Antimicrobial Resistance Pattern, Clustering Mechanisms and Correlation Matrix of Drug-Resistant Escherichia coli in Black Bengal Goats in West Bengal, India
Abstract
1. Introduction
2. Results
2.1. Drug Resistance Characteristics of the Caprine E. coli Isolates
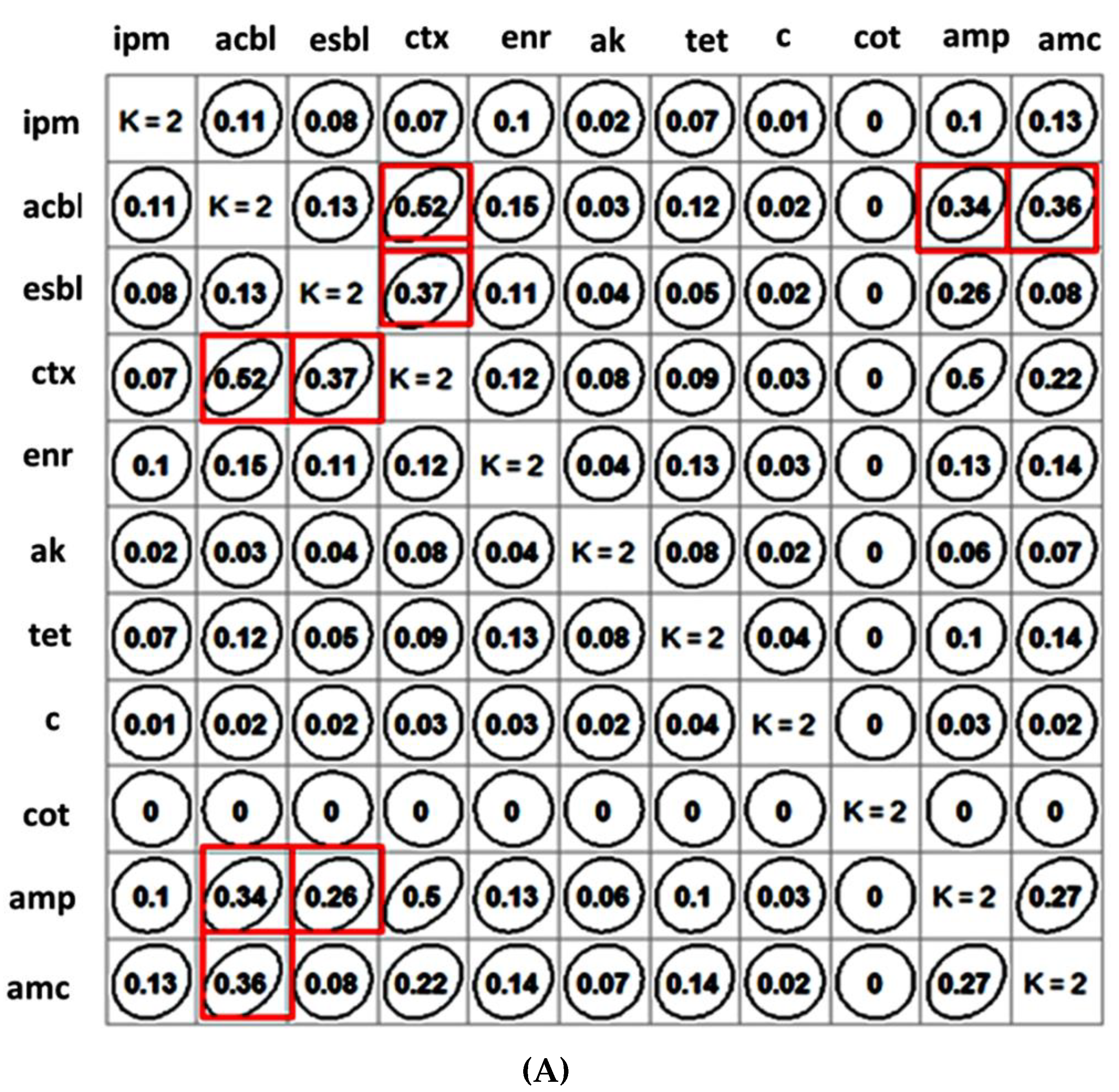
2.2. Clustering of Antibiotics and β-Lactam Resistance Mechanism and Their Correlation Matrix
2.3. Logistic Regression Analysis
2.4. Molecular Characterization of the MDR E. coli Isolates and Plasmid Incompatibility Types
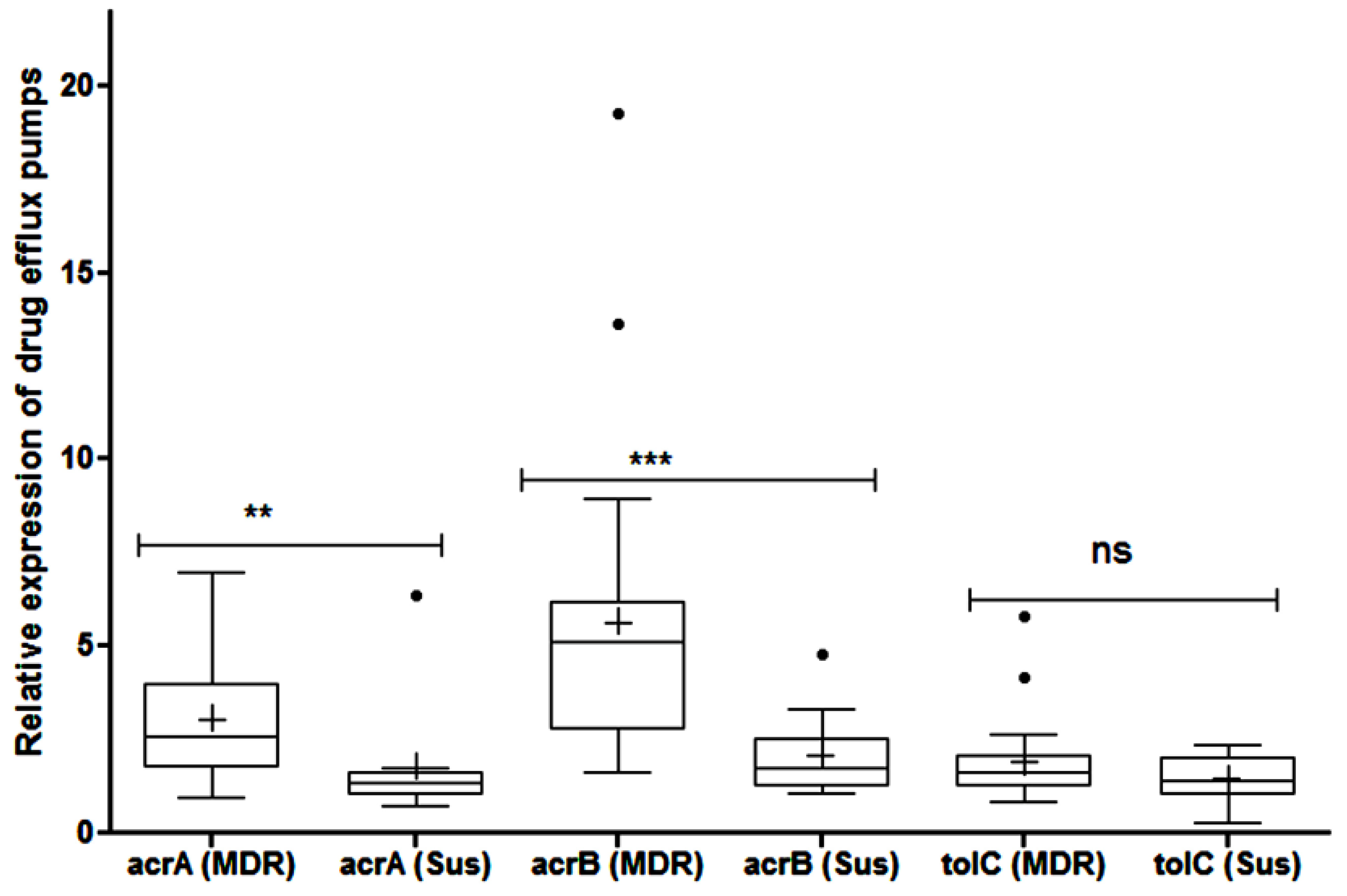
2.5. Efflux Pump Mediated Resistance
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Processing
4.2. Isolation and Confirmation of Pathogens
4.3. Antibiotic Resistance Profile of the Caprine E. coli Isolates
4.4. Clustering of Antibiotics and β-Lactam Resistance Mechanism and Their Correlation
4.5. Development of Logistic Regression Model for Prediction of Resistance Profile of the Caprine Isolates
4.6. Identification of Antibiotic Resistance Genes, Virulence Repertoire and Plasmid Incompatibility Types
4.7. Phenotypic Investigation of Efflux Pump Activity in the MDR Isolates
4.8. Transcriptional Expression of acrA, acrB and tolC Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abd-Allah, S.; Mohamed, M.I.; Shoukry, M.M.; Salman, F.M.; Abd- El Rahman, H.H. Assessment of the Traditional Goat Production Systems in Rural Areas of the Nile Delta in Egypt. Bull. Natl. Res. Cent. 2019, 43, 1–13. [Google Scholar] [CrossRef]
- Gilbert, W.; Thomas, L.F.; Coyne, L.; Rushton, J. Review: Mitigating the Risks Posed by Intensification in Livestock Production: The Examples of Antimicrobial Resistance and Zoonoses. Animal 2021, 15, 100123. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Samanta, I. Antimicrobial Resistance in Agri-Food Chain and Companion Animals as a Re-Emerging Menace in Post-COVID Epoch: Low-and Middle-Income Countries Perspective and Mitigation Strategies. Front. Vet. Sci. 2020, 7, 620. [Google Scholar] [CrossRef] [PubMed]
- Alegría, Á.; Arias-Temprano, M.; Fernández-Natal, I.; Rodríguez-Calleja, J.M.; García-López, M.-L.; Santos, J.A. Molecular Diversity of ESBL-Producing Escherichia coli from Foods of Animal Origin and Human Patients. Int. J. Environ. Res. Public Health 2020, 17, 1312. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, Y.; Jay-Russell, M.; Lemay, D.G.; Mills, D.A. Reservoirs of Antimicrobial Resistance Genes in Retail Raw Milk. Microbiome 2020, 8, 99. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Banerjee, J.; Bandyopadhyay, S.; Mondal, B.; Nanda, P.K.; Samanta, I.; Mahanti, A.; Das, A.K.; Das, G.; Dandapat, P.; et al. First Report on Vancomycin-Resistant Staphylococcus aureus in Bovine and Caprine Milk. Microb. Drug Resist. 2016, 22, 675–681. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Banerjee, J.; Bhattacharyya, D.; Samanta, I.; Mahanti, A.; Dutta, T.K.; Ghosh, S.; Nanda, P.K.; Dandapat, P.; Bandyopadhyay, S. Genomic Identity of Fluoroquinolone-Resistant Bla CTX-M -15 -Type ESBL and PMAmpC β-Lactamase Producing Klebsiella pneumoniae from Buffalo Milk, India. Microb. Drug Resist. 2018, 24, 1345–1353. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Bhattacharyya, D.; Samanta, I.; Banerjee, J.; Habib, M.; Dutta, T.K.; Dutt, T. Characterization of Multidrug-Resistant Biofilm-Producing Escherichia coli and Klebsiella pneumoniae in Healthy Cattle and Cattle with Diarrhea. Microb. Drug Resist. 2021, 27, 1457–1469. [Google Scholar] [CrossRef]
- Tewari, R.; Mitra, S.; Ganaie, F.; Das, S.; Chakraborty, A.; Venugopal, N.; Shome, R.; Rahman, H.; Shome, B.R. Dissemination and Characterisation of Escherichia coli Producing Extended-Spectrum β-Lactamases, AmpC β-Lactamases and Metallo-β-Lactamases from Livestock and Poultry in Northeast India: A Molecular Surveillance Approach. J. Glob. Antimicrob. Resist. 2019, 17, 209–215. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Samanta, I. Emergence of Antimicrobial-Resistant Bacteria in Environment. In Antimicrobial Resistance in Agriculture 1st Edition Perspective, Policy and Mitigation; Academic Press Inc.—Elsevier: London, UK, 2019; pp. 39–45. [Google Scholar]
- Bhattacharyya, D.; Banerjee, J.; Habib, M.; Thapa, G.; Samanta, I.; Nanda, P.K.; Dutt, T.; Sarkar, K.; Bandyopadhyay, S. Elucidating the Resistance Repertoire, Biofilm Production and Phylogenetic Characteristics of Multidrug-resistant Escherichia coli Isolated from Community Ponds: A Study from West Bengal, India. Water Environ. Res 2021, 94, e1678. [Google Scholar] [CrossRef]
- Varga, C.; Rajić, A.; McFall, M.E.; Reid-Smith, R.J.; Deckert, A.E.; Pearl, D.L.; Avery, B.P.; Checkley, S.L.; McEwen, S.A. Comparison of Antimicrobial Resistance in Generic Escherichia coli and Salmonella Spp. Cultured from Identical Fecal Samples in Finishing Swine. Can. J. Vet. Res. 2008, 72, 181–187. [Google Scholar] [PubMed]
- Daniels, J.B.; Call, D.R.; Hancock, D.; Sischo, W.M.; Baker, K.; Besser, T.E. Role of Ceftiofur in Selection and Dissemination of BlaCMY-2- Mediated Cephalosporin Resistance in Salmonella enterica and Commensal Escherichia coli Isolates from Cattle. Appl. Environ. Microbiol. 2009, 75, 3648–3655. [Google Scholar] [CrossRef] [PubMed]
- Wadepohl, K.; Müller, A.; Seinige, D.; Rohn, K.; Blaha, T.; Meemken, D.; Kehrenberg, C. Association of Intestinal Colonization of ESBL-Producing Enterobacteriaceae in Poultry Slaughterhouse Workers with Occupational Exposure-A German Pilot Study. PLoS ONE 2020, 15. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, W.; Dorado-García, A.; Bonten, M.J.M.; Wagenaar, J.A.; Mevius, D.; Heederik, D.J.J. Risk Factors for ESBL-Producing Escherichia coli on Pig Farms: A Longitudinal Study in the Context of Reduced Use of Antimicrobials. PLoS ONE 2017, 12, e0174094. [Google Scholar] [CrossRef]
