isolates in South Asian countries.
| Country | Study Period | Isolation Site | No. of Positive VRSA/Total | No. of Positive VISA/Total | No. of Positive hVISA/Total | Genetic Marker (vanA/vanB/icaA) | Molecular Typing | Reference |
|---|---|---|---|---|---|---|---|---|
| Pakistan | 2020 | Pus, skin wound, CSF | 113/200 | --- | --- | --- | --- | [69] |
| Pakistan | February 2017–March 2018 | Pus swabs from diabetic foot ulcers, wounds, breast abscesses | 565/6780 | 792/6780 | --- | --- | --- | [67] |
| Pakistan | January 2010–December 2010 | Pus, urine, blood, vaginal swab, and other secretions | 1/174 | 4/174 | --- | --- | --- | [66] |
| Pakistan | 2016 | Pus | 22/110 | 5/110 | --- | --- | --- | [70] |
| Pakistan | 2017–2018 | Blood, body fluid, pus, skin wound | 14/100 | --- | --- | vanA | --- | [101] |
| Pakistan | 2016 | Pus from ear, skin wound | 11/150 | --- | --- | vanA | --- | [64] |
| Pakistan | 2011 | Blood | 1/1 | --- | --- | vanA, icaA | --- | [65] |
| Pakistan | 2015 | Wound, pus swab | 5/51 | --- | --- | --- | --- | [102] |
| Nepal | Blood, urine, sputum, catheter swab, pus, and body fluids | 2/57 | 31/57 | --- | --- | --- | [103] | |
| Nepal | 2010 | Lacrimal swabs from chronic dacryocystitis | 22/27 | --- | --- | --- | --- | [92] |
| Nepal | November 2011–May 2012 | Urine, blood, and body fluids | --- | 4/45 | --- | --- | --- | [93] |
| India | January 1997–March 2000 | Clinical isolates | --- | --- | 5/80 | --- | --- | [76] |
| India | August 2002–July 2005 | Pus, urine, wound swabs, catheters, blood, sputum, and CSF | 2/783 | 6/783 | --- | 0 | --- | [104] |
| India | 2016 | Dental caries | 27/150 | --- | --- | 13 (vanA) 2(vanB) | --- | [105] |
| India | July 2010–September 2012 | Pus, urine, wound swabs, catheters, blood, and sputum | 7/115 | 53/115 | --- | --- | --- | [74] |
| India | March 2008–October 2008 | Blood, urine and throat swabs, wounds, and ear swabs | 7/358 | 16/358 | --- | 6 (vanA) | --- | [106] |
| India | May 2013–October 2013 | Clinical samples | 3/100 | 12/100 | 6/100 | --- | --- | [107] |
| India | July 2009–December 2012 | Surgical site infection | 0/267 | 3/267 | --- | --- | --- | [108] |
| India | January 2014–December 2016 | Clinical samples | --- | --- | 66/500 | --- | SCCmec III (8%), SCCmec IV (17.7%), SCCmec V (50%) | |
| India | July 2015–June 2016 | Pus, respiratory tract, urine, blood, body fluids, and catheter tips | --- | 18/266 | 15/266 | --- | NA | [75] |
| India | February 2019–March 2020 | Pus, tissue, blood, catheter | --- | --- | 14/220 | --- | --- | [80] |
| Sri Lanka | April 2002 | Surgical site infection | --- | 1/1 | --- | --- | --- | [87] |
| Bangladesh | August 2010–July 2011 | Clinical samples | 114/122 | --- | --- | --- | ||
| Bangladesh | January 2010–December 2011 | Clinical samples | 3/38 | --- | --- | --- | --- | [109] |
| Bangladesh | July 2011–June 2012 | Wound swabs | 2/15 | --- | --- | vanB | --- | [81] |
| Bangladesh | April 2012–January 2013 | Burn wounds | --- | 16/29 | --- | --- | --- | [19] |
4. Conclusions
Infections due to VISA/VRSA are associated with high morbidity and mortality, and an increase in the MIC creep of vancomycin has been found among clinical isolates during recent decades in different countries of South Asia. The subsequent increase in the vancomycin MIC trend is quite alarming for clinicians in the treatment of infections. The epidemiological investigation should be implemented by clinical laboratories for the determination of vancomycin non-susceptibility with accurate precision. Furthermore, long-term studies are needed to explore the resistance mechanism and epidemiological patterns of VISA/VRSA. Vancomycin in a clinical setting should be cautiously used to curtail the emergence and spread of resistant strains of S. aureus.
