Antivirulence Agent as an Adjuvant of β-Lactam Antibiotics in Treating Staphylococcal Infections
Abstract
:1. Introduction
2. Results
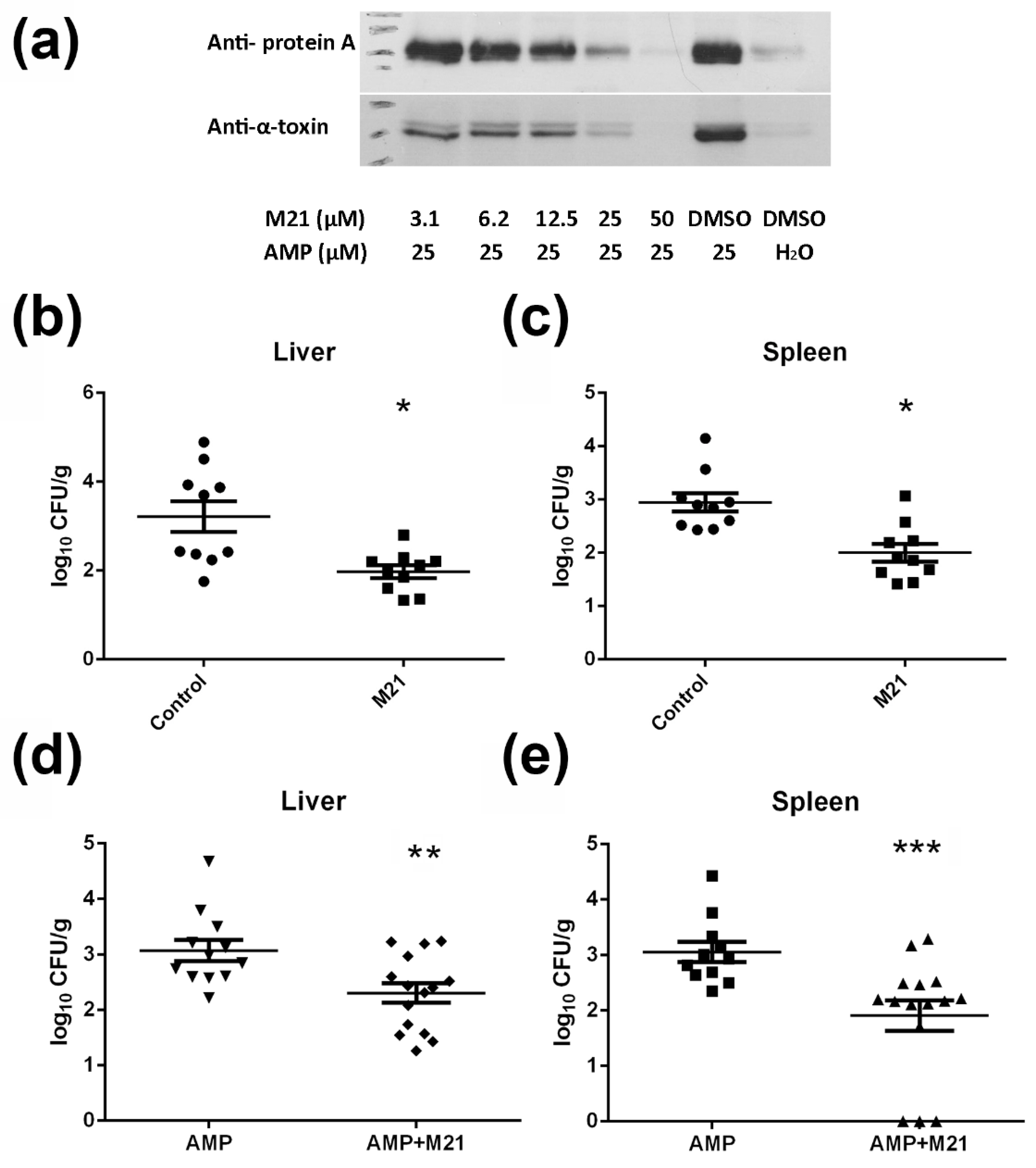
2.1. M21 Repressed the Virulence Gene Expression Induced by Ampicillin
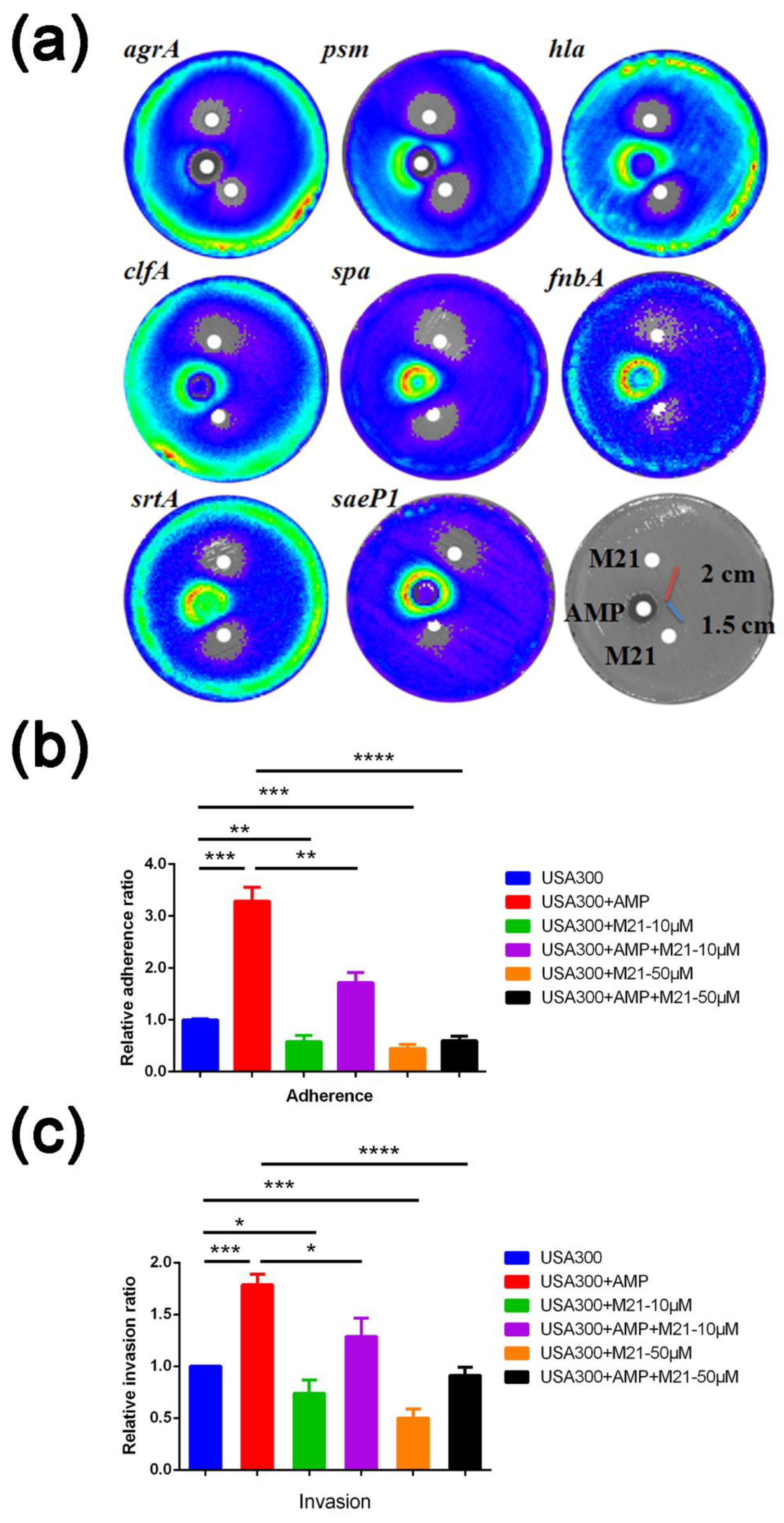
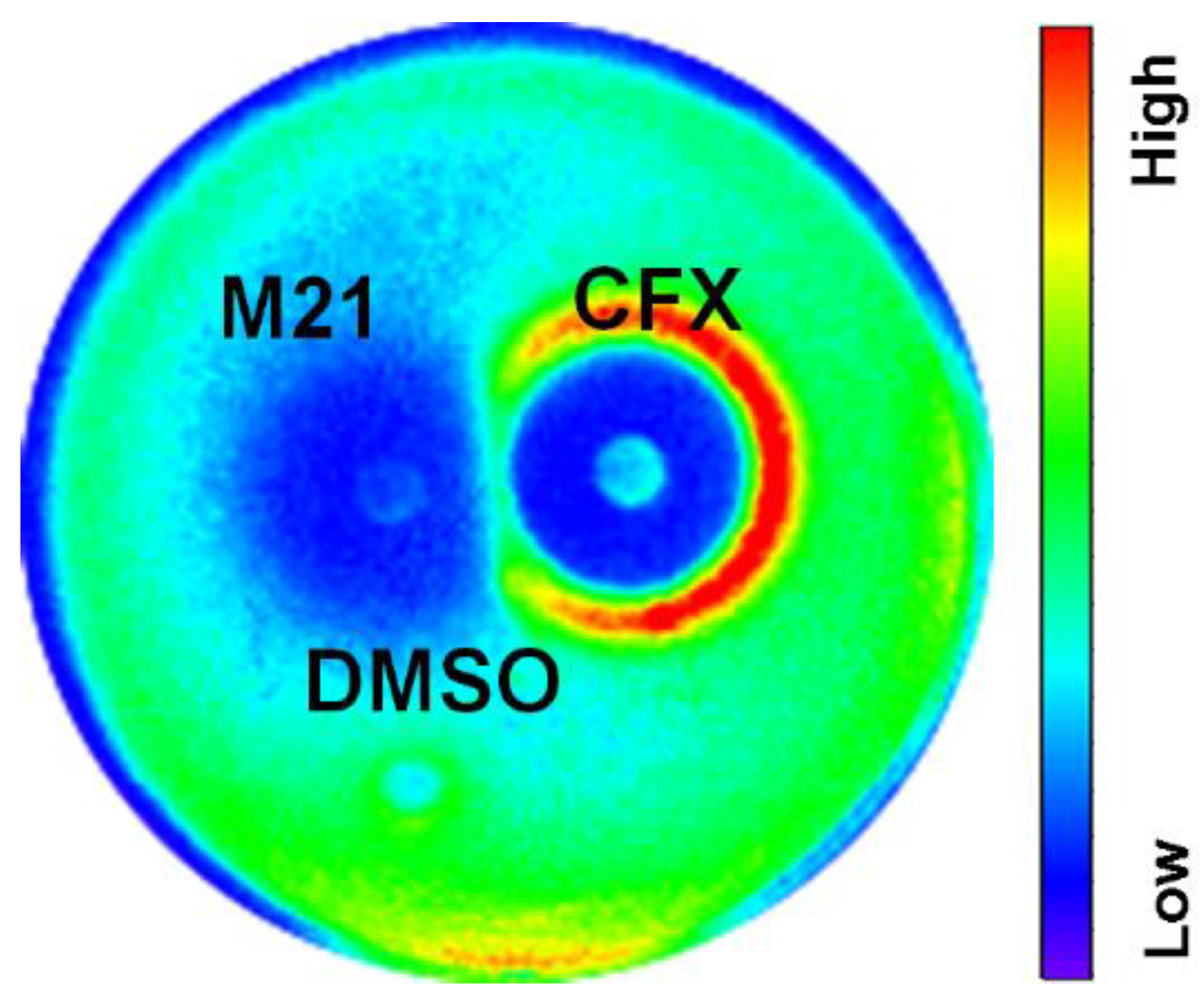
2.2. M21 Suppressed the S. aureus Virulence Induced by Different β-Lactam Antibiotics
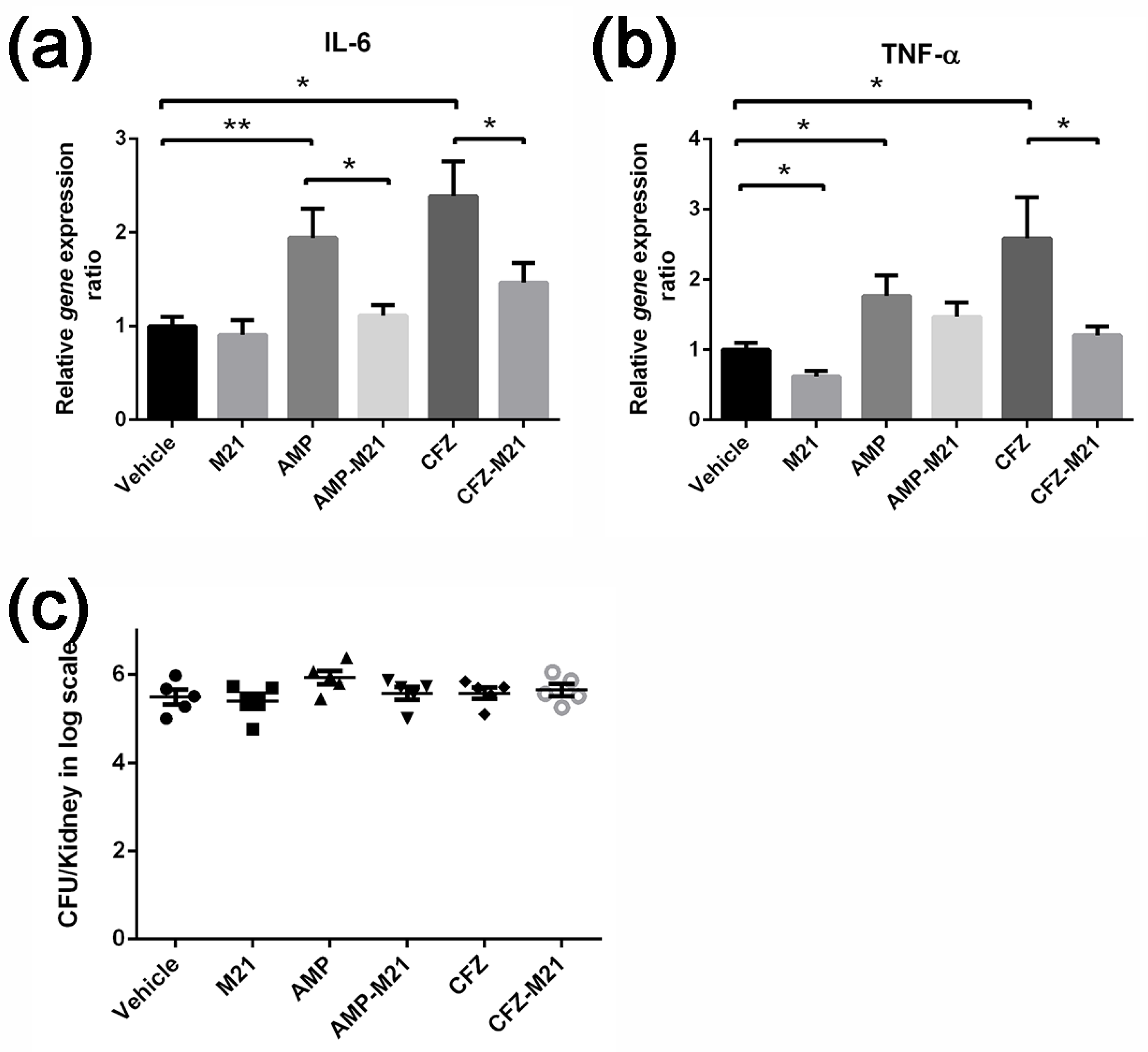
2.3. Combinatorial Treatment of Antivirulence Agent and Antibiotic Reduced Host Cytokine Expression
2.4. The Virulence Suppressing Effect of M21 Is Common in Different Strains
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Plasmids
4.2. Minimum Inhibitory Concentration (MIC) Tests
4.3. Disk Diffusion and Lux Assays
4.4. Real-Time PCR to Verify Expression Levels
4.5. Adherence Assay and Invasion Assay
4.6. Western Blot
4.7. Mouse Peritonitis Model
4.8. Mouse Bacteraemia Model
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balasubramanian, D.; Harper, L.; Shopsin, B.; Torres, V.J. Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 2017, 75, ftx005. [Google Scholar] [CrossRef] [Green Version]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Dancer, S.J. The effect of antibiotics on methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2008, 61, 246–253. [Google Scholar] [CrossRef]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.A.; Zhou, H.Y.; Huang, J.B.; Zhang, R.; Rao, X.C. Virulence alterations in staphylococcus aureus upon treatment with the sub-inhibitory concentrations of antibiotics. J. Adv. Res. 2021, 31, 165–175. [Google Scholar] [CrossRef]
