Abstract
Yersiniosis is the third most commonly reported foodborne zoonosis in the European Union. Here, we evaluated the prevalence of pathogenic Yersinia enterocolitica among healthy pigs (as a major reservoir) in a slaughterhouse in Bulgaria. A total of 790 tonsils and feces from 601 pigs were examined. Isolation and pathogenicity characterization was carried out by the ISO 10273:2003 protocol and Polymerase Chain Reaction (PCR), detecting the 16S rRNA gene, attachment and invasion locus (ail), Yersinia heat-stable enterotoxin (ystA), and Yersinia adhesion (yadA) genes. Genetic diversity was assessed by pulsed-field gel electrophoresis (PFGE), and antimicrobial resistance by the standard disk diffusion method. Of all the pigs tested, 6.7% were positive for Y. enterocolitica. All isolates belonged to Y. enterocolitica bioserotype 4/O:3. ail, and ystA genes were detected in all positive strains (n = 43), while the plasmid Yersinia virulence plasmid (pYV) was detected in 41. High homogeneity was observed among the strains, with all strains susceptible to ceftriaxone, amikacin and ciprofloxacin, and resistant to ampicillin. In conclusion, a low prevalence of Y. enterocolitica 4/O:3 was found in healthy pigs slaughtered in Bulgaria, not underestimating possible contamination of pork as a potential risk to consumer health.
1. Introduction
Yersinia enterocolitica is a Gram-negative rod within the genus Yersinia. The genus comprises 26 species, only 3 of which are known to be human pathogens [1]. Yersinia pestis is the causative agent of plague, while Y. enterocolitica and Yersinia pseudotuberculosis are significant foodborne pathogens associated with yersiniosis. Yersiniosis is an important zoonotic disease with a wide range of clinical symptoms. Symptoms may vary from mild self-limiting acute gastroenteritis to serious complications, systemic infection, and septicemia [2]. According to the European Food Safety Authority (EFSA), yersiniosis was the third most commonly reported zoonosis in Europe in 2021, an increase of 11.8% as compared to 2020, and a total of 6789 confirmed human cases [3]. Additionally, of the two pathogenic species, Y. enterocolitica caused the majority (98.1%) of human infections [3].
Y. enterocolitica is characterized by strong heterogeneity within the species. According to their biochemical activity, Y. enterocolitica strains are classified into 6 different groups: 1A, 1B, 2, 3, 4, and 5. The biotypes differ in terms of geographic distribution, ecological niches, and pathogenicity. Biotype 1A is widely distributed in the environment and regarded as non-pathogenic to animals and humans. Most of the human pathogenic strains belong to biotypes 1B, 2, 3, 4, and 5, of which 1B is considered highly pathogenic. In addition to biotypes, there are more than 70 serotypes of circulating Y. enterocolitica, some of which, such as O:3, O:5,27, O:8, and O:9, are shown to have been frequently associated with infection in humans [4]. Y. enterocolitica bioserotype 4/O:3, known as a “pig bio- and -serotype”, is the predominant causative strain of human yersiniosis over other bioserotypes [2,5]. Pigs are considered to be the major reservoir for human pathogenic strains [6]. Bacteria persist in the lymphatic tissue of healthy pigs and are frequently disseminated to porcine carcasses during the slaughter process [7,8,9,10,11]. Although Y. enterocolitica can be isolated from carcasses, tongues, and feces, the porcine tonsils are still the most important source for bacterial isolation [9,12].
The pathogenicity of Y. enterocolitica is due to different chromosomal- and plasmid-encoded virulence determinants. The most important determinants associated with clinical infection are the attachment and invasion locus (ail), Yersinia heat-stable enterotoxin (ystA), Yersinia adhesin (yadA) genes, invasin (invA), Yersinia outer membrane protein virulon (yop), low-calcium response regulon (Lcr), mucoid Yersinia factor (myfA), Yersinia enterocolitica chromosomal modulator (ymoA), and Yersinia virulence regulon (virF) genes [6,13]. Their detection provides reliable information about the risk of emerging infection in humans. However, detection of plasmid genes is always challenging because of the temperature dependence and easy loss of the plasmid during repeated cultivation reviewed by Bhaduri and Smith [14].
Among the molecular methods for genotyping of Y. enterocolitica, pulsed-field gel electrophoresis (PFGE) is commonly used as the gold standard in epidemiological investigations [6]. Moreover, research on the genetic relationship of the isolated strains is an opportunity to describe porcine and human Y. enterocolitica isolates, and/or the presence of cross contamination. Thus, this typing method can explain the circulation of strains and the correlation between them.
Relevant in vitro assays have proved that Y. enterocolitica strains isolated from slaughtered pigs are susceptible to many antibiotics, such as tetracycline, aminoglycosides, third generation cephalosporins, fluoroquinolones, and resistant to aminopenicillins and first-generation cephalosporins [7,10,15]. Heterogeneity of the antimicrobial resistance profile is shown to depend on the bioserotype and geographic distribution [16]. Multidrug-resistant Y. enterocolitica bioserotype 4/O:3 isolates from pigs have increased lately [17]. Their transmission highlights the importance of monitoring the resistance of circulating strains.
There are no data on the occurrence, virulence potential, genetic relationship, and antimicrobial resistance of Y. enterocolitica strains isolated from pigs of slaughter age in Bulgaria. The availability of such data would provide scientific outputs for the reports of EFSA. Thus, the aim of this study was to detect the prevalence of pathogenic Y. enterocolitica strains in healthy pigs from a slaughterhouse in Bulgaria and to determine their virulence-associated genes, genetic relationship, and susceptibility to different antibiotics.
2. Results
2.1. Detection of Yersinia enterocolitica
An overall of 601 slaughtered pigs were tested for the presence of Y. enterocolitica (Figure 1A). Of these, 790 samples were collected, including 601 tonsil samples (tonsil sample from each pig) and 189 fecal samples from 189 pigs (only). From the aforementioned samples (n = 790), 920 colonies were identified on Cefsulodin-Irgasan-Novobiocin (CIN) agar, comprising 666 colonies from tonsil samples and 254 colonies from fecal samples (Figure 1A). Morphological analysis confirmed that all colonies exhibited features of the genus Yersinia. After undergoing preliminary biochemical analysis, 136 isolated colonies were identified as presumptive Y. enterocolitica based on their degradation of urea, positive catalase test, negative oxidase test, and lack of tryptophan deaminase activity. Subsequently, 43 Y. enterocolitica isolates (38 from tonsil samples and 5 from fecal samples) originating from 40 pigs were confirmed using the 16S rRNA gene (Figure 1A).