Author Contributions
Conceptualization, M.E., M.A.S., M.S., and R.F.; methodology, M.E.; validation, M.E., M.A.S., C.R.J., M.S., and R.F.; formal analysis, M.E., M.A.S., C.R.J., M.S., and R.F.; investigation, M.E. and M.A.S.; resources, M.A.S. and C.R.J.; data curation, M.E.; writing—original draft preparation, M.E. and M.A.S.; writing—review and editing, M.E., M.A.S., C.R.J., M.S., and R.F.; visualization, M.E. and M.A.S.; supervision, M.A.S.; project administration, M.A.S.; funding acquisition, M.A.S. and C.R.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the U.S. Department of Agriculture-Agricultural internal project plan 6040-32000-079-00D.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The findings and conclusions in this report are those of the author(s) and should not be construed to represent any official USDA or U.S. Government determination or policy. The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
References
- Hemmadi, V.; Biswas, M. An overview of moonlighting proteins in Staphylococcus aureus infection. Arch. Microbiol. 2020, 203, 481–498. [Google Scholar] [CrossRef]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 2025–2027. [Google Scholar] [CrossRef]
- Abbasian, S.; Farahani, N.; Mir, Z.; Alinejad, F.; Haeili, M.; Dahmardehei, M.; Mirzaii, M.; Khoramrooz, S.; Nasiri, M.; Darban-Sarokhalil, D. Genotypic characterization of Staphylococcus aureus isolated from a burn centre by using agr, spa and SCCmec typing methods. New Microbes New Infect. 2018, 26, 15–19. [Google Scholar] [CrossRef]
- Mitchell, D.H.; Howden, B. Diagnosis and management of Staphylococcus aureus bacteraemia. Intern. Med. J. 2005, 35, S17–S24. [Google Scholar] [CrossRef]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Focus: Infectious diseases: Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269. [Google Scholar]
- Huttner, A.; Harbarth, S.; Carlet, J.; Cosgrove, S.; Goossens, H.; Holmes, A.; Jarlier, V.; Voss, A.; Pittet, D. Antimicrobial resistance: A global view from the 2013 World Healthcare-Associated Infections Forum. Antimicrob. Resist. Infect. Control 2013, 2, 31. [Google Scholar] [CrossRef]
- Bamigboye, B.T.; Olowe, O.A.; Taiwo, S.S. Phenotypic and molecular identification of vancomycin resistance in clinical Staphylococcus aureus isolates in Osogbo, Nigeria. Eur. J. Microbiol. Immunol. 2018, 8, 25–30. [Google Scholar] [CrossRef]
- Makgotlho, P.E.; Kock, M.; Hoosen, A.; Lekalakala, R.; Omar, S.V.; Dove, M.; Ehlers, M.M. Molecular identification and genotyping of MRSA isolates. FEMS Immunol. Med. Microbiol. 2009, 57, 104–115. [Google Scholar] [CrossRef]
- Chen, C.-J.; Huang, Y.-C. New epidemiology of Staphylococcus aureus infection in Asia. Clin. Microbiol. Infect. 2014, 20, 605–623. [Google Scholar] [CrossRef]
- Nelwan, E.J.; Andayani, D.; Clarissa, G.; Pramada, T. Vancomycin-resistant Staphylococcus aureus infection post-liposuction in South Korea. Cureus 2021, 13, e14357. [Google Scholar] [CrossRef]
- Hanaki, H.; Hososaka, Y.; Yanagisawa, C.; Nakae, T.; Sunakawa, K.; Otsuka, Y.; Nagasawa, Z. Occurrence of vancomycin-intermediate-resistant Staphylococcus aureus in Japan. J. Infect. Chemother. 2007, 13, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Berglee, R. World Regional Geography: People, Places and Globalization; University of Minnesota Libraries Publishing: Minneapolis, MN, USA, 2012. [Google Scholar]
- Worldometer. Southern Asia Population. 4 April 2021. Available online: https://www.worldometers.info/world-population/southern-asia-population/ (accessed on 4 April 2022).