- Gao, P.; Wang, Y.L.; Villanueva, I.; Ho, P.L.; Davies, J.; Kao, R.Y.T. Construction of a Multiplex Promoter Reporter Platform to Monitor Staphylococcus aureus Virulence Gene Expression and the Identification of Usnic Acid as a Potent Suppressor of psm Gene Expression. Front. Microbiol. 2016, 7, 1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.R.; Otto, M.; Cheung, A.L.; Edwards, B.S.; et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 2014, 10, e1004174. [Google Scholar] [CrossRef]
- Gao, P.; Ho, P.L.; Yan, B.; Sze, K.H.; Davies, J.; Kao, R.Y.T. Suppression of Staphylococcus aureus virulence by a small-molecule compound. Proc. Natl. Acad. Sci. USA 2018, 115, 8003–8008. [Google Scholar] [CrossRef] [Green Version]
- Shang, W.; Rao, Y.; Zheng, Y.; Yang, Y.; Hu, Q.; Hu, Z.; Yuan, J.; Peng, H.; Xiong, K.; Tan, L.; et al. Beta-Lactam Antibiotics Enhance the Pathogenicity of Methicillin-Resistant Staphylococcus aureus via SarA-Controlled Lipoprotein-Like Cluster Expression. mBio 2019, 10, e00880-19. [Google Scholar] [CrossRef] [Green Version]
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the pharmaceutical industry: New estimates of R&D costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar] [CrossRef] [Green Version]
- Douafer, H.; Andrieu, V.; Phanstiel, O.; Brunel, J.M. Antibiotic Adjuvants: Make Antibiotics Great Again! J. Med. Chem. 2019, 62, 8665–8681. [Google Scholar] [CrossRef] [PubMed]