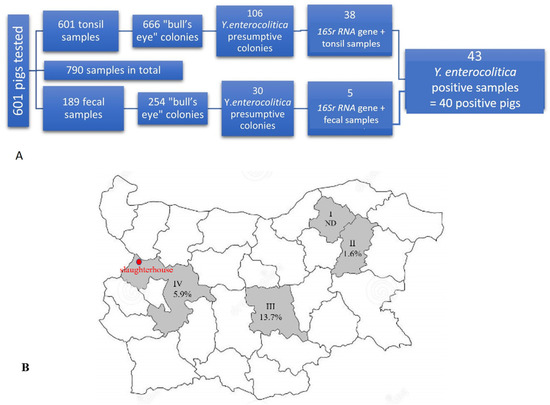
Figure 1.
Numbers of pigs and samples tested for Yersinia enterocolitica (A) and the distribution of the positive pigs per region in Bulgaria (B). Regions are presented in roman numerals I: Razgrad region (Farm I), II: Shumen region (Farm II), III: Stara Zagora region (Farm III), and IV: Sofia region (Farms IV, V, VI, and VII). The red dot indicates the location of the slaughterhouse (Kostinbrod city).
Total prevalence of Y. enterocolitica in slaughter age pigs was calculated to be 6.7% (40/601 pigs). As shown in Figure 1B, the frequency of Y. enterocolitica positive pigs varied according to the region of origin. It was higher among the pigs derived from Stara Zagora region (13.7%, 19/139, Farm III), followed by those from Sofia region (5.9%, 19/324, Farms IV, V, VI, and VII), and the lowest frequency was detected in pigs from Shumen region (1.6%, 2/126, Farm II). There were no positive pigs from Razgrad region (0/12, Farm I). In general, the positive animals originated from six farms (Farms II–VII), located in Stara Zagora, Sofia, and Shumen. All Y. enterocolitica strains were isolated only during the cold season (of all sampling periods)—from October to March (2016–2021).
2.2. Biotyping and Serotyping
Bioserotyping was performed on the 43 confirmed isolates of Y. enterocolitica. All isolates were determined as Y. enterocolitica serotype O:3 and biotype 4 based on trehalose utilization, detection of indole, tween esterase and pyrazinamidase activity, and esculin and salicin hydrolysis.
2.3. Detection of Virulence Genes
PCR analysis revealed that all Y. enterocolitica 4/O:3 isolates (n = 43) were positive for the chromosomally encoded virulence genes ail and ystA. In 41 isolates, the pYV-coded yadA gene was detected by PCR analysis, while 21 were proved positive for pYV by phenotypic assay on Congo red-magnesium oxalate (CR-MOX) agar. The prevalence of ail- and ystA-positive Y. enterocolitica isolates among pigs was calculated to be 6.7% (40/601), while the prevalence of the yadA gene was 6.3% (38/601).
2.4. Genetic Diversity of Y. enterocolitica Strains Determined by PFGE
Forty-three Y. enterocolitica strains were examined by pulsed-field gel electrophoresis, using SpeI as a restriction enzyme. Five pulsotypes (I, II, II, IV, and V) with minor differences were detected as shown in Figure 2.
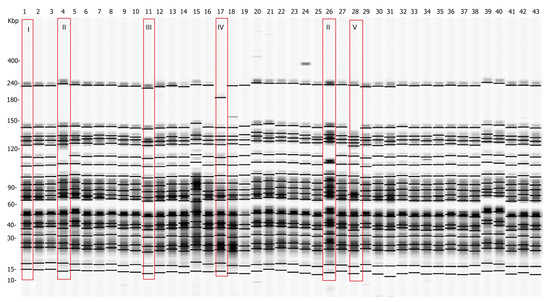
Figure 2.
Pulsed-field gel electrophoresis pulsotypes of 43 Y. enterocolitica isolated strains after restriction with SpeI. Five pulsotypes were observed (surrounded by red rectangles). Pulsotypes II, III, IV, and V differ from the major pulsotype I, which is representative of all analyzed strains except strains 4, 11, 17, 26, and 28. The number of each lane corresponds to the number of the analyzed Y. enterocolitica strain (1–43). The pulse marker used was 50–1000 kb.
Additional clustering was performed at 97% similarity. The strains were organized in two clusters, labeled S01 and S02, and three single pulsotypes, SP1, SP2, and SP3 (Figure 3). The number of fragments within the pulsotypes obtained after restriction varied between 19 and 20, and the size ranged from 15 kb to 240 kb (Figure 2). As depicted in Figure 3, all strains proved to be closely related, sharing a high percentage of genetic similarity (over 92%). Cluster S01 included 38 (88.4%) of the 43 strains, being the predominant macro-restriction pulsotype (Figure 3). All Y. enterocolitica strains clustered within S01 were of serotype O:3 and were isolated from the tonsils and feces of pigs originating from the “positive” farms (Figure 1B, Farms II–VII). Two Y. enterocolitica strains with identical pulsotypes clustered in S02 (Figure 3). These originated from farms III and VI, which are located in different geographic regions (Figure 1B).

Figure 3.
Dendrogram obtained derived by digesting Y. enterocolitica genomic DNA with SpeI. The DNA of 43 examined strains formed two different clusters, assigned S01 and S02, and three single pulsotypes, named SP1, SP2, and SP3. Clustering was calculated by the Unweighted pair group method with arithmetic mean (UPGMA) with 97% similarity. Dice correlation was with tolerance of 2.5% and optimization setting of 0.5. Additional information about dates of isolation, farm distribution, and virulence profile of the strains is given.
As shown in Figure 3, the strains isolated from tonsils and feces showed minor genomic differences. Clusters S01 and S02 were closely related to each other with 97.4% genetic similarity. One tonsil-derived strain, of a pig (originating) from farm II showed a single pulsotype SP1 (Figure 3). Pulsotype SP1 was closely related to S01 and S02 with 97.4% similarity. Single pulsotypes SP2 and SP3 were assigned to the remaining two Y. enterocolitica strains. Both strains originated from different farms and regions and were isolated from tonsils. Pulsotypes SP2 and SP3 were closely related with 97.4% similarity (Figure 3). Additionally, pulsotypes SP1 and SP2 were at equal distance from both S01 and S02 clusters (97.4% and 94.7%, respectively). SP3 was determined to be the most distant from other, and clustered as follows: to SP1 with 92.3% similarity, to SP2 with 97.4% similarity, to S01 cluster with 95% similarity, and to S02 cluster with 92.3% similarity (Figure 3).
