Foodborne Pathogen Dynamics in Meat and Meat Analogues Analysed Using Traditional Microbiology and Metagenomic Sequencing
Abstract
:1. Introduction
2. Material and Methods
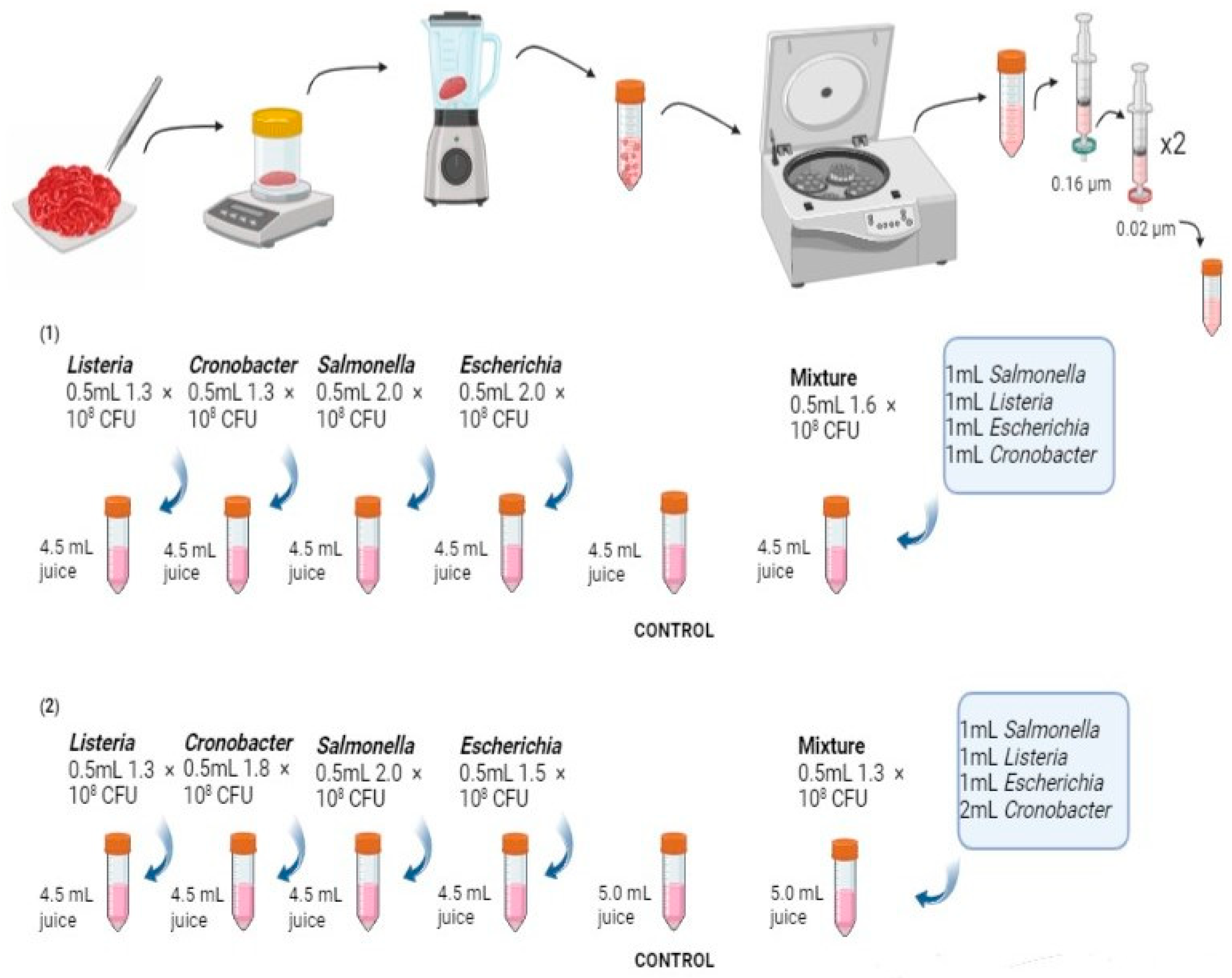
2.1. Sample Collection and Preparation
2.2. Bacterial Pathogen Preparation
2.3. Bacterial Inoculation and Culturomics
2.4. DNA Extraction and Metagenomic Sequencing
2.5. Bioinformatic Analyses
3. Results
3.1. Viability of Bacterial Pathogens in Various Meat and Meat Analogues Using Culture-Dependent Approach