- Boyd, N.K.; Teng, C.W.; Frei, C.R. Brief Overview of Approaches and Challenges in New Antibiotic Development: A Focus on Drug Repurposing. Front. Cell. Infect. Microbiol. 2021, 11, 684515. [Google Scholar] [CrossRef]
- Gill, E.E.; Franco, O.L.; Hancock, R.E. Antibiotic adjuvants: Diverse strategies for controlling drug-resistant pathogens. Chem. Biol. Drug Des. 2015, 85, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.Y.; Yang, Y.Y.; Tay, S.B.; Chua, K.L.; Yew, W.S. Disruption of Biofilm Formation by the Human Pathogen Acinetobacter baumannii Using Engineered Quorum-Quenching Lactonases. Antimicrob. Agents Chemother. 2014, 58, 1802–1805. [Google Scholar] [CrossRef] [Green Version]
- Smeltzer, M.S.; Lee, C.Y.; Harik, N.; Hart, M.E. Molecular Basis of Pathogenicity. In Staphylococci in Human Disease; Wiley-Blackwell: Chichester, UK; Hoboken, NJ, USA, 2009; pp. 65–108. [Google Scholar]
- Buroni, S.; Chiarelli, L.R. Antivirulence compounds: A future direction to overcome antibiotic resistance? Future Microbiol. 2020, 15, 299–301. [Google Scholar] [CrossRef]
- Muhlen, S.; Dersch, P. Anti-virulence Strategies to Target Bacterial Infections. Curr. Top. Microbiol. Immunol. 2016, 398, 147–183. [Google Scholar] [CrossRef]