2.5. Antimicrobial Susceptibility of Y. enterocolitica Strains
All 43 strains were tested for susceptibility to 15 antibiotics, belonged to 8 classes antibiotics and 1 unclassified antibiotic. The results are shown in Table 1. All strains were sensitive to ceftriaxone, amikacin, gentamicin, and ciprofloxacin. None of them was susceptible to ampicillin, novobiocin, cefamandole, and bacitracin. Forty-one strains were also sensitive to tetracycline, nalidixic acid, chloramphenicol, streptomycin, levofloxacin, trimethoprim/sulfamethoxazole, and doxycycline (Table 1). Two strains were observed to be multidrug resistant, demonstrating resistance to three other antibiotics: tetracycline, nalidixic acid, and chloramphenicol. One of them was also resistant to streptomycin and levofloxacin, and the other one to trimethoprim/sulfamethoxazole and doxycycline, respectively. In general, three resistance profiles were observed (Table 1). The most common profile was ampicillin/cefamandole/novobiocin/bacitracin resistance, detected in 95.3% of the strains. One strain (2.3%) demonstrated resistance to ampicillin/cefamandole/novobiocin/bacitracin/tetracycline/nalidixic acid/chloramphenicol/streptomycin/levofloxacin, and one strain (2.3%) was resistant to ampicillin/cefamandole/novobiocin/bacitracin/tetracycline/nalidixic acid/chloramphenicol/trimethoprim/sulfamethoxazole/doxycycline.

Table 1.
Antimicrobial susceptibility of Y. enterocolitica strains isolated from the slaughtered pigs. Three profiles of resistance to the selected antibiotics were observed.
3. Discussion
The study revealed three main findings: (1) low prevalence of pathogenic Y. enterocolitica in healthy pigs from a slaughterhouse in Bulgaria with predominant Y. enterocolitica bioserotype 4/O:3; (2) high genetic similarity of the isolated Y. enterocolitica strains; and (3) three antimicrobial resistance profiles of the isolated Y. enterocolitica strains.
To the best of our knowledge, this is the first five-year study presenting data on the prevalence of Y. enterocolitica in pigs slaughtered in Bulgaria. The tested samples were collected in a single slaughterhouse, and originated from seven farms, located in four regions of Bulgaria. These data could be used for detailed epidemiological analysis of distribution patterns of pathogenic Y. enterocolitica in animals and humans in Bulgaria.
The estimated 6.7% prevalence of pathogenic Y. enterocolitica in healthy slaughtered pigs in this study is relatively lower compared to other European countries, such as Serbia (10.4%) [8], Italy (14% and 27.4%) [19,20], Belgium (23.5%) [9], Latvia and Lithuania (35%) [7], Croatia (43%) [17], Finland (60%) [21], Switzerland (85%) [12], and Spain (93%) [22]. The high prevalence in some countries could be explained by the different isolation methods, different age of the pigs, technical parameters, etc. It is well known that molecular methods are more sensitive and precise in comparison to culture methods [12,23]. Conventional microbiological methods of isolation with enrichment step followed by a PCR method for confirmation reduce the likelihood of false positive results due to dead cells [23,24]. Here, we combined microbiological, biochemical, and PCR methods with enrichment step in Peptone Sorbitol Bile Broth (PSB broth) to ensure the most successful possible isolation and detection of pathogenic Y. enterocolitica strains. To enhance recovery of pathogenic Y. enterocolitica, the enrichment period was reduced from five to two days, and isolation was performed on selective agar after alkali treatment of PSB broth [17,25]. Although we believe that the choice of farm type can also affect the detection of pathogenic Y. enterocolitica, specific types of farms that could be responsible for the occurrence of Y. enterocolitica among pigs were not explored herein and remain to be further researched. Porcine tonsils are well recognized as a source of pathogenic Y. enterocolitica, in view of their lymphoid tissue tropism. In line with previous studies [9,15,21,23], we primarily detected pathogenic Y. enterocolitica in pig tonsils, as well as in pig feces. Different studies have demonstrated that shedding of yersiniae in the feces increases in piglets younger than 30 days and decreases when pigs reach slaughter age [15,26].
Y. enterocolitica is a psychrophilic bacterium and can withstand cold temperatures over long periods of time [2]. Indeed, in the current study Y. enterocolitica strains were isolated during the cold period of the year similarly to other studies reporting isolation of Y. enterocolitica from tonsil samples mostly during cold months [27,28]. However, periodic recurrences of this pathogen (from March to August) cannot be excluded as it has been detected in pigs originated from the Saharan region [29] and from northern regions of Europe [21]. The latter author reported the highest prevalence of Y. enterocolitica in pig feces during July and August, assumed to be related to the higher consumption of pork, and an increased number of yersiniosis cases among humans during the warm period [21].
Sequencing data analysis revealed different similarity of our strains to the 16S rRNA nucleotide sequence of Y. enterocolitica strain KNG22703, complete genome (GenBank accession number: CP011286.1), and the 16S rRNA gene, partial sequence originated from uncultured bacteria clone 05-951_IBD.37307 (GenBank accession number: GQ965064.1). A very high similarity (98–99%) was established for most isolated strains in this study. Some of the isolated strains, such as Y. enterocolitica strains 18, 28, and 43, demonstrated similarities to KNG22703 of 97%, 93%, and 95%, respectively. Although small, the deviation of similarity among our isolates and the reference strain KNG22703 may be due to the different origin of the isolates, as KNG22703 has been adapted to humans.
Like other authors, we have shown that the most commonly isolated biotype among pigs is Y. enterocolitica biotype 4 [9,15,19,21,30], with the O:3 serotype confirmed in all 43 isolates. Moreover, in line with similar studies, we support the hypothesis of serotype O:3 predominance within biotype 4 of pig isolates. Y. enterocolitica 4/O:3 seems to be the most frequently detected and isolated bioserotype from healthy pigs in European counties [8,19,21], with the exception of the United Kingdom [27]. Our study identified Y. enterocolitica 4/O:3 as the only bioserotype isolated among healthy pigs in a slaughterhouse in Bulgaria. The prevalence of bio/serotype 4/O:3 poses a risk to human health, as this pathogen can easily enter the food chain through meat processing, spread to consumers, and eventually cause yersiniosis in humans [3]. Thus, our findings underline the importance of pigs in the epidemiology of yersiniosis.
The ail protein and yersiniabactin are important factors of virulence in Y. enterocolitica, both chromosomally encoded. Genes responsible for their expression—ail and ystA, respectively—are one of the most commonly used chromosomal targets for determination of pathogenicity. In our study, all genetically proven Y. enterocolitica strains harbored the genes of virulence ail and ystA, indicating a high pathogenic potential of the pig Y. enterocolitica isolates. Similarly, other studies identified ystA and ail genes with a high rate of presence among isolated Y. enterocolitica [8,19,20,30]. Of note, the presence of pYV is based on the detection of different genes, and the complete virulence of Y. enterocolitica depends on plasmid availability as well. The pYV is unstable and its detection is faced with some difficulties. However, many authors use yadA gene detection for plasmid confirmation [20,29]. We detected the plasmid-borne gene yadA by both PCR and culture methods, and, as expected, the PCR method revealed a higher detection rate.
