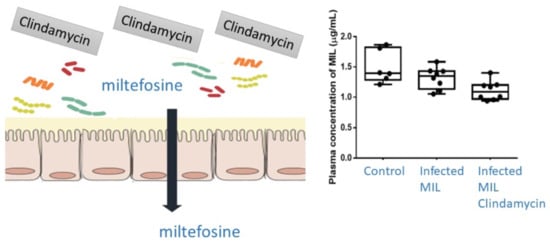
Effect of Clindamycin on Intestinal Microbiome and Miltefosine Pharmacology in Hamsters Infected with Leishmania infantum
Abstract
1. Introduction
2. Results
2.1. Orally Administered Miltefosine (MIL) Does Not Significantly Modify the Main Composition of the Intestinal Microbiome of Syrian Hamsters Infected with Leishmania infantum
2.2. Clindamycin Elicits a Deep Although Transient Modification of the Intestinal Microbiome of Hamsters Infected with Leishmania infantum and Treated with Miltefosine (MIL)
2.3. Pharmacokinetics (PK) and Biodistribution of Miltefosine (MIL)
3. Discussion
4. Materials and Methods
4.1. Chemicals and Drugs
4.2. Parasites and Hamsters
4.3. Follow-Up and Assessment of Infection
4.4. Determination of Miltefosine in Plasma and Target Organs’ Samples
4.5. Genomic Analysis of Microbiota
4.5.1. Fecal Samples, DNA Extraction, 16S Metagenome Library Construction and NGS Sequencing
4.5.2. Sequence Data Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Expert Committee on the Control of the Leishmaniases & World Health Organization. Control of the Leishmaniases: Report of a Meeting of the WHO Expert Commitee on the Control of Leishmaniases, Geneva, Switzerland, 22–26 March 2010. Available online: https://apps.who.int/iris/handle/10665/44412 (accessed on 22 October 2022).
- Alvar, J.; Vélez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; den Boer, M.; WHO Leishmaniasis Control Team. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef] [PubMed]
- Pace, D. Leishmaniasis. J. Infect. 2014, 69, 510–518. [Google Scholar] [CrossRef] [PubMed]
- WHO. Leishmaniasis. 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 29 January 2023).
- Naucke, T.J.; Amelung, S.; Lorentz, S. First report of transmission of canine leishmaniosis through bite wounds from a naturally infected dog in Germany. Parasit. Vectors 2016, 9, 256. [Google Scholar] [CrossRef] [PubMed]
- Campos, R.; Santos, M.; Tunon, G.; Cunha, L.; Magalhães, L.; Moraes, J.; Ramalho, D.; Lima, S.; Pacheco, J.A.; Lipscomb, M.; et al. Epidemiological aspects and spatial distribution of human and canine visceral leishmaniasis in an endemic area in northeastern Brazil. Geospatial Health 2017, 12, 67–73. [Google Scholar] [CrossRef]
- Wamai, R.G.; Kahn, J.; McGloin, J.; Ziaggi, G. Visceral leishmaniasis: A global overview. J. Glob. Health Sci. 2020, 2, e3. [Google Scholar] [CrossRef]
- Morales-Yuste, M.; Martín-Sánchez, J.; Corpas-Lopez, V. Canine Leishmaniasis: Update on epidemiology, diagnosis, treatment, and prevention. Vet. Sci. 2022, 9, 387. [Google Scholar] [CrossRef]
- Koch, L.K.; Kochmann, J.; Klimpel, S.; Cunze, S. Modeling the climatic suitability of leishmaniasis vector species in Europe. Sci. Rep. 2017, 7, 13325. [Google Scholar] [CrossRef]
- Kholoud, K.; Denis, S.; Lahouari, B.; El Hidan, M.A.; Souad, B. Management of Leishmaniases in the era of climate change in Morocco. Int. J. Environ. Res. Public Health 2018, 15, 1542. [Google Scholar] [CrossRef]