- Shah, S.; Barton, G.; Fischer, A. Pharmacokinetic considerations and dosing strategies of antibiotics in the critically ill patient. J. Intensive Care Soc. 2015, 16, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Turner, C.E.; Sriskandan, S. Panton-Valentine leucocidin expression by Staphylococcus aureus exposed to common antibiotics. J. Infect. 2015, 71, 338–346. [Google Scholar] [CrossRef] [Green Version]
- Hodille, E.; Beraud, L.; Perian, S.; Berti, V.; Bes, M.; Tristan, A.; Blond, E.; Lina, G.; Dumitrescu, O. Sub-Inhibitory Concentrations of Oxacillin, but Not Clindamycin, Linezolid, or Tigecycline, Decrease Staphylococcal Phenol-Soluble Modulin Expression in Community-Acquired Methicillin-Resistant Staphylococcus aureus. Microbiol. Spectr. 2022, 10, e00808-21. [Google Scholar] [CrossRef]
- Rozgonyi, F.; Kocsis, E.; Kristof, K.; Nagy, K. Is MRSA more virulent than MSSA? Clin. Microbiol. Infect. 2007, 13, 843–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. 2017, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Rezzoagli, C.; Archetti, M.; Mignot, I.; Baumgartner, M.; Kümmerli, R. Combining antibiotics with antivirulence compounds can have synergistic effects and reverse selection for antibiotic resistance in Pseudomonas aeruginosa. PLoS Biol. 2020, 18, e3000805. [Google Scholar] [CrossRef]
- Damour, A.; Robin, B.; Deroche, L.; Broutin, L.; Bellin, N.; Verdon, J.; Lina, G.; Leclere, F.M.; Garcia, M.; Cremniter, J.; et al. Phenol-soluble modulins alpha are major virulence factors of Staphylococcus aureus secretome promoting inflammatory response in human epidermis. Virulence 2021, 12, 2474–2492. [Google Scholar] [CrossRef]