It seems that Y. enterocolitica 4/O:3 is a less genetically diverse bioserotype. Indeed, we found a high degree of similarity between the macro-restriction pulsotypes of the Y. enterocolitica strains, suggesting a pronounced genetic homogeneity among the population of this species with only few isolates distinguishable from the predominant genotype. Our study found a 100% similarity within two clusters. This finding is in accordance with other studies which indicate a high degree of similarity and a minor genetic variation among 4/O:3 bioserotype strains, restriction enzyme applied (SpeI, NotI, or XbaI) [8,19,30]. In previous studies, we found that the genome of Y. enterocolitica is the most stable compared to the other two pathogenic species, Y. pestis and Y. pseudotuberculosis [31]. Studies on the genomic stability of Y. enterocolitica strains isolated from different countries of the world (belonging to bioserotype 4/O:3 and using the NotI restriction enzyme) revealed higher homogeneity of this bioserotype compared to serotypes O:5 and O:9 [32]. Obviously, the geographic location is an important factor for the spread of a given pulsotype and enables the emergence of new branches. The high genetic similarity observed in our study suggests well pronounced homogeneity and conservation in the genome structure of the strains. The majority of strains belong to close pulsotypes, pointing to the wide distribution of one genotype of Y. enterocolitica 4/O:3. The results showed the predominance of this genotype for all observed farms, and its persistence over time, indicating low genetic variation. The few other PFGE pulsotypes detected could be contamination with these genotypes on a farm level. Clustering with 100% similarity of Y. enterocolitica 4/O:3 isolated from palatine tonsils confirmed the relevance of palatine tonsils for direct Y. enterocolitica contamination in carcasses. [10]. The observed high similarity could be a disadvantage when tracking a possible outbreak because of the impossibility to establish a relationship between the isolated strain and the epidemiological outbreak [33]. Nevertheless, future studies should take into account circulating porcine Y. enterocolitica 4/O:3 genotypes, applying more discriminatory analyses.
The overuse of antibiotics in veterinary medicine as growth promoters in farm animals, including pigs, amplifies the significance of antimicrobial resistant Y. enterocolitica. Three profiles of resistance were detected among the isolated Y. enterocolitica. Our results showed that all tested strains were resistant to ampicillin, novobiocin, cefamandole, and bacitracin. Resistance to ampicillin is commonly reported for Y. enterocolitica strains isolated from pigs [8,10,15]. Since ampicillin belongs to the β-lactam penicillines, the resistance of Y. enterocolitica depends either on β-lactamase enzyme (BlaA and BlaB) production or on mutations of genes responsible for affinity to penicillin binding protein [34]. However, neither ampicillin, nor first-generation cephalosporin resistance of Y. enterocolitica O:3 is associated with plasmid presence [34], but probably with a mutation in the chromosome-coding genes, blaA and blaB, which deserves further investigation in detail. In 2 out of the 43 isolated Y. enterocolitica strains, we detected resistance also to tetracycline, nalidixic acid, and chloramphenicol, and only in 1 (out of 43) to streptomycin, trimethoprim/sulfamethoxazole, levofloxacin, and doxycycline. Unlike our data, resistance to sulfamethoxazole and streptomycin of Y. enterocolitica isolated from pigs and humans is continuously reported [10,17,20]. In line with our findings, resistance to chloramphenicol has not been frequently found [35], which contradicts other reports on the high rate of Y. enterocolitica resistance to chloramphenicol among slaughtered pigs in Northern Italy and Croatia [17,20,36], and among Y. enterocolitica clinical isolates that appeared during the time of the Swedish outbreaks [37]. There is some controversy regarding the resistance to tetracycline as well. Some authors reported none or a low proportion of isolates resistant to tetracycline [7,10,15,38], while others confirmed higher resistance to tetracycline of Y. enterocolitica strains [39]. A possible explanation could be associated with both chromosomal and plasmid-encoded mechanisms [37]. Nalidixic acid-resistant Y. enterocolitica has been frequently reported among pig isolates with resistance rates higher than the rates in our study, varying between 31% in Croatia, 49.1% in Northern Italy, and 62.5% in Malaysia [17,20,39]. It is known that the resistance to levofloxacin and to fluoroquinolones is rare [17] and could be reached by mutations of topoisomerase genes, which are targets of fluoroquinolones [40]. Overall, the resistance to quinolones is mediated by both chromosome- and plasmid-related mechanisms [16]. The resistance profiles against frequently used antibiotics found in our study are worrisome and should not be underestimated. Multi-resistant Y. enterocolitica strains can be a serious threat to human health after transmission in the food chain and food contamination.
4. Materials and Methods
4.1. Animals and Samples
The present study was conducted over a period of five years, spanning from January 2016 to December 2021. A total of 601 pigs were examined during the course of the sampling periods. Specifically, palatine tonsils (veli palatini) were collected from each of the 601 pigs. Both tonsils were aseptically removed immediately after evisceration using a sterile surgical blade. Additionally, a total of 189 fecal samples were collected from a subset of the slaughtered pigs (189 pigs only) and were aseptically harvested after colon incision. All samples were placed in sterile plastic bags, transported to the laboratory in a cooler bag at 4 °C and processed within 4 h after collection. Sampling was carried out during the slaughtering process in a single slaughterhouse located in Kostinbrod city (Sofia region), which serves pig farms across the country. The number of samples collected per visit in the slaughterhouse varied depending on the batch, with a target of collecting 50% of the total number of slaughtered pigs for the day. Importantly, all samples collected per day originated from a single farm. The study population comprised pigs from seven fattening pig farms (I, II, III, IV, V, VI, and VII) distributed across four different regions of Bulgaria, including Razgrad, Shumen, Stara Zagora, and Sofia Region. Specifically, farms I, II, and III were located in Razgrad, Shumen, and Stara Zagora, respectively, while farms IV, V, VI, and VII were located in the Sofia Region,. Sampling was conducted during two distinct periods: a cold period and a warm period. A total of 18 visits were conducted during the cold period and 8 visits during the warm period. The sampling period was chosen after taking into account the minimal and maximal monthly temperature of the cold period from October to March, and of the warm period from April to September.