- Ndegwa, E.; Almehmadi, H.; Chyer, K.; Kaseloo, P.; Ako, A.A. Longitudinal Shedding Patterns and Characterization of Antibiotic Resistant E. coli in Pastured Goats Using a Cohort Study. Antibiotics 2019, 8, 136. [Google Scholar] [CrossRef]
- Salvia, M.-V.; Fieu, M.; Vulliet, E. Determination of Tetracycline and Fluoroquinolone Antibiotics at Trace Levels in Sludge and Soil. Appl. Environ. Soil Sci. 2015, 2015, 435741. [Google Scholar] [CrossRef]
- Koovapra, S.; Bandyopadhyay, S.; Das, G.; Bhattacharyya, D.; Banerjee, J.; Mahanti, A.; Samanta, I.; Nanda, P.K.; Kumar, A.; Mukherjee, R.; et al. Molecular Signature of Extended Spectrum β-Lactamase Producing Klebsiella pneumoniae Isolated from Bovine Milk in Eastern and North-Eastern India. Infect. Genet. Evol. 2016, 44, 395–402. [Google Scholar] [CrossRef]
- Kar, D.; Bandyopadhyay, S.; Bhattacharyya, D.; Samanta, I.; Mahanti, A.; Nanda, P.K.; Mondal, B.; Dandapat, P.; Das, A.K.; Dutta, T.K.; et al. Molecular and Phylogenetic Characterization of Multidrug Resistant Extended Spectrum Beta-Lactamase Producing Escherichia coli Isolated from Poultry and Cattle in Odisha, India. Infect. Genet. Evol. 2015, 29, 82–90. [Google Scholar] [CrossRef]
- Antão, E.-M.; Wieler, L.H.; Ewers, C. Adhesive Threads of Extraintestinal Pathogenic Escherichia coli. Gut Pathog. 2009, 1, 22. [Google Scholar] [CrossRef]
- Franz, E.; Veenman, C.; van Hoek, A.H.A.M.; Husman, A.D.R.; Blaak, H. Pathogenic Escherichia coli Producing Extended-Spectrum β-Lactamases Isolated from Surface Water and Wastewater. Sci. Rep. 2015, 5, 14372. [Google Scholar] [CrossRef]
- Das, L.; Borah, P.; Sharma, R.K.; Malakar, D.; Saikia, G.K.; Sharma, K.; Tamuly, S.; Dutta, R. Phenotypic and Molecular Characterization of Extended Spectrum β-Lactamase Producing 2 Escherichia coli and Klebsiella pneumoniae Isolates from Various Samples of Animal Origin 3 from Assam, India. bioRxiv 2020. [Google Scholar] [CrossRef]
- Shabana, I.I.; Al-Enazi, A.T. Investigation of Plasmid-Mediated Resistance in E. Coli Isolated from Healthy and Diarrheic Sheep and Goats. Saudi J. Biol. Sci. 2020, 27, 788. [Google Scholar] [CrossRef] [PubMed]
- Subramanya, S.H.; Bairy, I.; Metok, Y.; Baral, B.P.; Gautam, D.; Nayak, N. Detection and Characterization of ESBL-Producing Enterobacteriaceae from the Gut of Subsistence Farmers, Their Livestock and the Surrounding Environment in Rural Nepal. Sci. Rep. 2021, 11, 2091. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Xu, X.; Zhang, Z.; Shen, H.; Chen, J.; Zhang, K. Molecular Characterization of Clinical Multidrug-Resistant Klebsiella pneumoniae Isolates. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 16. [Google Scholar] [CrossRef]