- Onogawa, T. Local delivery of soluble interleukin-6 receptors to improve the outcome of alpha-toxin producing Staphylococcus aureus infection in mice. Immunobiology 2005, 209, 651–660. [Google Scholar] [CrossRef]
- Ledo, C.; Gonzalez, C.D.; Garofalo, A.; Sabbione, F.; Keitelman, I.A.; Giai, C.; Stella, I.; Trevani, A.S.; Gomez, M.I. Protein A Modulates Neutrophil and Keratinocyte Signaling and Survival in Response to Staphylococcus aureus. Front. Immunol. 2020, 11, 524180. [Google Scholar] [CrossRef] [PubMed]
- Kao, R.Y.; Yang, D.; Lau, L.S.; Tsui, W.H.; Hu, L.; Dai, J.; Chan, M.P.; Chan, C.M.; Wang, P.; Zheng, B.J.; et al. Identification of influenza A nucleoprotein as an antiviral target. Nat. Biotechnol. 2010, 28, 600–605. [Google Scholar] [CrossRef]

| Isolates | Cefoxitin Induced Virulence | M21 Antivirulence Effect | Interaction |
|---|---|---|---|
| Isolate 14 | 5 | −6 | 0 |
| Isolate 86 | 1 | −5 | 0 |
| Isolate 22 | 5 | −6 | 0 |
| Isolate 24 | 9 | −6 | 0 |
| Isolate 25 | 4 | −3 | 1 |
| Isolate 34 | 2 | −8 | 0 |
| Isolate 42 | 3 | −7 | 0 |
| Isolate 43 | 5 | −8 | 0 |
| Isolate 44 | 2 | −3 | 1 |
| Isolate 45 | 4 | −5 | 0 |
| Isolate 46 | 5 | −3 | 0 |
| Isolate 63 | 2 | −2 | 1 |
| Isolate 64 | 2 | −2 | 1 |
| Isolate 65 | 6 | −4 | 1 |
| Isolate 72 | 3 | −2 | 1 |
| Isolate 73 | 6 | −5 | 1 |
| Isolate 76 | 9 | −5 | −1 |
| Isolate 83 | 1 | −1 | −2 |
| Isolate 84 | 7 | −3 | 0 |
| Isolate 85 | 1 | −8 | 0 |
| Strain | Phenotype | Source |
|---|---|---|
| Lab strains | ||
| USA300 FPR 3757 | CA-MRSA, Agr+ | ATCC ABB1776 |