4.2. Microbiological and Biochemical Tests for Detection of Yersinia enterocolitica
The presence of pathogenic Y. enterocolitica was detected according to ISO 10273:2003 [41] with some modifications, aiming to increase the detection efficacy of the test [25]. Briefly, tonsil tissue from each pig was aseptically cut into small pieces. A piece of each sample (tonsils and feces), approximately 12 g in weight from each, was suspended in peptone sorbitol bile salt (PSB) broth (Himedia, Mumbai, India) in a mass/volume ratio of 1:10. The suspension was homogenized for 4 min in a stomacher (Stomacher 80 Biomaster Lab, Seward, Worthing, UK). The enrichment period in PSB broth was reduced to 48 h at 28 °C. After alkali treatment with 4.5 mL 0.05% KOH solution and 0.5 mL PSB homogenate, for 20 s, a 10 µL aliquot of each sample was streaked onto Yersinia selective base agar (Oxoid, Hampshire, UK) supplemented with Cefsulodin-Irgasan-Novobiocin (CIN agar) (Oxoid, UK). The plates were incubated at 28 °C for 48 h. Small, smooth, flat colonies exhibiting a transparent appearance with a red central zone, commonly known as “bull’s eye” were selected for subculturing on Tryptic Soy agar (TSA, Difco, Atlanta, GA, USA) for further analysis. Preliminary identification included tests for urea hydrolysis and phenylalanine deamination on Christensen agar (Merck, Darmstadt, Germany) and Phenylalanine agar (Himedia, India), respectively, and tests for presence of catalase and cytochrome C oxidase. Briefly, urea positive, phenylalanine negative, catalase positive, and oxidase negative colonies were deemed presumptive Y. enterocolitica. Presumptive Y. enterocolitica were further identified biochemically by Microlatest ENTEROtest 24N (Erba Lacherna, Brno, Czech Republic), according to manufacturer’s instruction. The identification scheme included a test for: determination of arginine, lysine and ornithine decarboxylation, fermentation of carbohydrates, such as sucrose, lactose, trehalose, rhamnose, rafinose, melibiose, hydrolysis of citrate, production of hydrogen sulfide and utilization of manithol, arabitol, inositol. To confirm their identity, the colonies underwent 16S rRNA gene identification.
4.3. DNA Isolation
Pure colonies of biochemically identified Y. enterocolitica were collected after overnight cultivation on TSA, and DNA extraction was completed using GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA). Extracted DNA was quantified by a spectrophotometer (Quawell, Labgene Scientific SA, Châtel-Saint-Denis, Switzerland) and analyzed by gel electrophoresis. The DNA eluate in an appropriate amount was used as a template in PCR assays.
4.4. 16S rRNA Gene Identification
The Yersinia 16S rRNA gene was detected by genus-specific primers followed by DNA sequencing performed to all biochemically identified Y. enterocolitica isolates (n = 136). PCR was performed using Phire Green Hot start II PCR Master Mix (Thermo Fisher Scientific Baltics, Vilnius, Lithuania). The total reaction volume was 30 µL, containing 1.5 μL of each primer (with a final concentration of 0.5 µM), nuclease-free water (Thermo Fisher Scientific Baltics, Lithuania) and 25 ng/µL DNA. The amplifications were performed in a Thermal Cycler (Bio-Rad, Hercules, CA, USA), with details of the primer sequences, annealing temperature, and PCR conditions, as described in Table 2. DNA extracted from Y. enterocolitica 8081 (O:8) was used as a positive control, while master mix with HPLC water was used as a negative control. The PCR products (8 µL) were subjected to electrophoresis in a 1.5% agarose gel in 1% TBE and stained with PeqGreen (Peqlab Biotech, Hafenstr, Germany). For the positive control, DNA extracted from Y. enterocolitica 8081 (O:8) was used, and the negative control was a master mix with HPLC water. The positive PCR products for the 16S rRNA gene were sequenced in both directions (Macrogene, Amsterdam, The Netherlands) and the sequenced data were compared with reference sequences of Y. enterocolitica in the database of the National Center for Biotechnology Information (NCBI) amplicons using Basic Local Alignment Search Tool (BLAST).
4.5. Biotyping and Serotyping Methods
Yersinia enterocolitica isolates confirmed by 16S rRNA analysis were biotyped according to the biotyping scheme of ISO 10273:2003 using the reactions of: trehalose, tween esterase/lipase, pyrazinamidase, esculin/salicin, and indole [41]. Esculin/salicin hydrolysis and trehalose utilization were detected on Microlatest ENTEROtest 24N (Erba Lacherna, Czech Republic). Pyrazinamidase activity and tween esterase/lipase activity were assessed using pyrazinamidase agar (Himedia, India) and tween esterase test agar base (Himedia, India), supplemented with Tween 80 (Sigma Aldrich, Taufkirchen, Germany). Indole production was detected from trypthophan deamination using Kovacs’ Indole Reagent (Himedia, India). The serotype was determined by slide agglutination with the use of antisera for somatic antigens O:3 (Sifin, Berlin, Germany), O:5, O:8, and O:9 (BB NCIPD Ltd., Sofia, Bulgaria). Physiological saline was used as a negative control.
4.6. Phenotypic Test for Detection of Virulent Plasmid
The presence of pYV (plasmid Yersinia virulence) was studied by both absorptions of congo red in the Congo red-magnesium oxalate agar (CR-MOX) (Himedia, India) and autoagglutination on TSB (Difco, USA). CR-MOX agar plates were incubated at 28 °C and 37 °C for 24 h to 48 h and autoagglutination was performed at 28 °C and 37 °C as well.
4.7. PCR for Detection of Y. enterocolitica Virulence Genes
The genes encoding virulence determinants, such as the attachment-invasion locus (ail), and Yersinia heat-stable enterotoxin (ystA) and the plasmid-borne Yersinia adhesin (yadA) were analyzed by a PCR assay. Reactions were carried out using 2× PCR Buffer EURx Taq PCR Master Mix (containing 1.25 U Taq DNA polymerase, 1.5 mM MgCl2, 0.2 mM of each dNTPs, EURx, Poland), with a final concentration of 0.5 µM of each primer, 0.2 µg of DNA, and nuclease-free water. The primer sequences, annealing temperature, and PCR conditions are shown in Table 2. DNA extracted from Y. enterocolitica 8081 (O:8) pYV+ was used as a positive control and a master mix with HPLC water as a negative control.