- Zaidi, S.; Saligram, P.; Ahmed, S.; Sonderp, E.; Sheikh, K. Expanding access to healthcare in South Asia. Br. Med. J. 2017, 357, j1645. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Yang, S.; Rao, X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J. Adv. Res. 2020, 21, 169–176. [Google Scholar] [CrossRef]
- Saha, B.; Singh, A.K.; Ghosh, A.; Bal, M. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J. Med. Microbiol. 2008, 57, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Sabokroo, N.; Wang, Y.; Hashemian, M.; Karamollahi, S.; Kouhsari, E. Systematic review and meta-analysis of the epidemiology of vancomycin-resistance Staphylococcus aureus isolates. Antimicrob. Resist. Infect. Control 2021, 10, 101. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.; Hussain, A.; Sohail, M.; Rehman, S.U.; Javed, N.; Abbas, Z. Isolation and characterization of Vancomycin-Resistant Staphylococcus aureus (VRSA) from Intensive Care Units (ICU) of different hospitals in Lahore, Pakistan. Adv. Life Sci. 2021, 8, 339–344. [Google Scholar]
- Hasan, R.; Acharjee, M.; Noor, R. Prevalence of Vancomycin-Resistant Staphylococcus aureus (VRSA) in Methicillin-Resistant S. aureus (MRSA) strains isolated from burn wound infections. Tzu Chi Med. J. 2016, 28, 49–53. [Google Scholar] [CrossRef]
- Shariati, A.; Dadashi, M.; Moghadam, M.T.; van Belkum, A.; Yaslianifard, S.; Darban-Sarokhalil, D. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 12698. [Google Scholar] [CrossRef]
- Stepanović, S.; Dakić, I.; Djukić, S.; Lozuk, B.; Svabić-Vlahović, M. Surgical wound infection associated with Staphylococcus sciuri. Scand. J. Infect. Dis. 2002, 34, 685–686. [Google Scholar] [CrossRef]
- Gill, S.R.; Fouts, D.E.; Archer, G.L.; Mongodin, E.F.; DeBoy, R.T.; Ravel, J.; Paulsen, I.T.; Kolonay, J.F.; Brinkac, L.; Beanan, M.; et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Rammelkamp, C.H.; Maxon, T. Resistance of Staphylococcus aureus to the action of penicillin. Proc. Soc. Exp. Biol. Med. 1942, 51, 386–389. [Google Scholar] [CrossRef]
- Olsen, J.E.; Christensen, H.; Aarestrup, F.M. Diversity and evolution of blaZ from Staphylococcus aureus and coagulase-negative staphylococci. J. Antimicrob. Chemother. 2006, 57, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Jevons, M.P. “Celbenin”-resistant staphylococci. Br. Med. J. 1961, 1, 124. [Google Scholar] [CrossRef]
- Baede, V.O.; David, M.Z.; Andrasevic, A.T.; Blanc, D.S.; Borg, M.; Brennan, G.; Catry, B.; Chabaud, A.; Empel, J.; Enger, H.; et al. MRSA surveillance programmes worldwide: Moving towards a harmonised international approach. Int. J. Antimicrob. Agents 2022, 59, 106538. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.; Smith, D.L.; Laxminarayan, R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 2007, 13, 1840. [Google Scholar] [CrossRef]
- Tang, J.; Hu, J.; Kang, L.; Deng, Z.; Wu, J.; Pan, J. The use of vancomycin in the treatment of adult patients with methicillin-resistant Staphylococcus aureus (MRSA) infection: A survey in a tertiary hospital in China. Int. J. Clin. Exp. Med. 2015, 8, 19436. [Google Scholar]
- Klevens, R.M.; Morrison, M.A.; Nadle, J.; Petit, S.; Gershman, K.; Ray, S.; Harrison, L.H.; Lynfield, R.; Dumyati, G.; Townes, J.M.; et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. J. Am. Med. Assoc. 2007, 298, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, S.; Matsunaga, N.; Yahara, K.; Gu, Y.; Hayakawa, K.; Hirabayashi, A.; Kajihara, T.; Sugai, M.; Shibayama, K.; Ohmagari, N. National trend of blood-stream infection attributable deaths caused by Staphylococcus aureus and Escherichia coli in Japan. J. Infect. Chemother. 2020, 26, 367–371. [Google Scholar] [CrossRef]