- Hu, L.; Zhong, Q.; Shang, Y.; Wang, H.; Ning, C.; Li, Y.; Hang, Y.; Xiong, J.; Wang, X.; Xu, Y.; et al. The Prevalence of Carbapenemase Genes and Plasmid-Mediated Quinolone Resistance Determinants in Carbapenem-Resistant Enterobacteriaceae from Five Teaching Hospitals in Central China. Epidemiol. Infect. 2014, 142, 1972–1977. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.; le Devendec, L.; Lucas, P.; Larvor, E.; Jové, T.; Kempf, I. Characterisation of Plasmids Harbouring Extended-Spectrum Cephalosporin Resistance Genes in Escherichia Coli from French Rivers. Vet. Microbiol. 2020, 243, 108619. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids Carrying Antimicrobial Resistance Genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef]
- de Lagarde, M.; Larrieu, C.; Praud, K.; Lallier, N.; Trotereau, A.; Sallé, G.; Fairbrother, J.M.; Schouler, C.; Doublet, B. Spread of Multidrug-Resistant IncHI1 Plasmids Carrying ESBL Gene blaCTX-M-1 and Metabolism Operon of Prebiotic Oligosaccharides in Commensal Escherichia coli from Healthy Horses, France. Int. J. Antimicrob. Agents 2020, 55, 105936. [Google Scholar] [CrossRef]
- Camp, J.; Schuster, S.; Vavra, M.; Schweigger, T.; Rossen, J.W.A.; Reuter, S.; Kern, W.V. Limited Multidrug Resistance Efflux Pump Overexpression among Multidrug-Resistant Escherichia coli Strains of ST131. Antimicrob. Agents Chemother. 2021, 65, e01735-20. [Google Scholar] [CrossRef]
- Hora, P.I.; Pati, S.G.; McNamara, P.J.; Arnold, W.A. Increased Use of Quaternary Ammonium Compounds during the SARS-CoV-2 Pandemic and Beyond: Consideration of Environmental Implications. Environ. Sci. Technol. Lett. 2020, 7, 622–631. [Google Scholar] [CrossRef]
- Molina, F.; López-Acedo, E.; Tabla, R.; Roa, I.; Gómez, A.; Rebollo, J.E. Improved Detection of Escherichia coli and Coliform Bacteria by Multiplex PCR. BMC Biotechnol. 2015, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Siegel, J.D.; Rhinehart, E.; Jackson, M.; Chiarello, L. Healthcare Infection Control Practices Advisory Committee (2007). Management of multidrug-resistant organisms in health care settings. Am. J. Infect. Control 2006, 35 (Suppl. 2), S165–S193. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J. Detection of Extended-Spectrum b-Lactamases (ESBLs) in Escherichia coli and Klebsiella species. J. Clin. Microbiol. 1996, 34, 908–911. [Google Scholar] [CrossRef]
- Polsfuss, S.; Bloemberg, G.V.; Giger, J.; Meyer, V.; Böttger, E.C.; Hombach, M. Practical Approach for Reliable Detection of AmpC Beta-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2011, 49, 2798–2803. [Google Scholar] [CrossRef] [PubMed]