- Trájer, A.J.; Sebestyén, V. The changing distribution of Leishmania infantum Nicolle, 1908 and its Mediterranean sandfly vectors in the last 140 kys. Sci. Rep. 2019, 9, 11820. [Google Scholar] [CrossRef]
- Moirano, G.; Zanet, S.; Giorgi, E.; Battisti, E.; Falzoi, S.; Acquaotta, F.; Fratianni, S.; Richiardi, L.; Ferroglio, E.; Maule, M. Integrating environmental, entomological, animal, and human data to model the Leishmania infantum transmission risk in a newly endemic area in Northern Italy. One Health 2020, 10, 100159. [Google Scholar] [CrossRef]
- Pintado, V.; López-Vélez, R. HIV-associated visceral leishmaniasis. Clin. Microbiol. Infect. 2001, 7, 291–300. [Google Scholar] [CrossRef]
- Antinori, S.; Cascio, A.; Parravicini, C.; Bianchi, R.; Corbellino, M. Leishmaniasis among organ transplant recipients. Lancet. Infect. Dis. 2008, 8, 191–199. [Google Scholar] [CrossRef]
- Monge-Maillo, B.; Norman, F.F.; Cruz, I.; Alvar, J.; López-Vélez, R. Visceral leishmaniasis and HIV coinfection in the Mediterranean region. PLoS Negl. Trop. Dis. 2014, 8, e3021. [Google Scholar] [CrossRef]
- Van Griensven, J.; Carrillo, E.; López-Vélez, R.; Moreno, J. Leishmaniasis in immunosuppressed individuals. Clin. Microbiol. Infect. 2014, 20, 286–299. [Google Scholar] [CrossRef]
- Gajurel, K.; Dhakal, R.; Deresinski, S. Leishmaniasis in solid organ and hematopoietic stem cell transplant recipients. Clin. Transplant. 2017, 31, e12867. [Google Scholar] [CrossRef]
- Carrillo, E.; Carrasco-Antón, N.; López-Medrano, F.; Salto, E.; Fernández, L.; San Martín, J.V.; Alvar, J.; Aguado, J.M.; Moreno, J. Cytokine release assays as tests for exposure to Leishmania, and for confirming cure from leishmaniasis, in solid organ transplant recipients. PLoS Negl. Trop. Dis. 2015, 9, e0004179. [Google Scholar] [CrossRef]
- Dhaliwal, A.; Chauhan, A.; Aggarwal, D.; Davda, P.; David, M.; Amel-Kashipaz, R.; Brown, R.; Dedicoat, M.; Clark, F.; Shah, T.; et al. Donor acquired visceral leishmaniasis following liver transplantation. Frontline Gastroenterol. 2021, 12, 690–694. [Google Scholar] [CrossRef]
- Busutti, M.; Deni, A.; De Pascali, A.M.; Ortalli, M.; Attard, L.; Granozzi, B.; Fabbrizio, B.; La Manna, G.; Comai, G.; Varani, S. Updated diagnosis and graft involvement for visceral leishmaniasis in kidney transplant recipients: A case report and literature review. Infection 2022, 1–12. [Google Scholar] [CrossRef]
- Clemente, W.T.; Orlandi Mourão, P.H.; Aguado, J.M. Current approaches to visceral leishmaniasis treatment in solid organ transplant recipients. Expert Rev. Anti Infect. Ther. 2018, 16, 391–397. [Google Scholar] [CrossRef]
- Lindoso, J.A.L.; Moreira, C.H.V.; Cunha, M.A.; Queiroz, I.T. Visceral leishmaniasis and HIV coinfection: Current perspectives. HIV AIDS 2018, 15, 193–201. [Google Scholar] [CrossRef]
- Burki, T. Guidelines for visceral leishmaniasis and HIV co-infection. Lancet Infect. Dis. 2022, 22, 124–1125. [Google Scholar] [CrossRef] [PubMed]
- WHO. 2022. Available online: https://www.who.int/news/item/08-06-2022-visceral-leishmaniasis-and-HIV-coinfection-WHO-publishes-new-guideline-with-region-specific-treatment-recommendations (accessed on 26 January 2023).