4.8. Pulsed-Field Gel Electrophoresis
Macro-digestion of isolated pathogenic Y. enterocolitica strains (n = 43) was performed using the restriction enzyme SpeI (Sigma-Aldrich, Schnelldorf, Germany). PFGE was performed according to the PulseNet protocol for Y. pestis [42]. Bacteria grown for 24 h on TSB were centrifuged at 10,000× g for 10 min. Pellet was re-suspended in 2 mL Cell Suspension Buffer (100 mM Tris:100 mM EDTA, pH 8.0, Sigma-Aldrich, Schnelldorf, Germany) and bacterial concentration was adjusted by a spectrophotometer (Cole-Parmer, Vernon Hills, IL, USA) to optical density 1.3–1.4, measured at 610 nm wavelength. Agarose plugs were prepared with 200 µL bacterial suspensions, 10 µL proteinase K (0.5 mg/mL) (Sigma Aldrich, Germany), and 200 µL melted agarose (1% SeaKem Gold:1% SDS, Lonza, Bend, OR, USA and Thermo Fisher Scientific, Waltham, MA, USA, respectively). Cell lysis was performed with Cell Lysis/Proteinase K Buffer (50 mM Tris:50 mM EDTA, pH 8.0 + 1% Sarcosyl, Sigma Aldrich, Germany) and Proteinase K (final concentration of 0.1 mg/mL), followed by incubation at 54 °C for 2 h on a shaker water bath with constant and vigorous agitation (180 rpm). In order to inactivate proteinase K, the plugs were washed twice with sterile ultrapure water containing 1 mM Phenyl-methyl-sulphonyl-fluoride (PMSF) (Sigma-Aldrich, USA), followed by three washes in TE buffer (10 mM Tris-borate and 1 mM EDTA, pH 8.0). Washing steps were completed at 54 °C on a shaker bath. Plugs were subjected to individual restriction with SpeI (20 U/100 µL) for 18 h at 37 °C. Restriction fragments were separated by using Contour-clamped homogeneous electric field—Chef DR II system (Bio-Rad, USA) with 0.5× TBE buffer (45 mM Tris-borate and 1 mM EDTA, pH 8.3, Serva, Heidelberg, Germany) for 24 h at 16 °C, initial pulse of 2 s, final pulse 20 s with a voltage of 5 V/cm. Afterwards, the gels were stained with ethidium bromide (0.5 µg/mL) for 45 min, then de-stained in water for 1 h. Pulse marker (50–1000 kb DNA ladder, Sigma-Aldrich, USA) was applied as a molecular size standard. PFGE pulsotypes were analyzed using GelCompar software (Applied Maths, Keistraat, Brussels, Belgium). Clustering of strains was performed by unweighted pair-group method considering using arithmetic averages (UPGMA) with Dice bands correlation tolerance set on 2.5% and optimization of 0.5%.
4.9. Determination of the Antimicrobial Susceptibility of Y. enterocolitica Strains
Antimicrobial susceptibility tests were carried out by the standard disk diffusion method according to CLSI recommendations [18]. Standardized to 0.5 McFarland bacterial suspensions were plated on Muller-Hinton agar (Himedia, India) and commercial antimicrobial disks (BB-NCPID Ltd., Sofia, Bulgaria) were applied. Pathogenic Y. enterocolitica strains were tested for susceptibility to 15 antibiotics, belonging to different pharmacological classes (8 classes antibiotics and 1 unclassified antibiotic, according to [18]): tetracycline (30 µg), doxycycline (30 µg), amikacin (30 µg), gentamicin (10 µg), streptomycin (10 µg), ampicillin (10 µg), cefamandole (30 µg), ceftriaxone (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), nalidixic acid (30 µg), chloramphenicol (30 µg), trimethoprim/sulfamethoxazole (1.25/23.75 µg), bacitracin (0.07 U), and novobiocin (5 µg). The diameter of Inhibition zone for each antibiotic was measured after incubation of the plates at 28 °C for 24 h. Results were interpreted as susceptible, intermediate, or resistant strains [43].

Table 2.
Primers are used for PCR amplification.
Table 2.
Primers are used for PCR amplification.
| Target Gene | Primers | Tm °C | Sequence (5′ to 3′) | Amplicon Length (bp) | Reference | PCR Conditions |
|---|---|---|---|---|---|---|
| 16S rRNA Yersinia enterocolitica | YeI-6SrRNA YeII-6SrRNA | 48 °C 45 °C | ATACCGCATAACGTCTTCG TTCTTCTGCGAGTAACGTC | 328 | [44] | 95 °C for 5 min 35 cycles 94 °C for 30 s 47 °C for 30 s 72 °C for 1 min 72 °C for 10 min |
| ail | F-real 10A R-real 9A | 55 °C 55 °C | ATGATAACTGGGGAGTAATAGGTTCG CCCAGTAATCCATAAAGGCTAACATAT | 163 | [45] | 95 °C for 5 min 35 cycles 94 °C for 30 s 55 °C for 45 s 72 °C for 1 min 72 °C for 10 min |
| ystA | ystA-F ystA-R | 63 °C 57 °C | AATGCTGTCTTCATTTGGAGC ATCCCAATCACTACTGACTTC | 145 | [29] | 95 °C for 5 min 35 cycles 94 °C for 30 s 60 °C for 45 s 72 °C for 1 min 72 °C for 10 min |
| yadA | yadA-F yadA-R | 63 °C 63 °C | CTTCAGATACTGGTGTCGCTGT ATGCCTGACTAGAGCGATATCC | 849 | [29] | 95 °C for 5 min 35 cycles 94 °C for 30 s 63 °C for 45 s 72 °C for 1 min 72 °C for 10 min |
5. Conclusions
Our results showed that 6.7% of the tested pigs slaughtered in Bulgaria were infected with pathogenic Y. enterocolitica strains with predominance of bioserotype 4/O:3. The isolated strains had high genetic similarity and most of them were sensitive to clinically important antibiotics. Our study indicated the significance of tonsils as predilection sites of pathogenic Y. enterocolitica, and the role of pigs as carriers of this zoonotic pathogen, suggesting the need of a surveillance system for monitoring Y. enterocolitica at the slaughterhouse level.
Author Contributions
Conceptualization, M.A. and H.N.; methodology, M.A., M.M.Z., L.L.D., T.D. and I.G.; software, Z.U.; investigation, M.A., M.M.Z., L.L.D., T.D., I.G., Y.I., M.D.K., T.C.K., S.N. and Z.D.; data curation, M.A.; writing—original draft preparation, M.A.; writing—review and editing, M.A., T.D., Z.U. and H.N.; supervision, H.N. All authors have read and agreed to the published version of the manuscript.