- Dallenne, C.; da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important β-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Bertini, A.; Falbo, V.; Carattoli, A.; Hopkins, K.L.; Threlfall, E.J.; Villa, L. Identification of Plasmids by PCR-Based Replicon Typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Osei Sekyere, J.; Amoako, D.G. Carbonyl Cyanide M-Chlorophenylhydrazine (CCCP) Reverses Resistance to Colistin, but Not to Carbapenems and Tigecycline in Multidrug-Resistant Enterobacteriaceae. Front. Microbiol. 2017, 8, 228. [Google Scholar] [CrossRef]
- Vera-Leiva, A.; Carrasco-Anabalón, S.; Lima, C.A.; Villagra, N.; Domínguez, M.; Bello-Toledo, H.; González-Rocha, G. The Efflux Pump Inhibitor Phenylalanine-Arginine β-Naphthylamide (PAβN) Increases Resistance to Carbapenems in Chilean Clinical Isolates of KPC-Producing Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2018, 12, 73–76. [Google Scholar] [CrossRef]
- Helaly, G.F.; Shawky, S.; Amer, R.; El-sawaf, G.; Kholy, M.A. Expression of AcrAB Efflux Pump and Role of Mefloquine as Efflux Pump Inhibitor in MDR E. coli. Am. J. Infect. Dis. 2016, 4, 6–13. [Google Scholar] [CrossRef]

| Resistance Characteristics | Risk Factors | Proportion (%) with 95% CI | Odds Ratio with 95% CI | Significance |
|---|---|---|---|---|
| ESBL positivity | Farming practice | |||
| Intensive * | 11.8 (8.8–14.8) | 1.1 (0.6–2.0) | ns | |
| Semi-intensive * | 14.6 (9.5–19.9) | |||
| Health condition | ||||
| With diarrhoea ** | 14.6 (9.5–20) | 1.4 (0.8–2.4) | ns | |
| Without diarrhoea * | 10.9 (8–13.9) | |||
| Agro-climatic zones | ||||
| Coastal saline zone * | 12.2 (7.7–17.2) | 1.1 (0.6–2.2) | ns | |
| Red and laterite zone * | 13.0 (8.2–18.4) | 0.8 (0.4–1.8) | ||
| New alluvial zone * | 10.3 (6–16) | 1.0 (0.5–2.1) | ||
| Old alluvial zone * | 12.2 (7–18) | |||
| ACBL positivity | Farming practice | |||
| Intensive * | 19.1 (12.7–25.8) | 1.3 (0.7–2.1) | ns | |
| Semi-intensive * | 15.7 (12.5–19.3) | |||
| Health condition | ||||
| With diarrhoea ** | 22.9 (16.6–29.3) | 1.9 (1.2–3.0) | 0.001 | |
| Without diarrhoea * | 13.8 (10.6–17.3) | |||
| Agro-climatic zones | ||||
| Coastal saline zone * | 17.3 (12.2–23.5) | |||
| Red and laterite zone * | 16.4 (11.0–22.4) | 1.0 (0.5–1.8) | ns | |
| New alluvial zone * | 15.4 (9.4–1.6) | 0.9 (0.5–1.7) | ||
| Old alluvial zone * | 16.5 (10.4–23.2) | 0.9 (0.5–1.8) | ||
| Ampicillin resistance | Farming practice | |||
| Intensive ** | 29.4 (22.2–37.9) | 1.4 (0.9–2.2) | ns | |
| Semi-intensive ** | 22.6 (18.6–26.7) | |||
| Health condition | ||||
| With diarrhoea ** | 26.8 (20.4–34.0) | 1.2 (0.8–1.9) | ns | |
| Without diarrhoea ** | 23.1 (19.1–27.5) | |||
| Agro-climatic zones | ||||
| Coastal saline zone ** | 25.0 (18.6–31.9) | |||
| Red and laterite zone ** | 24.7 (18.5–32.1) | 1.0 (0.6–1.7) | ns | |