- Ruhe, J.J.; Smith, N.; Bradsher, R.W.; Menon, A. Community-onset methicillin-resistant Staphylococcus aureus skin and soft-tissue infections: Impact of antimicrobial therapy on outcome. Clin. Infect. Dis. 2007, 44, 777–784. [Google Scholar] [CrossRef]
- Foster, T.J.; Geoghegan, J.A. Staphylococcus aureus. In Molecular Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch: Galveston, TX, USA, 2015; pp. 655–674. [Google Scholar]
- Japoni, A.; Jamalidoust, M.; Farshad, S.; Ziyaeyan, M.; Alborzi, A.; Japoni, S.; Rafaatpour, N. Characterization of SCCmec types and antibacterial susceptibility patterns of methicillin-resistant Staphylococcus aureus in Southern Iran. Jpn. J. Infect. Dis. 2011, 64, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, D.; Peters, B.M.; Li, L.; Li, B.; Xu, Z.; Shirliff, M.E. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb. Pathog. 2016, 101, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Chongtrakool, P.; Ito, T.; Ma, X.X.; Kondo, Y.; Trakulsomboon, S.; Tiensasitorn, C.; Hiramatsu, K. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: A proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 2006, 50, 1001–1012. [Google Scholar] [CrossRef]
- Peng, H.; Liu, D.; Ma, Y.; Gao, W. Comparison of community- and healthcare-associated methicillin-resistant Staphylococcus aureus isolates at a Chinese tertiary hospital, 2012–2017. Sci. Rep. 2018, 8, 17916. [Google Scholar] [CrossRef] [PubMed]
- Kateete, D.P.; Bwanga, F.; Seni, J.; Mayanja, R.; Kigozi, E.; Mujuni, B.; Ashaba, F.K.; Baluku, H.; Najjuka, C.F.; Källander, K.; et al. CA-MRSA and HA-MRSA coexist in community and hospital settings in Uganda. Antimicrob. Resist. Infect. Control 2019, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Cheng, H.; Yuan, W.; Zeng, F.; Shang, W.; Tang, D.; Xue, W.; Fu, J.; Zhou, R.; Zhu, J.; et al. Panton-Valentine Leukocidin (PVL)-positive health care-associated methicillin-resistant Staphylococcus aureus isolates are associated with skin and soft tissue infections and colonized mainly by infective PVL-encoding bacteriophages. J. Clin. Microbiol. 2015, 53, 67–72. [Google Scholar] [CrossRef]
- Rehm, S.J.; Tice, A. Staphylococcus aureus: Methicillin-susceptible S. aureus to methicillin-resistant S. aureus and vancomycin-resistant S. aureus. Clin. Infect. Dis. 2010, 51 (Suppl. S2), S176–S182. [Google Scholar]
- Hiramatsu, K.; Hanaki, H.; Ino, T.; Yabuta, K.; Oguri, T.; Tenover, F.C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 1997, 40, 135–136. [Google Scholar] [CrossRef]
- Goldrick, B. First reported case of VRSA in the United States: An alarming development in microbial resistance. AJN Am. J. Nurs. 2002, 102, 17. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Update: Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. MMWR. Morb. Mortal. Wkly. Rep. 1997, 46, 813–815.
- Hallin, M.; Friedrich, A.W.; Struelens, M.J. spa typing for epidemiological surveillance of Staphylococcus aureus. Mol. Epidemiol. Microorg. Methods Protocols 2009, 551, 189–202. [Google Scholar]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontéen, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Shekarabi, M.; Hajikhani, B.; Chirani, A.S.; Fazeli, M.; Goudarzi, M. Molecular characterization of vancomycin-resistant Staphylococcus aureus strains isolated from clinical samples: A three-year study in Tehran, Iran. PLoS ONE 2017, 12, e0183607. [Google Scholar] [CrossRef]
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D.; et al. A Field Guide to Pandemic, Epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 2011, 6, e17936. [Google Scholar]