| Mu3 | MRSA, Agr+ | ATCC700698 |
| Clinical isolates | ||
| Isolate 14 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 22 | Clinical isolate from patient blood, MSSA | This study |
| Isolate 24 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 25 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 34 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 42 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 43 | Clinical isolate from patient blood, MSSA | This study |
| Isolate 44 | Clinical isolate from patient blood, MSSA | This study |
| Isolate 45 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 46 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 63 | Clinical isolate from patient blood, MSSA | This study |
| Isolate 64 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 65 | Clinical isolate from patient blood, MSSA | This study |
| Isolate 72 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 73 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 83 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 84 | Clinical isolate from patient blood, MSSA | This study |
| Isolate 85 | Clinical isolate from patient blood, MRSA | This study |
| Isolate 86 | Clinical isolate from patient blood, MRSA | This study |
| Plasmid | ||
| pGL | gfp-luxABCDE dual reporter plasmid | Lab stock |
| pGLhla | gfp-luxABCDE dual reporter driven by hla promoter | Lab stock |
| Gene | Primer for Real-Time PCR |
|---|---|
| rt-hprt-f | CTGGTGAAAAGGACCTCTCG |
| rt-hprt-r | TGAAGTACTCATTATAGTCAAGGGCA |
| rt-Tnf-α-f | CTCCAGGCGGTGCCTATGT |
| rt-Tnf-α-r | GAAGAGCGTGGTGGCCC |
| rt-Il-6-f | CCAGAAACCGCTATGAAGTTCC |
| rt-Il-6-r | TCACCAGCATCAGTCCCAAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, P.; Wei, Y.; Tai, S.S.C.; Halebeedu Prakash, P.; Iu, H.T.V.; Li, Y.; Yam, H.C.B.; Chen, J.H.K.; Ho, P.L.; Davies, J.; et al. Antivirulence Agent as an Adjuvant of β-Lactam Antibiotics in Treating Staphylococcal Infections. Antibiotics 2022, 11, 819. https://doi.org/10.3390/antibiotics11060819
Gao P, Wei Y, Tai SSC, Halebeedu Prakash P, Iu HTV, Li Y, Yam HCB, Chen JHK, Ho PL, Davies J, et al. Antivirulence Agent as an Adjuvant of β-Lactam Antibiotics in Treating Staphylococcal Infections. Antibiotics. 2022; 11(6):819. https://doi.org/10.3390/antibiotics11060819
Chicago/Turabian StyleGao, Peng, Yuanxin Wei, Sherlock Shing Chiu Tai, Pradeep Halebeedu Prakash, Ho Ting Venice Iu, Yongli Li, Hin Cheung Bill Yam, Jonathan Hon Kwan Chen, Pak Leung Ho, Julian Davies, and et al. 2022. "Antivirulence Agent as an Adjuvant of β-Lactam Antibiotics in Treating Staphylococcal Infections" Antibiotics 11, no. 6: 819. https://doi.org/10.3390/antibiotics11060819
APA StyleGao, P., Wei, Y., Tai, S. S. C., Halebeedu Prakash, P., Iu, H. T. V., Li, Y., Yam, H. C. B., Chen, J. H. K., Ho, P. L., Davies, J., & Kao, R. Y. T. (2022). Antivirulence Agent as an Adjuvant of β-Lactam Antibiotics in Treating Staphylococcal Infections. Antibiotics, 11(6), 819. https://doi.org/10.3390/antibiotics11060819