Funding
Studies on the prevalence of Yersinia enterocolitica in the intestinal tract of pigs and antimicrobial resistance were funded by Bulgarian National Science Fund, Ministry of Education and Science, Republic of Bulgaria, grant number KP-06-N36/7. The genetic studies were partially financed by The National Scientific Program “Healthy Food for Strong Bioeconomy and Quality of Life”, Ministry of Education and Science, Republic of Bulgaria, contract number: 577/17.08.2018.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The research team acknowledges the project BG05M2OP001-1.002-0019: “Clean Technologies for Sustainable Environment—Waters, Waste, Energy for Circular Economy” funded by the Operational Program “Science and Education for Smart Growth”, co-financed by the European Union through the European Structural and Investment Funds, for providing the PCR equipment used in this study. A special acknowledgement should be given to Iva Tsvetkova and Trayana Draganova for their technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Le Guern, A.-S.; Savin, C.; Angermeier, H.; Brémont, S.; Clermont, D.; Mühle, E.; Orozova, P.; Najdenski, H.; Pizarro-Cerdá, J. Yersinia artesiana sp. nov., Yersinia proxima sp. nov., Yersinia alsatica sp. nov., Yersina vastinensis sp. nov., Yersinia thracica sp. nov. and Yersinia occitanica sp. nov., isolated from humans and animals. Int. J. Syst. Evol. Microbiol. 2020, 70, 5363. [Google Scholar] [CrossRef] [PubMed]
- Bottone, E.J. Yersinia enterocolitica: Revisitation of an enduring human pathogen. Clin. Microbiol. Newsl. 2015, 37, 1–8. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- Virdi, J.S.; Sachdeva, P. Molecular heterogeneity in Yersinia enterocolitica and ‘Y. enterocolitica-like’species—Implications for epidemiology, typing and taxonomy. FEMS Immunol. Med. Microbiol. 2005, 45, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Marimon, J.; Figueroa, R.; Idigoras, P.; Gomariz, M.; Alkorta, M.; Cilla, G.; Pérez-Trallero, E. Thirty years of human infections caused by Yersinia enterocolitica in northern Spain: 1985–2014. Epidemiol. Infect. 2017, 145, 2197–2203. [Google Scholar] [CrossRef] [PubMed]
- Morka, K.; Wałecka-Zacharska, E.; Schubert, J.; Dudek, B.; Woźniak-Biel, A.; Kuczkowski, M.; Wieliczko, A.; Bystroń, J.; Bania, J.; Bugla-Płoskońska, G. Genetic diversity and distribution of virulence-associated genes in Y. enterocolitica and Y. enterocolitica-like isolates from humans and animals in Poland. Pathogens 2021, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Terentjeva, M.; Ķibilds, J.; Gradovska, S.; Alksne, L.; Streikiša, M.; Meistere, I.; Valciņa, O. Prevalence, virulence determinants, and genetic diversity in Yersinia enterocolitica isolated from slaughtered pigs and pig carcasses. Int. J. Food Microbiol. 2022, 376, 109756. [Google Scholar] [CrossRef]
- Arsić, M.; Vićić, I.; Galić, N.; Dmitrić, M.; Kureljušić, J.; Dimitrijević, M.; Petrović, M.; Šarić, L.; Karabasil, N. Risk factors and the overall characterization of Yersinia enterocolitica as an initial model of pathogen surveillance in the pig production system in Serbia. Res. Vet. Sci. 2022, 152, 167–174. [Google Scholar] [CrossRef]
- Van Damme, I.; Berkvens, D.; Vanantwerpen, G.; Baré, J.; Houf, K.; Wauters, G.; De Zutter, L. Contamination of freshly slaughtered pig carcasses with enteropathogenic Yersinia spp.: Distribution, quantification and identification of risk factors. Int. J. Food Microbiol. 2015, 204, 33–40. [Google Scholar] [CrossRef]
- Martins, B.T.F.; Botelho, C.V.; Silva, D.A.L.; Lanna, F.G.P.A.; Grossi, J.L.; Campos-Galvão, M.E.M.; Yamatogi, R.S.; Falcão, J.P.; dos Santos Bersot, L.; Nero, L.A. Yersinia enterocolitica in a Brazilian pork production chain: Tracking of contamination routes, virulence and antimicrobial resistance. Int. J. Food Microbiol. 2018, 276, 5–9. [Google Scholar] [CrossRef]
- Vanantwerpen, G.; Van Damme, I.; De Zutter, L.; Houf, K. Within-batch prevalence and quantification of human pathogenic Yersinia enterocolitica and Y. pseudotuberculosis in tonsils of pigs at slaughter. Vet. Microbiol. 2014, 169, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M.; Stolle, A.; Stephan, R. Prevalence of pathogenic Yersinia enterocolitica in pigs slaughtered at a Swiss abattoir. Int. J. Food Microbiol. 2007, 119, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Bancerz-Kisiel, A.; Pieczywek, M.; Łada, P.; Szweda, W. The most important virulence markers of Yersinia enterocolitica and their role during infection. Genes 2018, 9, 235. [Google Scholar] [CrossRef] [PubMed]
- Bhaduri, S.; Smith, J.L. Virulence plasmid (pYV)-associated expression of phenotypic virulent determinants in pathogenic Yersinia species: A convenient method for monitoring the presence of pYV under culture conditions and its application for isolation/detection of Yersinia pestis in food. J. Pathog. 2011, 2011, 727313. [Google Scholar] [CrossRef] [PubMed]
- Fois, F.; Piras, F.; Torpdahl, M.; Mazza, R.; Ladu, D.; Consolati, S.G.; Spanu, C.; Scarano, C.; De Santis, E.P. Prevalence, bioserotyping and antibiotic resistance of pathogenic Yersinia enterocolitica detected in pigs at slaughter in Sardinia. Int. J. Food Microbiol. 2018, 283, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fàbrega, A.; Ballesté-Delpierre, C.; Vila, J. Antimicrobial resistance in Yersinia enterocolitica. In Antimicrobial resistance and food safety; Elsevier: Amsterdam, The Netherlands, 2015; pp. 77–104. [Google Scholar]
- Zdolec, N.; Kiš, M.; Jankuloski, D.; Blagoevska, K.; Kazazić, S.; Pavlak, M.; Blagojević, B.; Antić, D.; Fredriksson-Ahomaa, M.; Pažin, V. Prevalence and Persistence of Multidrug-Resistant Yersinia enterocolitica 4/O: 3 in Tonsils of Slaughter Pigs from Different Housing Systems in Croatia. Foods 2022, 11, 1459. [Google Scholar] [CrossRef]
- Clinical and Labortory Standards Institute (CLSI). CLSI Supplement M100. In Performance Standard for Antimicrobial Susceptibility Testing, 31st ed.; Clinical and Labortory Standards Institute (CLSI): Wayne, PA, USA, 2021. [Google Scholar]
- Sacchini, L.; Garofolo, G.; Di Serafino, G.; Marotta, F.; Ricci, L.; Di Donato, G.; Miracco, M.G.; Perletta, F.; Di Giannatale, E. The prevalence, characterisation, and antimicrobial resistance of Yersinia enterocolitica in pigs from Central Italy. Vet. Ital. 2018, 54, 115–123. [Google Scholar] [CrossRef]