- Bozdogan, B.; Ednie, L.; Credito, K.; Kosowska, K.; Appelbaum, P.C. Derivatives of a vancomycin-resistant Staphylococcus aureus strain isolated at Hershey Medical Center. Antimicrob. Agents Chemother. 2004, 48, 4762–4765. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Seemann, T.; Harrison, P.F.; McEvoy, C.R.; Stanton JA, L.; Rand, C.J.; Stinear, T.P. Complete genome sequence of Staphylococcus aureus strain JKD6008, an ST239 clone of methicillin-resistant Staphylococcus aureus with intermediate-level vancomycin resistance. J. Bacteriol. 2010, 192, 5848–5849. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Davies, J.K.; Johnson, P.D.R.; Stinear, T.P.; Grayson, M.L. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar] [CrossRef]
- Arthur, M.; Molinas, C.; Depardieu, F.; Courvalin, P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 1993, 175, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Périchon, B.; Courvalin, P. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009, 53, 4580–4587. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Clewell, D.B.; Ike, Y.; Shankar, N. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Weinstein, R.A.; Fridkin, S.K. Vancomycin-intermediate and -resistant Staphylococcus aureus: What the infectious disease specialist needs to know. Clin. Infect. Dis. 2001, 32, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.K.; Wennersten, C.; Venkataraman, L.; Eliopoulos, G.M.; Moellering, J.R.C.; Karchmer, A.W. Development of reduced vancomycin susceptibility in methicillin-susceptible Staphylococcus aureus. Clin. Infect. Dis. 2009, 49, 1169–1174. [Google Scholar] [CrossRef] [PubMed]
- Gardete, S.; Kim, C.; Hartmann, B.M.; Mwangi, M.; Roux, C.M.; Dunman, P.M.; Chambers, H.F.; Tomasz, A. Genetic pathway in acquisition and loss of vancomycin resistance in a Methicillin Resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLoS Pathog. 2012, 8, e1002505. [Google Scholar]
- Wang, W.-Y.; Lee, S.-Y.; Chiueh, T.-S.; Lu, J.-J. Molecular and phenotypic characteristics of methicillin-resistant and vancomycin-intermediate Staphylococcus aureus isolates from patients with septic arthritis. J. Clin. Microbiol. 2009, 47, 3617–3623. [Google Scholar]
- Holmes, N.E.; Turnidge, J.D.; Munckhof, W.J.; Robinson, J.O.; Korman, T.; O’Sullivan, M.; Anderson, T.L.; Roberts, S.A.; Gao, W.; Christiansen, K.J.; et al. Antibiotic choice may not explain poorer outcomes in patients with Staphylococcus aureus bacteremia and high vancomycin minimum inhibitory concentrations. J. Infect. Dis. 2011, 204, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Haripur District Demographics. 17 May 2022. Available online: https://kp.gov.pk/page/haripurdistrictdemographics (accessed on 17 May 2022).
- Wang, G.; Hindler, J.F.; Ward, K.W.; Bruckner, D.A. Increased vancomycin MICs for Staphylococcus aureus clinical isolates from a university hospital during a 5-Year Period. J. Clin. Microbiol. 2006, 44, 3883–3886. [Google Scholar] [CrossRef]
- Tenover, F.C.; Moellering, R.C., Jr. The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin. Infect. Dis. 2007, 44, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Edition, A.S.N. CLSI Document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Azhar, A.; Rasool, S.; Haque, A.; Shan, S.; Saeed, M.; Ehsan, B.; Haque, A. Detection of high levels of resistance to linezolid and vancomycin in Staphylococcus aureus. J. Med. Microbiol. 2017, 66, 1328–1331. [Google Scholar] [CrossRef]
- Mirani, Z.A.; Jamil, N. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J. Basic Microbiol. 2011, 51, 191–195. [Google Scholar] [CrossRef]
- Taj, Y.; Abdullah, F.E.; Kazmi, S.U. Current pattern of antibiotic resistance in Staphylococcus aureus clinical isolates and the emergence of vancomycin resistance. J. Coll. Physicians Surg. Pak. 2010, 20, 728–732. [Google Scholar]
- Ahmad, S.; Ahmed, S.; Sabir, M.; Khan, H.; Rehman, M.; Niaz, Z. Frequency and comparison among antibiotic resistant Staphylococcus aureus strains in selected hospitals of Peshawar, Pakistan. J. Pak. Med. Assoc. 2020, 70, 1199–1202. [Google Scholar] [CrossRef]