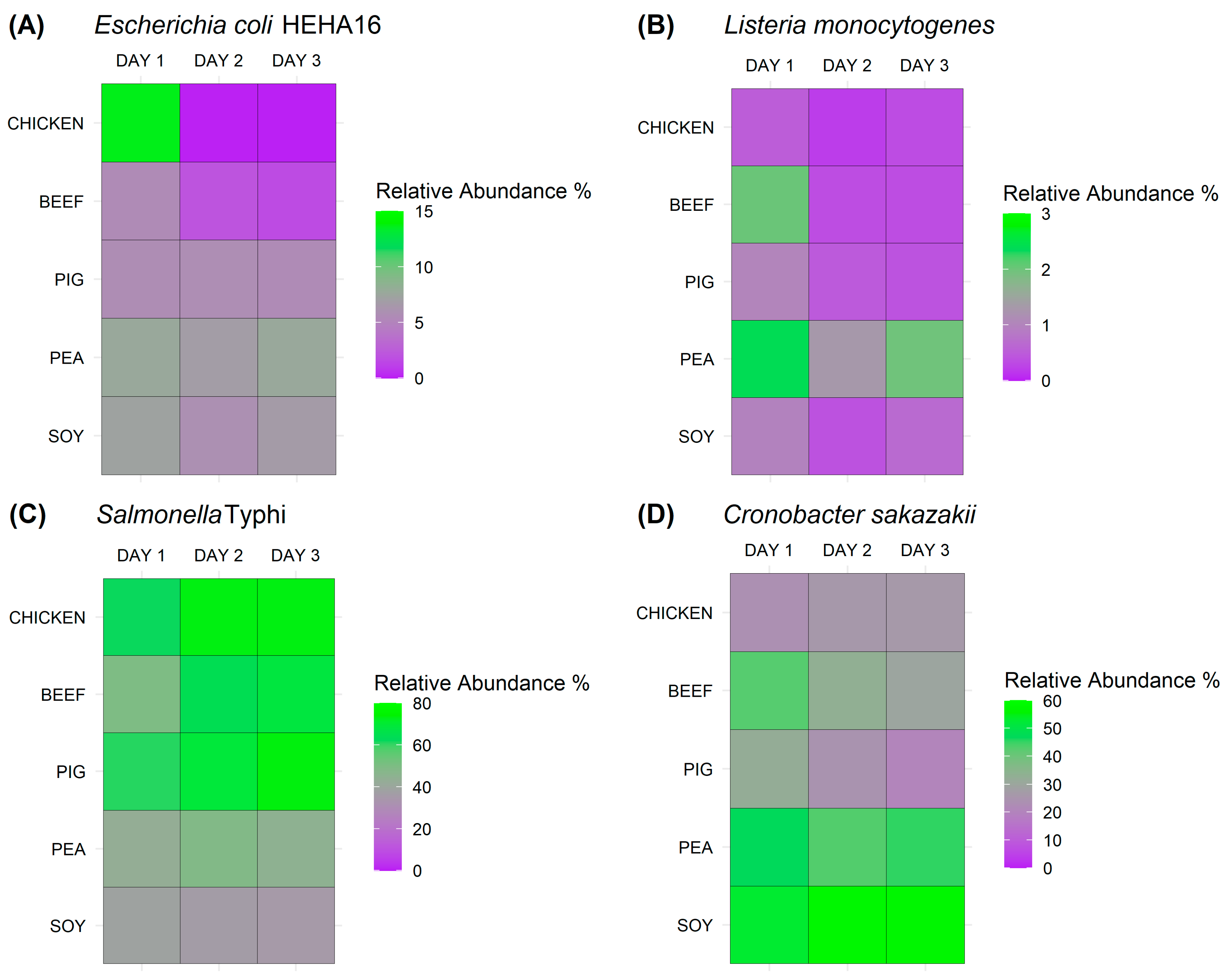
3.2. In-Depth Detection of Bacterial Pathogens in Various Meat and Meat Analogues Using Metagenomic Sequencing Approach
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Steps up Action to Improve Food Safety and Protect People from Disease. 2021. Available online: https://www.who.int/news/item/07-06-2021-who-steps-up-action-to-improve-food-safety-and-protect-people-from-disease (accessed on 27 November 2023).