- Alvar, J.; den Boer, M.; Dagne, D.A. Towards the elimination of visceral leishmaniasis as a public health problem in east Africa: Reflections on an enhanced control strategy and a call for action. Lancet Glob. Health 2021, 9, e1763–e1769. [Google Scholar] [CrossRef] [PubMed]
- Velez, R.; Gállego, M. Commercially approved vaccines for canine leishmaniosis: A review of available data on their safety and efficacy. Trop. Med. Int. Health 2020, 25, 540–557. [Google Scholar] [CrossRef] [PubMed]
- Croft, S.L.; Yardley, V. Chemotherapy of leishmaniasis. Curr. Pharm. Des. 2002, 8, 319–342. [Google Scholar] [CrossRef]
- Corral-Caridad, M.J.; Alunda, J.M. Chemotherapy of leishmaniasis: A veterinary perspective. In Trypanosomatid Diseases; Molecular Routes to Drug Discovery; Jäger, T., Koch, O., Flohé, L., Eds.; Wiley-VCH: Weinheim, Germany, 2013; pp. 17–36. [Google Scholar]
- Uliana, S.; Trinconi, C.; Coelho, A. Chemotherapy of leishmaniasis: Present challenges. Parasitology 2018, 145, 464–480. [Google Scholar] [CrossRef]
- Ponte-Sucre, A.; Gamarro, F.; Dujardin, J.C.; Barrett, M.P.; López-Vélez, R.; García-Hernández, R.; Pountain, A.W.; Mwenechanya, R.; Papadopoulou, B. Drug resistance and treatment failure in leishmaniasis: A 21st century challenge. PLoS Negl. Trop. Dis. 2017, 11, e0006052. [Google Scholar] [CrossRef]
- Singh, O.P.; Singh, B.; Chakravarty, J.; Sundar, S. Current challenges in treatment options for visceral leishmaniasis in India: A public health perspective. Infect. Dis. Poverty 2016, 5, 19. [Google Scholar] [CrossRef]
- Bastos, D.S.S.; Silva, A.C.; Novaes, R.D.; Souza, A.C.F.; Santos, E.C.; Gonçalves, R.V.; Marques da Silva, E.A. Could combination chemotherapy be more effective than monotherapy in the treatment of visceral leishmaniasis? A systematic review of preclinical evidence. Parasitology 2022, 9, 1–14. [Google Scholar] [CrossRef]
- Kumar, P.; Kumar, P.; Singh, N.; Khajuria, S.; Patel, R.; Rajana, V.K.; Mandal, D.; Velayutham, R. Limitations of current chemotherapy and future of nanoformulation-based AmB delivery for visceral leishmaniasis—An updated review. Front. Bioeng. Biotechnol. 2022, 10, 1016925. [Google Scholar] [CrossRef]
- Musa, A.M.; Mbui, J.; Mohammed, R.; Olobo, J.; Ritmeijer, K.; Alcoba, G.; Muthoni Ouattara, G.; Egondi, T.; Nakanwagi, P.; Omollo, T.; et al. Paromomycin and miltefosine combination as an alternative to treat patients with visceral leishmaniasis in eastern Africa: A randomized, controlled, multicountry trial. Clin. Infect. Dis. 2022, ciac643. [Google Scholar] [CrossRef]
- Reguera, R.M.; Pérez-Pertejo, Y.; Gutiérrez-Corbo, C.; Domínguez-Asenjo, B.; Ordóñez, C.; García-Estrada, C.; Martínez-Valladares, M.; Balaña-Fouce, R. Current and promising novel drug candidates against visceral leishmaniasis. Pure Appl. Chem. 2019, 91, 1385–1404. [Google Scholar] [CrossRef]
- eBioMedicine. Leishmania: An urgent need for new treatments. eBioMedicine 2023, 87, 104440. [Google Scholar] [CrossRef]
- Sinderman, H.; Engel, J. Development of miltefosine as an oral treatment for leishmaniasis. Trans. R. Soc. Trop. Med. Hyg. 2006, 100, S17–S20. [Google Scholar] [CrossRef]
- Loreto, E.S.; Tondolo, J.S.M.; Oliveira, D.C.; Santurio, J.M.; Alves, S.H. In vitro activities of miltefosine and antibacterial agents from the macrolide, oxazolidinone, and pleuromutilin classes against Pythium insidiosum and Pythium aphanidermatum. Antimicrob. Agents Chemother. 2018, 62, e01678-17. [Google Scholar] [CrossRef]
- Fiester, S.E.; Arivett, B.A.; Beckett, A.C.; Wagner, B.R.; Ohneck, E.J.; Schmidt, R.E.; Grier, J.T.; Actis, L.A. Miltefosine reduces the cytolytic activity and virulence of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2019, 63, e01409-18. [Google Scholar] [CrossRef]