- Hakim, S.T.; Arshed, S.; Iqbal, M.; Javaid, S.G. Vancomycin sensitivity of Staphylococcus aureus isolates from hospital patients in Karachi, Pakistan. Libyan J. Med. 2007, 2, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.; Ahsan, F.; Nawaz, M.; Iqbal, K.; Rehman, K.U.; Ijaz, T. Incidence of vancomycin resistant phenotype of the Methicillin Resistant Staphylococcus aureus isolated from a tertiary care hospital in Lahore. Antibiotics 2019, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Ghias, W.; Sharif, M.; Yazdani, F.A.; Rabbani, M. Isolation and identification of methicillin and vancomycin resistance Staphylococcus aureus from pus samples of injured skin patients in Lahore, Pakistan. Biomed. Lett. 2016, 2, 103–112. [Google Scholar]
- Hanif, E.; Hassan, S.A. Evaluation of antibiotic resistance pattern in clinical isolates of Staphylococcus aureus. Pak. J. Pharm. Sci. 2019, 32, 1749–1753. [Google Scholar] [PubMed]
- Banerjee, T.; Anupurba, S. Colonization with vancomycin-intermediate Staphylococcus aureus strains containing the vanA resistance gene in a tertiary-care center in North India. J. Clin. Microbiol. 2012, 50, 1730–1732. [Google Scholar] [CrossRef] [PubMed]
- Goud, R.; Gupta, S.; Neogi, U.; Agarwal, D.; Naidu, K.; Chalannavar, R.; Subhaschandra, G. Community prevalence of methicillin and vancomycin resistant Staphylococcus aureus in and around Bangalore, southern India. Rev. da Soc. Bras. de Med. Trop. 2011, 44, 309–312. [Google Scholar] [CrossRef]
- Moses, V.K.; Kandi, V.; Rao, S.K.D. Minimum inhibitory concentrations of vancomycin and daptomycin against methicillin-resistant Staphylococcus aureus isolated from various clinical specimens: A study from South India. Cureus 2020, 12, e6749. [Google Scholar] [CrossRef]
- Mohanty, S.; Behera, B.; Sahu, S.; Praharaj, A.K. Recent pattern of antibiotic resistance in Staphylococcus aureus clinical isolates in Eastern India and the emergence of reduced susceptibility to vancomycin. J. Lab. Physicians 2019, 11, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-H.; Hiramatsu, K.; Suh, J.Y.; Ko, K.S.; Ito, T.; Kapi, M.; Kiem, S.; Kim, Y.-S.; Oh, W.S.; Peck, K.R.; et al. Emergence in Asian countries of Staphylococcus aureus with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 2004, 48, 4926–4928. [Google Scholar] [CrossRef]
- Chung, D.R.; Lee, C.; Kang, Y.R.; Baek, J.Y.; Kim, S.H.; Ha, Y.E.; Kang, C.-I.; Peck, K.R.; Lee, N.Y.; Song, J.-H. Genotype-specific prevalence of heterogeneous vancomycin-intermediate Staphylococcus aureus in Asian countries. Int. J. Antimicrob. Agents 2015, 46, 338–341. [Google Scholar] [CrossRef]
- Sistla, S.; Amberpet, R.; Sugumar, M.; Nagasundaram, N.; Manoharan, M.; Parija, S.C. Detection of heterogeneous vancomycin-intermediate Staphylococcus aureus: A preliminary report from south India. Indian J. Med. Res. 2019, 150, 194–198. [Google Scholar] [CrossRef]
- Selvabai, A.P.; Sattar, S.B.A.; Jayaraman, P.; Shanmugam, P. Detection and characterisation of heteroresistant Vancomycin Intermediate Staphylococcus aureus (hVISA) using phenotypic and genotypic methods. J. Clin. Diagn. Res. 2019, 13, 1–5. [Google Scholar] [CrossRef]
- Sreejisha, M.; Mulki, S.S.; Shenoy, S.; Dhanashree, B.; Chakrapani, M.; Bhat, G. Heterogeneous vancomycin intermediate Staphylococcus aureus infections in diabetic and non-diabetic patients–A hospital-based comparative study. Infect. Drug Resist. 2023, 16, 9–17. [Google Scholar]
- Islam, T.A.B.; Shamsuzzaman, S. Prevalence and antimicrobial susceptibility pattern of methicillin-resistant, vancomycin-resistant, and Panton-Valentine Leukocidin positive Staphylococcus aureus in a tertiary care hospital Dhaka, Bangladesh. Tzu Chi Med. J. 2015, 27, 10–14. [Google Scholar] [CrossRef]