| New alluvial zone ** | 21.4 (14.5–28.8) | 0.9 (0.5–1.5) | ||
| Old alluvial zone ** | 25.2 (18.3–33.7) | 1.1 (0.6–1.9) | ||
| Amoxycillin-clavulanic acid resistance | Farming practice | |||
| Intensive * | 15.1(9.5–1.3) | 1.3 (0.7–2.4) | ns | |
| Semi-intensive * | 11.5 (8.6–4.5) | |||
| Health condition | ||||
| With diarrhoea * | 14.0 (9.6–9.7) | 1.2 (0.7–2.2) | ns | |
| Without diarrhoea * | 11.7 (8.8–14.9) | |||
| Agro-climatic zones | ||||
| Coastal saline zone * | 12.8 (8.3–18.1) | |||
| Red and laterite zone * | 13.7 (8.9–19.4) | 1.1 (0.6–2.2) | ns | |
| New alluvial zone * | 10.3 (6.0–16.0) | 0.8 (0.4–1.8) | ||
| Old alluvial zone * | 12.2 (7.0–18.0) | 1.0 (0.5–2.0) | ||
| Cefotaxime resistance | Farming practice | |||
| Intensive ** | 28.6 (21.4–37.0) | 1.50 (0.97–2.31) | 0.05 | |
| Semi-intensive ** | 21.1 (17.4–25.2) | |||
| Health condition | ||||
| With diarrhoea ** | 28.0 (21.7–35.5) | 1.50 (0.94–2.38) | 0.05 | |
| Without diarrhoea ** | 20.7 (16.7–24.8) | |||
| Agro-climatic zones | ||||
| Coastal saline zone ** | 25.0 (18.6–31.9) | |||
| Red and laterite zone ** | 20.6 (14.4 -27.0) | 0.81 (0.47–1.40) | ns | |
| New alluvial zone ** | 22.2 (15.4 -29.9) | 0.92 (0.51–1.64) | ||
| Old alluvial zone ** | 23.5 (16.5–31.5) | 0.95 (0.53–1.69) | ||
| Amikacin resistance | Farming practice | |||
| Intensive ** | 22.2 (15.9–29.9) | 1.05 (0.63–1.70) | ns | |
| Semi-intensive ** | 21.0 (17.0- 25.1) | |||
| Health condition | 21.1 (17.4–25.2) | |||
| With diarrhoea ** | 22.3 (16.6–29.2) | 1.09 (0.69–1.71) | ns | |
| Without diarrhoea ** | 21.0 (17.0- 25.1) | |||
| Agro-climatic zones | ||||
| Coastal saline zone ** | 22.4 (16.7–29.4) | |||
| Red and laterite zone ** | 21.9 (15.8–28.8) | 0.98 (0.56–1.69) | ns | |
| New alluvial zone ** | 20.5 (13.7–27.7) | 0.90 (0.49–1.62) | ||
| Old alluvial zone ** | 20.0 (13.9–27.8) | 0.86 (0.47–1.57) | ||
| Tetracycline resistance | Farming practice | |||
| Intensive ** | 26.98 (19.84–35.04) | 1.4 (0.8–2.1) | ns | |
| Semi-intensive ** | 22.06 (18.14–26.09) | |||
| Health condition | ||||
| With diarrhoea ** | 28.66(22.29–36.26) | 1.5 (1.0–2.3) | ns | |
| Without diarrhoea ** | 20.95(16.98–25.06) | |||
| Agro-climatic zones | ||||
| Coastal saline zon e ** | 22.44(16.67–29.36) | |||
| Red and laterite zone ** | 22.6(16.44–29.61) | 1.0 (0.6–1.8) | ns | |
| New alluvial zone ** | 23.93(17.09–32.07) | 1.1 (0.6–2.0) | ||
| Old alluvial zone ** | 24.35 (17.39–32.6) | 1.1 (0.6–2.0) | ||
| Enrofloxacin resistance | Farming practice | |||
| Intensive ** | 31.8(23.8–39.9) | 1.7 (1.0–2.6) | 0.01 | |
| Semi-intensive ** | 22.3 (18.4–26.4) | |||
| Health condition | ||||
| With diarrhoea ** | 31.2 (24.2–38.6) | 1.6 (1.1–2.5) | 0.01 | |
| Without diarrhoea ** | 21.8 (17.8–26.0) | |||
| Agro-climatic zones | ||||
| Coastal saline zone ** | 25.0 (18.6–31.9) | |||