- Llull, D.; Rivas, L.; García, E. In vitro bactericidal activity of the antiprotozoal drug miltefosine against Streptococcus pneumoniae and other pathogenic Streptococci. Antimicrob. Agents Chemother. 2007, 51, 1844–1848. [Google Scholar] [CrossRef]
- Sundar, S.; Olliaro, P.L. Miltefosine in the treatment of leishmaniasis: Clinical evidence for informed clinical risk management. Ther. Clin. Risk Manag. 2007, 3, 733–740. [Google Scholar]
- Sundar, S.; Makharia, A.; More, D.K.; Agrawal, G.; Voss, A.; Fischer, C.; Bachmann, P.; Murray, H.W. Short-course of oral miltefosine for treatment of visceral leishmaniasis. Clin. Infect. Dis. 2000, 31, 1110–1113. [Google Scholar] [CrossRef]
- Monge-Maillo, B.; López-Vélez, R. Miltefosine for visceral and cutaneous leishmaniasis: Drug characteristics and evidence-based treatment recommendations. Clin. Infect. Dis. 2015, 60, 1398–1404. [Google Scholar] [CrossRef]
- Berman, J. Miltefosine to treat leishmaniasis. Expert. Opin. Pharmacother. 2005, 6, 1381–1388. [Google Scholar] [CrossRef]
- Chakravarty, J.; Sundar, S. Drug resistance in leishmaniasis. J. Glob. Infect. Dis. 2010, 2, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Breiser, A.; Kim, D.J.; Fleer, E.A.M.; Damenz, W.; Drube, A.; Berger, M.; Nagel, G.A.; Eibl, H.; Unger, C. Distribution and metabolism of hexadecylphosphocholine in mice. Lipids 1987, 22, 925–926. [Google Scholar] [CrossRef] [PubMed]
- Valicherla, G.R.; Tripathi, P.; Singh, S.K.; Syed, A.A.; Riyazuddin, M.; Husain, A.; Javia, D.; Italiya, K.S.; Mishra, P.R.; Gayen, J.R. Pharmacokinetic and bioavailability assessment of miltefosine in rats. Pharmacokinetic and bioavailability assessment of miltefosine in rats using high performance liquid chromatography tandem mass spectrometry. J. Chromatogr. B 2016, 1031, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Sharma, A.; Shukla, M.; Lal, J. LC-coupled ESI MS for quantification of miltefosine in human and hamster plasma. Bioanalysis 2016, 8, 533–545. [Google Scholar] [CrossRef]
- Dorlo, T.P.C.; Rijal, S.; Ostyn, B.; de Vries, P.J.; Singh, R.; Bhattarai, N.; Uranw, S.; Dujardin, J.C.; Boelaert, M.; Beijnen, J.H.; et al. Failure of miltefosine in visceral leishmaniasis is associated with low drug exposure. J. Infect. Dis. 2014, 210, 146–153. [Google Scholar] [CrossRef]
- Wasunna, M.; Njenga, S.; Balasegaram, M.; Alexander, N.; Omollo, R.; Edwards, T.; Dorlo, T.P.; Musa, B.; Ali, M.H.; Elamin, M.Y.; et al. Efficacy and safety of AmBisome in combination with sodium stibogluconate or miltefosine and miltefosine monotherapy for African visceral leishmaniasis: Phase II randomized trial. PLoS Negl. Trop. Dis. 2016, 10, e0004880. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.M.; Fakiola, M.; Singh, O.P. Genetics, transcriptomics and meta-taxonomics in visceral leishmaniasis. Front. Cell. Infect. Microbiol. 2020, 5, 590888. [Google Scholar] [CrossRef]
- Lappan, R.; Classon, C.; Kumar, S.; Singh, O.P.; de Almeida, R.V.; Chakravarty, J.; Kumari, P.; Kansal, S.; Sundar, S.; Blackwell, J.M. Meta-taxonomic analysis of prokaryotic and eukaryotic gut flora in stool samples from visceral leishmaniasis cases and endemic controls in Bihar State India. PLoS Negl. Trop. Dis. 2019, 13, e0007444. [Google Scholar] [CrossRef]
- Jayasena Kaluarachchi, T.D.; Campbell, P.M.; Wickremasinghe, R.; Ranasinghe, S.; Wickremasinghe, R.; Yasawardene, S.; De Silva, H.; Menike, C.; Jayarathne, M.C.K.; Jayathilake, S.; et al. Distinct microbiome profiles and biofilms in Leishmania donovani-driven cutaneous leishmaniasis wounds. Sci. Rep. 2021, 11, 23181. [Google Scholar] [CrossRef]