- Shahriar, M.; Shahid, S.; Katha, K.K.; Nasreen, W.; Bhuiyan, M.A. Vancomycin sensitivity of clinical isolates of Staphylococcus aureus from patients in Dhaka City, Bangladesh. Bangladesh Pharm. J. 2012, 15, 159–163. [Google Scholar] [CrossRef]
- Haque, N.; Aung, M.S.; Paul, S.K.; Bari, S.; Ahmed, S.; Sarkar, S.R.; Roy, S.; Nasreen, S.A.; Mahmud, M.C.; Hossain, M.A.; et al. Molecular epidemiological characterization of methicillin-susceptible and -resistant Staphylococcus aureus isolated from skin and soft tissue infections in Bangladesh. Microb. Drug Resist. 2019, 25, 241–250. [Google Scholar] [CrossRef]
- Taz, K.A.; Jobayer, M.; Shamsuzzaman, S.M. Nasal colonization of methicillin resistant Staphylococcus aureus among healthcare providers in a tertiary care hospital, Bangladesh. Mymensingh Med. J. MMJ 2019, 28, 627–633. [Google Scholar] [PubMed]
- Roy, S.; Barman, T.K.; Hossain, M.A.; Paul, S.K.; Haque, N.; Ahmed, S.; Nasreen, S.A.; Hossain, M.S.; Sarkar, S.R.; Kubayashi, N.; et al. Molecular-characterization of Methicillin-Resistance Staphylococcus aureus (MRSA) from different tertiary care hospitals in Bangladesh. Mymensingh Med. J. 2017, 26, 37–44. [Google Scholar]
- Parvez, M.A.K.; Ferdous, R.N.; Rahman, M.S.; Islam, S. Healthcare-associated (HA) and community-associated (CA) methicillin resistant Staphylococcus aureus (MRSA) in Bangladesh–Source, diagnosis and treatment. J. Genet. Eng. Biotechnol. 2018, 16, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-H.; Hsueh, P.-R.; Chung, D.R.; Ko, K.S.; Kang, C.-I.; Peck, K.R.; Yeom, J.-S.; Kim, S.-W.; Chang, H.-H.; Kim, Y.-S.; et al. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: An ANSORP study. J. Antimicrob. Chemother. 2011, 66, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Jayaweera, J.; Karunarathne, M.; Kumbukgolla, W.W. The importance of timely introduction of vancomycin therapy against methicillin-resistant Staphylococcus aureus (MRSA) bacteremia and severity of MRSA bacteremia at teaching hospital, Anuradhapura, Sri Lanka. Int J. One Health 2017, 3, 7–11. [Google Scholar] [CrossRef]
- Samaranayake, W.; Karunanayake, L.; Patabendige, C. Characteristics of community acquired and hospital acquired methicillin resistant Staphylococcus aureus isolates in the National Hospital of Sri Lanka. Sri Lankan J. Infect. Dis. 2019, 9, 24. [Google Scholar] [CrossRef]
- McTavish, S.M.; Snow, S.J.; Cook, E.C.; Pichon, B.; Coleman, S.; Coombs, G.W.; Pang, S.; Arias, C.A.; Díaz, L.; Boldock, E.; et al. Genomic and epidemiological evidence of a dominant Panton-Valentine Leucocidin-positive methicillin resistant Staphylococcus aureus lineage in Sri Lanka and presence among isolates from the United Kingdom and Australia. Front. Cell. Infect. Microbiol. 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Wijitha Weerakoon, S.D.A.; Gamage, D.S.; Wijeratne, M. Staphylococcus aureus with reduced susceptibility to vancomycin. Ceylon Med. J. 2003, 48, 58–59. [Google Scholar] [CrossRef]
- Chaudhary, M.; Bhattarai, A.; Adhikari, S.K.; Bhatta, D.R. Bacteriology and antimicrobial susceptibility of adult chronic dacryocystitis. Nepal. J. Ophthalmol. 2010, 2, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Pahadi, P.C.; Shrestha, U.T.; Adhikari, N.; Shah, P.K.; Amatya, R. Growing resistance to vancomycin among methicillin resistant Staphylococcus aureus isolates from different clinical samples. J. Nepal Med. Assoc. 2014, 52, 977–981. [Google Scholar] [CrossRef]
- Raut, S.; Bajracharya, K.; Adhikari, J.; Pant, S.S.; Adhikari, B. Prevalence of methicillin resistant Staphylococcus aureus in Lumbini Medical College and Teaching Hospital, Palpa, Western Nepal. BMC Res. Notes 2017, 10, 187. [Google Scholar] [CrossRef]
- Shrestha, B.; Pokhrel, B.M.; Mohapatra, T.M. Phenotypic characterization of nosocomial isolates of Staphylococcus aureus with reference to MRSA. J. Infect. Dev. Ctries. 2009, 3, 554–560. [Google Scholar] [CrossRef]