| Red and laterite zone ** | 24.0 (17.8–31.3) | 1.0 (0.6–1.7) | ns | |
| New alluvial zone ** | 22.2 (15.4–29.9) | 0.9 (0.5–1.7) | ||
| Old alluvial zone ** | 27.0 (19.1–35.0) | 1.2 (0.7–2.0) | ||
| Cotrimoxazole resistance | Farming practice | |||
| Intensive ** | 14.29 (8.73–20.12) | 1.3 (0.7–2.3) | ns | |
| Semi-intensive ** | 11.52 (8.58–14.46) | |||
| Health condition | ||||
| With diarrhoea ** | 13.38 (8.92–18.88) | 1.2 (0.7–2.1) | ns | |
| Without diarrhoea ** | 11.67 (8.75–14.92) | |||
| Agro-climatic zones | ||||
| Coastal saline zone ** | 13.46 (8.97–19) | |||
| Red and laterite zone ** | 11.64 (7.53–17.12) | 0.9 (0.4–1.7) | ns | |
| New alluvial zone ** | 12.82 (7.69–18.9) | 1.0 (0.5–2.0) | ||
| Old alluvial zone ** | 10.43 (6.09–16.24) | 0.8 (0.3–1.6) | ||
| Imipenem resistance | Farming practice | |||
| Intensive * | 12.7 (7.94–18.7) | 1.6 (0.8–2.9) | ns | |
| Semi-intensive | 8.82 (6.37–11.54) | |||
| Health condition | ||||
| With diarrhoea * | 11.46 (7.01–16.24) | 1.3 (0.7–2.4) | ns | |
| Without diarrhoea | 9.02 (6.37–11.79) | |||
| Agro-climatic zones | ||||
| Coastal saline zone * | 10.26 (6.41–15.16) | |||
| Red and laterite zone | 8.22 (4.79–12.83) | 0.8 (0.4–1.8) | ns | |
| New alluvial zone * | 11.97 (6.84–17.66) | 1.3 (0.6–2.8) | ||
| Old alluvial zone | 8.7 (4.35–13.6) | 0.9 (0.4–2.0) |
| Isolates | Resistance Genotype | ESBL | AmpC-βL | MBL | BF | MEM-CCCP | EP (CIP) | Plasmid Replicon |
|---|---|---|---|---|---|---|---|---|
| CaEC1 | blaCTXM-1-qnrB-aac(6′)-Ib-cr-tetA | P | N | N | MP | N | N | HI1, FIA, FrepB |
| CaEC2 | blaCTXM-1-qnrB-tetA | P | P | N | SP | N | P | FIA, I1, FrepB |
| CaEC3 | blaCTXM-1-qnrS-tetA | P | P | N | SP | P | N | FIA, I1, FrepB |
| CaEC4 | blaCTXM-1-blaSHV12-qnrB-tetA | P | P | N | MP | N | N | FIA, I1, FrepB |
| CaEC5 | blaAmpC-tetA | N | P | N | SP | N | P | N |
| CaEC6 | blaCTXM-1-qnrB | P | N | N | SP | N | N | I1, FrepB |
| CaEC8 | blaCTXM-1-qnrB-aac(6’)-Ib-cr-tetA | P | P | N | SP | N | P | HI1, FIB, FrepB |
| CaEC9 | blaAmpC-blaCITM | N | N | N | WP | N | N | HI1 |
| CaEC10 | blaAmpC-qnrS-tetA | N | P | N | MP | P | N | N, HI1 |
| CaEC12 | blaCTXM-1-qnrS-aac(6’)-Ib-cr-tetA | P | P | N | MP | N | N | FIA, N, FrepB |
| CaEC13 | blaCTXM-1-blaSHV12-qnrB-tetA | P | P | N | MP | N | N | FIA, FIB, FrepB |
| CaEC15 | blaCTXM-1-qnrS-tetB | P | P | N | MP | N | N | FIA, FIC |
| CaEC16 | blaCTXM1-tetA | P | N | N | MP | N | N | FIB, N |
| CaEC18 | blaCTXM-1 | P | P | N | MP | N | N | FIA |
| CaEC19 | blaCTXM-1-blaSHV12-qnrB-tetA | N | P | N | MP | N | P | FIA, I1, FrepB, N |
| CaEC20 | blaCTXM-1-blaSHV12-qnrS | P | P | N | WP | N | P | FIA, FIB |
| CaEC22 | blaAmpC-tetA | N | P | N | WP | N | P | N |
| CaEC23 | blaAmpC-blaCITM-tetB | P | P | N | MP | N | N | HI1 |