- Correia Passos, F.; Biondaro Gois, M.; Duranes Sousa, A.; Lima de Marinho, A.I.; Corvo, L.; Soto, M.; Barral-Netto, M.; Barral, A.; Baccan, G.C. Investigating associations between intestinal alterations and parasite load according to Bifidobacterium spp. and Lactobacillus spp. abundance in the gut microbiota of hamsters infected by Leishmania infantum. Mem. Inst. Oswaldo Cruz Rio de Janeiro 2020, 115, e200377. [Google Scholar] [CrossRef]
- Olías-Molero, A.I.; Botías, P.; Cuquerella, M.; García-Cantalejo, J.; Barcia, E.; Torrado, S.; Torrado, J.J.; Alunda, J.M. Leishmania infantum infection does not affect the main composition of the intestinal microbiome of the Syrian hamster. Parasit. Vectors 2022, 15, 468. [Google Scholar] [CrossRef]
- Ruppé, E.; Burdet, C.; Grall, N.; de Lastours, V.; Lescure, F.X.; Andremont, A.; Armand-Lefèvre, L. Impact of antibiotics on the intestinal microbiota needs to be re-defined to optimize antibiotic usage. Clin. Microbiol. Infect. 2018, 24, 3–5. [Google Scholar] [CrossRef]
- Walsh, J.; Griffin, B.T.; Clarke, G.; Hyland, N.P. Drug-gut microbiota interactions: Implications for neuropharmacology. Br. J. Pharmacol. 2018, 175, 4415–4429. [Google Scholar] [CrossRef]
- Toepp, A.J.; Monteiro, G.R.G.; Coutinho, J.F.V.; Lima, A.L.; Larson, M.; Wilson, G.; Grinnage-Pulley, T.; Bennett, C.; Mahachi, K.; Anderson, K.; et al. Comorbid infections induce progression of visceral leishmaniasis. Parasit. Vectors 2019, 23, 54. [Google Scholar] [CrossRef]
- Khayeka-Wandabwa, C.; Anjili, C.O.; Nyambati, V.C.; Kutima, L.H.; Choge, J.K.; Karani, L.K.; Kemei, W.K. Leishmaniases and schistosomiasis comorbidity potential in Kenya: The need for follow up studies. Preprints 2020, 2020100268. [Google Scholar] [CrossRef]
- Oliveira-Sena, I.V.; Werneck, G.L. Risk factors for in-hospital mortality from visceral leishmaniasis: A case-control study. J. Infect. Public Health 2020, 13, 538–543. [Google Scholar] [CrossRef]
- Palma, D.; Mercuriali, L.; Figuerola, J.; Montalvo, T.; Bueno-Marí, R.; Millet, J.P.; Simón, P.; Masdeu, E.; Rius, C. Trends in the epidemiology of Leishmaniasis in the city of Barcelona (1996–2019). Front. Vet. Sci. 2021, 8, 653999. [Google Scholar] [CrossRef]
- Zimmermann, P.; Curtis, N. The effect of antibiotics on the composition of the intestinal microbiota-a systematic review. J. Infect. 2019, 79, 471–489. [Google Scholar] [CrossRef]
- Ramirez, J.; Guarner, F.; Bustos-Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as major disruptors of gut microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef]
- Dorlo, T.P.C.; Balasegaram, M.; Beijnen, J.H.; de Vries, P.J. Miltefosine: A review of its pharmacology and therapeutic efficacy in the treatment of leishmaniasis. J. Antimicrob. Chemother. 2012, 67, 2576–2597. [Google Scholar] [CrossRef]
- Vincent, I.M.; Weidt, S.; Rivas, L.; Burgess, K.; Smith, T.K.; Ouellette, M. Untargeted metabolomic analysis of miltefosine action in Leishmania infantum reveals changes to the internal lipid metabolism. Int. J. Parasitol. Drugs Drug Resist. 2013, 4, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Knuplez, E.; Kienzl, M.; Trakaki, A.; Schicho, R.; Heinemann, A.; Sturm, E.M.; Marsche, G. The anti-parasitic drug miltefosine suppresses activation of human eosinophils and ameliorates allergic inflammation in mice. Br. J. Pharmacol. 2021, 178, 1234–1248. [Google Scholar] [CrossRef] [PubMed]