- Bonardi, S.; Bruini, I.; D’incau, M.; Van Damme, I.; Carniel, E.; Brémont, S.; Cavallini, P.; Tagliabue, S.; Brindani, F. Detection, seroprevalence and antimicrobial resistance of Yersinia enterocolitica and Yersinia pseudotuberculosis in pig tonsils in Northern Italy. Int. J. Food Microbiol. 2016, 235, 125–132. [Google Scholar] [CrossRef]
- Ibañez, T.R.; Laukkanen-Ninios, R.; Hakkinen, M.; Johansson, T.; Vilar, M.; Korkeala, H. Prevalence of pathogenic Yersinia enterocolitica in finnish slaughter pigs. J. Food Prot. 2016, 79, 677–681. [Google Scholar] [CrossRef]
- Martinez, P.O.; Fredriksson-Ahomaa, M.; Pallotti, A.; Rosmini, R.; Houf, K.; Korkeala, H. Variation in the prevalence of enteropathogenic Yersinia in slaughter pigs from Belgium, Italy, and Spain. Foodborne Pathog. Dis. 2011, 8, 445–450. [Google Scholar] [CrossRef]
- Mazzette, R.; Fois, F.; Consolati, S.G.; Salza, S.; Tedde, T.; Soro, P.; Collu, C.; Ladu, D.; Virgilio, S.; Piras, F. Detection of pathogenic Yersinia enterocolitica in slaughtered pigs by cultural methods and real-time polymerase chain reaction. Ital. J. Food Saf. 2015, 4, 4579. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M.; Korkeala, H. Low occurrence of pathogenic Yersinia enterocolitica in clinical, food, and environmental samples: A methodological problem. Clin. Microbiol. Rev. 2003, 16, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, I.; Habib, I.; De Zutter, L. Yersinia enterocolitica in slaughter pig tonsils: Enumeration and detection by enrichment versus direct plating culture. Food Microbiol. 2010, 27, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Nesbakken, T.; Iversen, T.; Eckner, K.; Lium, B. Testing of pathogenic Yersinia enterocolitica in pig herds based on the natural dynamic of infection. Int. J. Food Microbiol. 2006, 111, 99–104. [Google Scholar] [CrossRef]
- Martínez, P.O.; Mylona, S.; Drake, I.; Fredriksson-Ahomaa, M.; Korkeala, H.; Corry, J.E. Wide variety of bioserotypes of enteropathogenic Yersinia in tonsils of English pigs at slaughter. Int. J. Food Microbiol. 2010, 139, 64–69. [Google Scholar] [CrossRef]
- Bonardi, S.; Bassi, L.; Brindani, F.; D’Incau, M.; Barco, L.; Carra, E.; Pongolini, S. Prevalence, characterization and antimicrobial susceptibility of Salmonella enterica and Yersinia enterocolitica in pigs at slaughter in Italy. Int. J. Food Microbiol. 2013, 163, 248–257. [Google Scholar] [CrossRef]
- Saraka, D.; Savin, C.; Kouassi, S.; Cissé, B.; Koffi, E.; Cabanel, N.; Brémont, S.; Faye-Kette, H.; Dosso, M.; Carniel, E. Yersinia enterocolitica, a neglected cause of human enteric infections in Cote d’Ivoire. PLoS Negl. Trop. Dis. 2017, 11, e0005216. [Google Scholar] [CrossRef]
- Martins, B.T.F.; de Azevedo, E.C.; Yamatogi, R.S.; Call, D.R.; Nero, L.A. Persistence of Yersinia enterocolitica bio-serotype 4/O: 3 in a pork production chain in Minas Gerais, Brazil. Food Microbiol. 2021, 94, 103660. [Google Scholar] [CrossRef]
- Najdenski, H.; Iteman, I.; Carniel, E. The genome of Yersinia enterocolitica is the most stable of the three pathogenic species. Contrib. Microbiol. Immunol. 1995, 13, 281–284. [Google Scholar]
- Najdenski, H.; Iteman, I.; Carniel, E. Efficient subtyping of pathogenic Yersinia enterocolitica strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 1994, 32, 2913–2920. [Google Scholar] [CrossRef]
- Raymond, P.; Houard, E.; Denis, M.; Esnault, E. Diversity of Yersinia enterocolitica isolated from pigs in a French slaughterhouse over 2 years. Microbiologyopen 2019, 8, e00751. [Google Scholar] [CrossRef] [PubMed]
- Bent, Z.W.; Young, G.M. Contribution of BlaA and BlaB β-lactamases to antibiotic susceptibility of Yersinia enterocolitica biovar 1B. Antimicrob. Agents Chemother. 2010, 54, 4000–4002. [Google Scholar] [CrossRef] [PubMed]
- Schneeberger, M.; Brodard, I.; Overesch, G. Virulence-associated gene pattern of porcine and human Yersinia enterocolitica biotype 4 isolates. Int. J. Food Microbiol. 2015, 198, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, S.; Alpigiani, I.; Pongolini, S.; Morganti, M.; Tagliabue, S.; Bacci, C.; Brindani, F. Detection, enumeration and characterization of Yersinia enterocolitica 4/O: 3 in pig tonsils at slaughter in Northern Italy. Int. J. Food Microbiol. 2014, 177, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, P.A.; Tano, E.; Jernberg, C.; Hickman, R.A.; Guy, L.; Järhult, J.D.; Wang, H. Molecular characterization of multidrug-resistant Yersinia enterocolitica from foodborne outbreaks in Sweden. Front. Microbiol. 2021, 12, 664665. [Google Scholar] [CrossRef] [PubMed]
- Gkouletsos, T.; Patas, K.; Lambrinidis, G.; Neubauer, H.; Sprague, L.; Ioannidis, A.; Chatzipanagiotou, S. Antimicrobial resistance of Yersinia enterocolitica and presence of plasmid pYV virulence genes in human and animal isolates. New Microbes New Infect. 2019, 32, 100604. [Google Scholar] [CrossRef] [PubMed]
- Thong, K.L.; Tan, L.K.; Ooi, P.T. Genetic diversity, virulotyping and antimicrobial resistance susceptibility of Yersinia enterocolitica isolated from pigs and porcine products in Malaysia. J. Sci. Food Agric. 2018, 98, 87–95. [Google Scholar] [CrossRef]
- Hooper, D.C. Mechanisms of action of antimicrobials: Focus on fluoroquinolones. Clin. Infect. Dis. 2001, 32, S9–S15. [Google Scholar] [CrossRef]
- ISO. Microbiology of Food and Animal Feeding Stuffs-Horizontal Method for the Detection of Presumptive Pathogenic Yersinia enterocolitica; ISO (International Organization for Standardization): London, UK, 2003. [Google Scholar]
- Centers for Disease Control and Prevention. One-Day (24–28 h) Standardized Laboratory. Protocol for Molecular Subtyping of Yersinia pestis by Pulsed Field Gel Electrophoresis (PFGE); Pulse Net: Atlanta, GA, USA, 2006. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint Tables for Interpretation of MICs and Zone Diameters. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_12.0_Breakpoint_Tables.pdf (accessed on 17 February 2023).
- Arnold, T.; Neubauer, H.; Nikolaou, K.; Roesler, U.; Hensel, A. Identification of Yersinia enterocolitica in minced meat: A comparative analysis of API 20E, Yersinia identification kit and a 16S rRNA-based PCR method. J. Vet. Med. Ser. B 2004, 51, 23–27. [Google Scholar] [CrossRef]
- Lambertz, S.T.; Nilsson, C.; Hallanvuo, S.; Lindblad, M. Real-time PCR method for detection of pathogenic Yersinia enterocolitica in food. Appl. Environ. Microbiol. 2008, 74, 6060–6067. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).