| CaEC24 | blaCTXM1-tetA | P | P | N | WP | N | N | FIB, N, FrepB |
| CaEC25 | blaCTXM1-tetA | P | P | N | WP | N | N | FIB, N, FrepB |
| CaEC26 | blaAmpC-blaCITM-tetB | N | N | N | SP | P | N | HI1, FIC |
| CaEC28 | blaAmpC-tetA | N | P | N | MP | N | N | N |
| CaEC29 | blaAmpC-tetA-qnrS | N | P | N | MP | N | N | FIA, N |
| CaEC30 | blaCTXM1-tetB-qnrB-CMY-6 | P | P | N | WP | N | N | FIA, FIB, HI1, FrepB |
| CaEC31 | blaCTXM-1-blaSHV12-qnrB-tetA | N | P | N | WP | N | P | I1, FIA, FrepB |
| CaEC32 | blaCTXM-1- blaSHV12- blaAmpC | N | P | N | MP | N | N | FIB, FepB |
| CaEC33 | blaNDM-5-blaTEM-sul1 | P | N | P | WP | N | N | FIB, A/C, FrepB |
| CaEC34 | blaCTXM-1-blaSHV12-ampC | P | P | N | SP | N | N | FIA, FrepB |
| CaEC36 | blaTEM-sul1 | N | N | N | WP | N | N | FIC, FrepB |
| CaEC37 | qnrB-qnrS-sul1 | N | N | N | MP | N | N | I1, HI1 |
| CaEC38 | blaAmpC | N | P | N | MP | N | N | Unknown |
| CaEC40 | blaCTXM-1-tetA | P | N | N | WP | N | P | FIB, FrepB |
| CaEC41 | blaCTXM-1-blaTEM-ampC | P | P | N | SP | N | P | I1, HI1, FrepB |
| CaEC42 | blaNDM-5 | N | N | P | MP | N | N | A/C, FrepB |
| CaEC44 | blaAmpC-sul1-tetA | N | P | N | MP | N | P | HI1, N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banerjee, J.; Bhattacharyya, D.; Habib, M.; Chaudhary, S.; Biswas, S.; Maji, C.; Nanda, P.K.; Das, A.K.; Dandapat, P.; Samanta, I.; et al. Antimicrobial Resistance Pattern, Clustering Mechanisms and Correlation Matrix of Drug-Resistant Escherichia coli in Black Bengal Goats in West Bengal, India. Antibiotics 2022, 11, 1344. https://doi.org/10.3390/antibiotics11101344
Banerjee J, Bhattacharyya D, Habib M, Chaudhary S, Biswas S, Maji C, Nanda PK, Das AK, Dandapat P, Samanta I, et al. Antimicrobial Resistance Pattern, Clustering Mechanisms and Correlation Matrix of Drug-Resistant Escherichia coli in Black Bengal Goats in West Bengal, India. Antibiotics. 2022; 11(10):1344. https://doi.org/10.3390/antibiotics11101344
Chicago/Turabian StyleBanerjee, Jaydeep, Debaraj Bhattacharyya, Md Habib, Siddharth Chaudhary, Suman Biswas, Chinmoy Maji, Pramod Kumar Nanda, Arun K. Das, Premanshu Dandapat, Indranil Samanta, and et al. 2022. "Antimicrobial Resistance Pattern, Clustering Mechanisms and Correlation Matrix of Drug-Resistant Escherichia coli in Black Bengal Goats in West Bengal, India" Antibiotics 11, no. 10: 1344. https://doi.org/10.3390/antibiotics11101344
APA StyleBanerjee, J., Bhattacharyya, D., Habib, M., Chaudhary, S., Biswas, S., Maji, C., Nanda, P. K., Das, A. K., Dandapat, P., Samanta, I., Lorenzo, J. M., Dutt, T., & Bandyopadhyay, S. (2022). Antimicrobial Resistance Pattern, Clustering Mechanisms and Correlation Matrix of Drug-Resistant Escherichia coli in Black Bengal Goats in West Bengal, India. Antibiotics, 11(10), 1344. https://doi.org/10.3390/antibiotics11101344