- Wieder, T.; Reutter, W.; Orfanos, C.E.; Geilen, C.C. Mechanisms of action of phospholipid analogs as anticancer compounds. Prog. Lipid. Res. 1999, 38, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Rakotomanga, M.; Blanc, S.; Gaudin, K.; Chaminade, P.; Loiseau, P.M. Miltefosine affects lipid metabolism in Leishmania donovani promastigotes. Antimicrob. Agents Chemother. 2007, 51, 1425–1430. [Google Scholar] [CrossRef] [PubMed]
- Moitra, S.; Basu, S.; Pawlowic, M.; Hsu, F.F.; Zhang, K. De novo synthesis of phosphatidylcholine is essential for the promastigote but not amastigote stage in Leishmania major. Front. Cell. Infect. Microbiol. 2021, 11, 647870. [Google Scholar] [CrossRef]
- Geiger, O.; López-Lara, I.M.; Sohlenkamp, C. Phosphatidylcholine biosynthesis and function in bacteria. Biochim. Biophys. Acta 2013, 1831, 503–513. [Google Scholar] [CrossRef]
- Woerly, V.; Maynard, L.; Sanquer, A.; Eun, H.M. Clinical efficacy and tolerance of miltefosine in the treatment of canine leishmaniosis. Parasitol. Res. 2009, 105, 463–469. [Google Scholar] [CrossRef]
- Iarussi, F.; Paradies, P.; Foglia Manzillo, V.; Gizzarelli, M.; Caratozzolo, M.F.; Navarro, C.; Greco, B.; Rubino, G.T.R.; Oliva, G.; Sasanelli, M. Comparison of two dosing regimens of miltefosine, both in combination with allopurinol, on clinical and parasitological findings of dogs with leishmaniosis: A pilot study. Front. Vet. Sci. 2020, 7, 577395. [Google Scholar] [CrossRef]
- La Reau, A.J.; Suen, G. The Ruminococci: Key symbionts of the gut ecosystem. J. Microbiol. 2018, 56, 199–208. [Google Scholar] [CrossRef]
- Smieja, M. Current indications for the use of clindamycin: A critical review. Can. J. Infect. Dis. 1998, 9, 22–28. [Google Scholar] [CrossRef]
- Spizek, J.; Novotna, J.; Rezanka, T. Lincosamides: Chemical structure, biosynthesis, mechanism of action, resistance, and applications. Adv. Appl. Microbiol. 2004, 56, 121–154. [Google Scholar] [CrossRef]
- Hertz, F.B.; Budding, A.E.; Van der Lugt-Degen, M.; Savelkoul, P.H.; Løbner-Olesen, A.; Frimodt-Møller, N. Effects of antibiotics on the intestinal microbiota of mice. Antibiotics 2020, 9, 191. [Google Scholar] [CrossRef]
- Lawley, T.D.; Clare, S.; Walker, A.W.; Goulding, D.; Stabler, R.A.; Croucher, N.; Mastroeni, P.; Scott, P.; Raisen, C.; Mottram, L.; et al. Antibiotic treatment of clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 2009, 77, 3661–3669. [Google Scholar] [CrossRef]
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 2012, 80, 62–73. [Google Scholar] [CrossRef]
- Peterfreund, G.L.; Vandivier, L.E.; Sinha, R.; Marozsan, A.J.; Olson, W.C.; Zhu, J.; Bushman, F.D. Succession in the gut microbiome following antibiotic and antibody therapies for Clostridium difficile. PLoS ONE 2012, 7, e46966. [Google Scholar] [CrossRef]
- Trejo, F.; De Antoni, G.; Pérez, P. Protective effect of bifidobacteria in an experimental model of Clostridium difficile associated colitis. J. Dairy Res. 2013, 80, 263–269. [Google Scholar] [CrossRef]
- Ferrer, M.; Méndez-García, C.; Rojo, D.; Barbas, C.; Moya, A. Antibiotic use and microbiome function. Biochem. Pharmacol. 2017, 134, 114–126. [Google Scholar] [CrossRef]
- Burdet, C.; Sayah-Jeanne, S.; Nguyen, T.T.; Hugon, P.; Sablier-Gallis, F.; Saint-Lu, N.; Corbel, T.; Ferreira, S.; Pulse, M.; Weiss, W.; et al. Antibiotic-induced dysbiosis predicts mortality in an animal model of Clostridium difficile infection. Antimicrob. Agents Chemother. 2018, 62, e00925-18. [Google Scholar] [CrossRef]
- Yang, L.; Bajinka, O.; Jarju, P.O.; Tan, Y.; Taal, A.M.; Ozdemir, G. The varying effects of antibiotics on gut microbiota. AMB Express 2021, 11, 116. [Google Scholar] [CrossRef]
- Liu, L.; Kirst, M.E.; Zhao, L.; Li, E.; Wang, G.P. Microbiome resilience despite a profound loss of minority microbiota following clindamycin challenge in humanized gnotobiotic mice. Microbiol. Spectr. 2022, 10, e01960-21. [Google Scholar] [CrossRef]