- Kshetry, A.O.; Pant, N.D.; Bhandari, R.; Khatri, S.; Shrestha, K.L.; Upadhaya, S.K.; Poudel, A.; Lekhak, B.; Raghubanshi, B.R. Minimum inhibitory concentration of vancomycin to methicillin resistant Staphylococcus aureus isolated from different clinical samples at a tertiary care hospital in Nepal. Antimicrob. Resist. Infect. Control 2016, 5, 27. [Google Scholar] [CrossRef]
- Maharjan, M.; Sah, A.K.; Pyakurel, S.; Thapa, S.; Maharjan, S.; Adhikari, N.; Rijal, K.R.; Ghimire, P.; Shrestha, U.T. Molecular Confirmation of vancomycin-resistant Staphylococcus aureus with vanA gene from a hospital in Kathmandu. Int. J. Microbiol. 2021, 2021, 3847347. [Google Scholar] [CrossRef]
- Naimi, H.M.; Rasekh, H.; Noori, A.Z.; Bahaduri, M.A. Determination of antimicrobial susceptibility patterns in Staphylococcus aureus strains recovered from patients at two main health facilities in Kabul, Afghanistan. BMC Infect. Dis. 2017, 17, 737. [Google Scholar] [CrossRef]
- Naimi, H.M.; André, C.; Bes, M.; Tristan, A.; Gustave, C.-A.; Vandenesch, F.; Nazari, Q.A.; Laurent, F.; Dupieux, C. Antibiotic resistance profile and molecular characterization of Staphylococcus aureus strains isolated in hospitals in Kabul, Afghanistan. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Calhoun, J.H.; Murray, C.K.; Manring, M.M. Multidrug-resistant organisms in military wounds from Iraq and Afghanistan. Clin. Orthop. Relat. Res. 2008, 466, 1356–1362. [Google Scholar] [CrossRef] [PubMed]
- Liaqat, F.; Sheikh, A.A.; Nazir, J.; Hussain, T.; Rabbani, M.; Shaheen, A.Y.; Muhammad, J. Report-Isolation, identification and control of vancomycin resistant Staphylococcus aureus. Pak. J. Pharm. Sci. 2015, 28, 997–1004. [Google Scholar]
- Lama, U.; Shah, D.; Shrestha, U.T. Vancomycin resistant Staphylococcus aureus reported from tertiary care hospital in Nepal. Tribhuvan Univ. J. Microbiol. 2017, 4, 63–72. [Google Scholar] [CrossRef][Green Version]
- Tiwari, H.K.; Sen, M.R. Emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. BMC Infect. Dis. 2006, 6, 156. [Google Scholar] [CrossRef] [PubMed]
- Vellappally, S.; Divakar, D.D.; Al Kheraif, A.A.; Ramakrishnaiah, R.; Alqahtani, A.; Dalati, M.H.N.; Anil, S.; Khan, A.A.; Varma, P.R.H. Occurrence of vancomycin-resistant Staphylococcus aureus in the oral cavity of patients with dental caries. Acta Microbiol. Immunol. Hung. 2017, 64, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Gaddad, S.M.; Thati, V.; Shivannavar, C.T. Vancomycin resistance among methicillin resistant Staphylococcus aureus isolates from intensive care units of tertiary care hospitals in Hyderabad. Indian J. Med. Res. 2011, 134, 704. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Bir, R.; Majumdar, T. Evaluation of multidrug resistant Staphylococcus aureus and their association with biofilm production in a tertiary care hospital, Tripura, Northeast India. J. Clin. Diagn. Res. JCDR 2015, 9, DC01. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Pal, K.; Jain, S.; Chatterjee, S.S.; Konar, J. Surgical site infection by methicillin resistant Staphylococcus aureus–On decline? J. Clin. Diagn. Res. JCDR 2016, 10, DC32. [Google Scholar] [CrossRef]
- Khanam, S.; Haq, J.A.; Shamsuzzaman, S.; Rahman, M.; Mamun, K.Z. Emergence of vancomycin resistant Staphylococcus aureus during hospital admission at a tertiary care hospital in Bangladesh. Bangladesh J. Infect. Dis. 2016, 3, 11–16. [Google Scholar] [CrossRef]
- Rehman, T.U.; Aslam, R.; Aqib, A.I.; Mohsin, M.; Manzoor, A.; Shoaib, M.; Naseer, M.A.; Hassan, A.; Sattar, H.; Kulyar, M.F.-E.; et al. Phylogeny of hospital acquired MRSA, and its comparative phenotypic clinico-epidemiology with vancomycin resistant S. aureus (VRSA). Microb. Pathog. 2020, 149, 104537. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).