- Kierzkowska, M.; Majewska, A.; Szymanek-Majchrzak, A.; Sawicka-Grzelak, A.; Mlynarczyk, A.; Mlynarczyk, G. In vitro effect of clindamycin against Bacteroides and Parabacteroides isolates in Poland. J. Glob. Antimicrob. Resist. 2018, 13, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kong, X.; Blachier, F.; Geng, N.N.; Guo-Qi, L.; Yulong, Y. Dietary supplementation of soybean-derived sterols regulates cholesterol metabolism and intestinal microbiota in hamsters. J. Funct. Foods 2019, 59, 242–250. [Google Scholar] [CrossRef]
- Hugenholtz, F.; de Vos, W.M. Mouse models for human intestinal microbiota research: A critical evaluation. Cell. Mol. Life Sci. 2018, 75, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.D.; Albeke, S.E.; Gigley, J.P.; Goldstein, A.M.; Ward, N.L. Microbiome composition in both wild-type and disease model mice is heavily influenced by mouse facility. Front. Microbiol. 2018, 9, 1598. [Google Scholar] [CrossRef]
- Jiménez-Antón, M.D.; García-Calvo, E.; Gutiérrez, C.; Escribano, M.D.; Kayali, N.; Luque-García, J.L.; Olías-Molero, A.I.; Corral, M.J.; Costi, M.P.; Torrado, J.; et al. Pharmacokinetics and disposition of miltefosine in healthy mice and hamsters experimentally infected with Leishmania infantum. Eur. J. Pharm. Sci. 2018, 121, 281–286. [Google Scholar] [CrossRef]
- Kip, A.E.; Schellens, J.; Beijnen, J.H.; Forlo, T. Clinical pharmacokinetics of systemically administered antileishmanial drugs. Clin. Pharmacokinet. 2018, 57, 151–176. [Google Scholar] [CrossRef]
- Bianciardi, P.; Brovida, C.; Valente, M.; Aresu, L.; Cavicchioli, L.; Vischer, C.; Giroud, L.; Castagnaro, M. Administration of miltefosine and meglumine antimoniate in healthy dogs: Clinicopathological evaluation of the impact on the kidneys. Toxicol. Pathol. 2009, 37, 770–775. [Google Scholar] [CrossRef]
- Bandana, K.; Jashandeep, K.; Jagdeep, K. Phospholipases in bacterial virulence and pathogenesis. Adv. Biotechnol. Microbiol. 2018, 10, 555798. [Google Scholar] [CrossRef]
- Sinha, A.K.; Dutta, A.; Chandravanshi, M.; Kanaujia, S.P. An insight into bacterial phospholipase C classification and their translocation through Tat and Sec pathways: A data mining study. Meta Gene 2019, 20, 100547. [Google Scholar] [CrossRef]
- Heinsen, F.A.; Knecht, H.; Neulinger, S.C.; Schmitz, R.A.; Knecht, C.; Kühbacher, T.; Rosenstiel, P.C.; Schreiber, S.; Friedrichs, A.K.; Ott, S.J. Dynamic changes of the luminal and mucosa-associated gut microbiota during and after antibiotic therapy with paromomycin. Gut Microbes 2015, 6, 243–254. [Google Scholar] [CrossRef]
- Manuzak, J.A.; Zevin, A.S.; Cheu, R.; Richardson, B.; Modesitt, J.; Hensley-McBain, T.; Miller, C.; Gustin, A.T.; Coronado, E.; Gott, T.; et al. Antibiotic-induced microbiome perturbations are associated with significant alterations to colonic mucosal immunity in rhesus macaques. Mucosal Immunol. 2020, 13, 471–480. [Google Scholar] [CrossRef]
- Jiménez-Antón, M.D.; Grau, M.; Corral, M.J.; Olías-Molero, A.I.; Alunda, J.M. Efficient infection of hamster with Leishmania donovani by retro-orbital inoculation. Virulence 2019, 10, 711–718. [Google Scholar] [CrossRef]
- Dorlo, T.P.C.; Hillebrand, M.J.; Rosing, H.; Eggelte, T.A.; de Vries, P.J.; Beijnen, J.H. Development and validation of a quantitative assay for the measurement of MIL in human plasma by liquid chromatography-tandem mass spectrometry. J. Chromatogr. B 2008, 865, 55–62. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Available online: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA843999 (accessed on 25 November 2022).


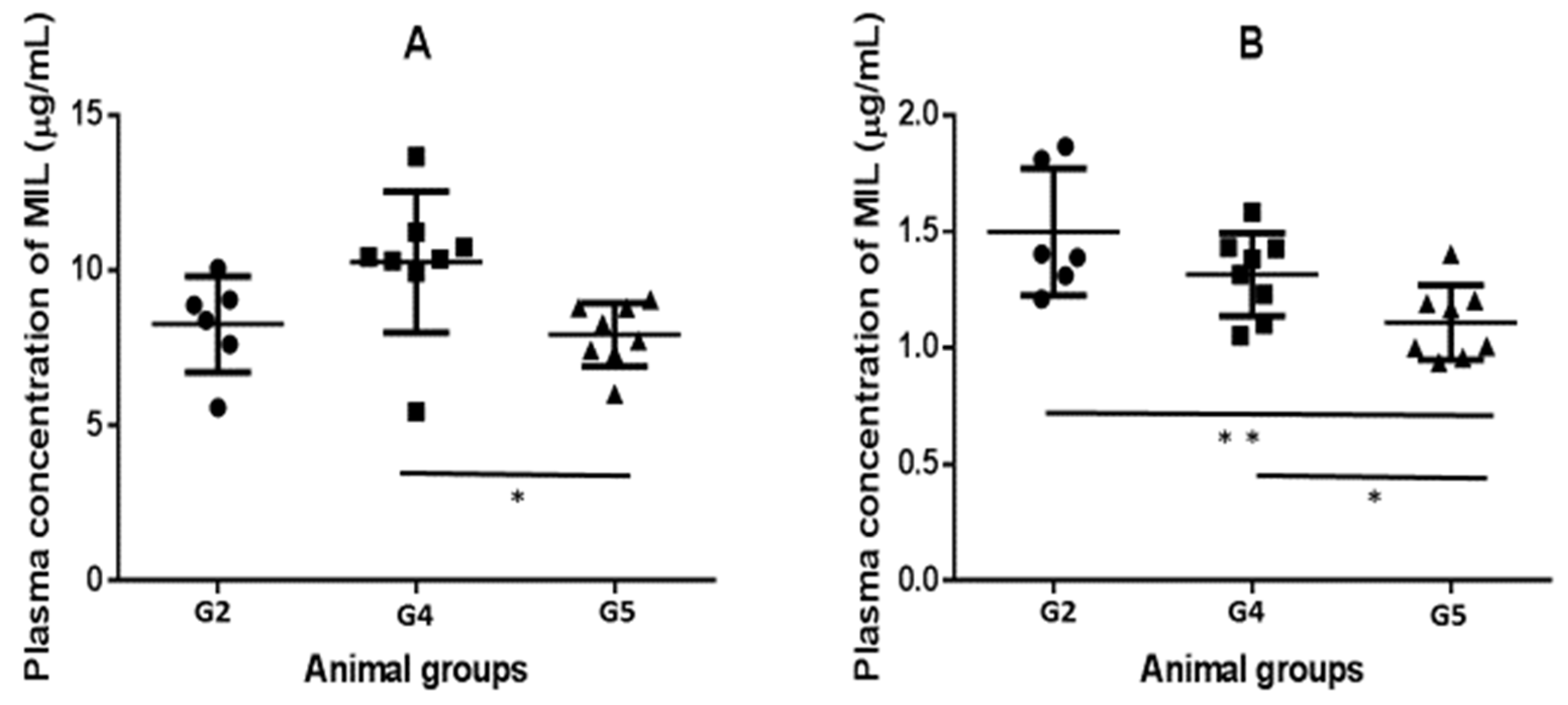
| Animal Group | Liver (µM) | Spleen (µM) | Liver/Plasma Ratio | Spleen/Plasma Ratio |
|---|---|---|---|---|
| G2 | 3.10 ± 0.70 | 0.92 ± 0.41 | 0.87 ± 0.30 | 0.27 ± 0.16 |
| G4 | 2.43 ± 0.70 | 0.85 ± 0.37 | 0.76 ± 0.24 | 0.27 ± 0.14 |
| G5 | 2.58 ± 0.54 | 0.71 ± 0.31 | 0.95 0.17 | 0.26 ± 0.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olías-Molero, A.I.; Botías, P.; Cuquerella, M.; García-Cantalejo, J.; Barcia, E.; Torrado, S.; Torrado, J.J.; Alunda, J.M. Effect of Clindamycin on Intestinal Microbiome and Miltefosine Pharmacology in Hamsters Infected with Leishmania infantum. Antibiotics 2023, 12, 362. https://doi.org/10.3390/antibiotics12020362
Olías-Molero AI, Botías P, Cuquerella M, García-Cantalejo J, Barcia E, Torrado S, Torrado JJ, Alunda JM. Effect of Clindamycin on Intestinal Microbiome and Miltefosine Pharmacology in Hamsters Infected with Leishmania infantum. Antibiotics. 2023; 12(2):362. https://doi.org/10.3390/antibiotics12020362
Chicago/Turabian StyleOlías-Molero, Ana Isabel, Pedro Botías, Montserrat Cuquerella, Jesús García-Cantalejo, Emilia Barcia, Susana Torrado, Juan José Torrado, and José María Alunda. 2023. "Effect of Clindamycin on Intestinal Microbiome and Miltefosine Pharmacology in Hamsters Infected with Leishmania infantum" Antibiotics 12, no. 2: 362. https://doi.org/10.3390/antibiotics12020362
APA StyleOlías-Molero, A. I., Botías, P., Cuquerella, M., García-Cantalejo, J., Barcia, E., Torrado, S., Torrado, J. J., & Alunda, J. M. (2023). Effect of Clindamycin on Intestinal Microbiome and Miltefosine Pharmacology in Hamsters Infected with Leishmania infantum. Antibiotics, 12(2), 362. https://doi.org/10.3390/antibiotics12020362