- Ge, H.; Wang, Y.; Zhao, X. Research on the drug resistance mechanism of foodborne pathogens. Microb. Pathog. 2022, 162, 105306. [Google Scholar] [CrossRef] [PubMed]
- Elbehiry, A.; Abalkhail, A.; Marzouk, E.; Elmanssury, A.E.; Almuzaini, A.M.; Alfheeaid, H.; Alshahrani, M.T.; Huraysh, N.; Ibrahem, M.; Alzaben, F.; et al. An Overview of the Public Health Challenges in Diagnosing and Controlling Human Foodborne Pathogens. Vaccines 2023, 11, 725. [Google Scholar] [CrossRef] [PubMed]
- Foddai, A.C.G.; Grant, I.R. Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281–4288. [Google Scholar] [CrossRef] [PubMed]
- Abebe, G.M. Cronobacter sakazakii in infant food contamination and its survival strategies in hostile conditions. Int. J. Pediatr. Res. 2020, 6, e067. [Google Scholar]
- Lianou, A.; Panagou, E.Z.; Nychas, G.J.E. Meat safety—I foodborne pathogens and other biological issues. In Lawrie’s Meat Science; Woodhead Publishing: Cambridge, UK, 2023; pp. 549–590. [Google Scholar]
- Lee, H.J.; Yong, H.I.; Kim, M.; Choi, Y.-S.; Jo, C. Status of meat alternatives and their potential role in the future meat market—A review. Asian-Australas. J. Anim. Sci. 2020, 33, 1533–1543. [Google Scholar] [CrossRef]
- Sha, L.; Xiong, Y.L. Plant protein-based alternatives of reconstructed meat: Science, technology, and challenges. Trends Food Sci. Technol. 2020, 102, 51–61. [Google Scholar] [CrossRef]
- Wild, F.; Czerny, M.; Janssen, A.M.; Kole, A.P.E.; Zunabovic, M.Z.; Domig, K.J. The evolution of a plant-based alternative to meat. Agro Food Ind. Hi Tech 2014, 25, 45–49. [Google Scholar]
- Sampson, G.L.; Ruelle, S.B.; Phan, L.; Williams-Hill, D.; Hellberg, R.S. Effectiveness of selected pre-enrichment broths for the detection of Salmonella spp. in meat analogs. Food Control 2023, 143, 109282. [Google Scholar] [CrossRef]
- Aladhadh, M. A Review of Modern Methods for the Detection of Foodborne Pathogens. Microorganisms 2023, 11, 1111. [Google Scholar] [CrossRef]
- Billington, C.; Kingsbury, J.M.; Rivas, L. Metagenomics Approaches for Improving Food Safety: A Review. J. Food Prot. 2022, 85, 448–464. [Google Scholar] [CrossRef]
- Cao, Y.; Fanning, S.; Proos, S.; Jordan, K.; Srikumar, S. A review on the applications of next generation sequencing technologies as applied to food-related microbiome studies. Front. Microbiol. 2017, 8, 1829. [Google Scholar] [CrossRef] [PubMed]
- Desdouits, M.; de Graaf, M.; Strubbia, S.; Munnink, B.B.O.; Kroneman, A.; Le Guyader, F.S.; Koopmans, M.P.G. Novel opportunities for NGS-based one health surveillance of foodborne viruses. One Health Outlook 2020, 2, 14. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.; Hudson, J.; Cook, N.; Barnes, J.; Haynes, E. Next-generation sequencing as a screening tool for foodborne pathogens in fresh produce. J. Microbiol. Methods 2020, 171, 105840. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Feng, J.; Ma, L.; Núñez, C.d.l.F.; Gölz, G.; Lu, X. Effects of meat juice on biofilm formation of Campylobacter and Salmonella. Int. J. Food Microbiol. 2017, 253, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Stanford, K.; Bie, X.M.; Niu, Y.D.; McAllister, T.A. Effects of beef juice on biofilm formation by Shiga toxin–producing Escherichia coli on stainless steel. Foodborne Pathog. Dis. 2020, 17, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Ligowska, M.; Cohn, M.T.; Stabler, R.A.; Wren, B.W.; Brøndsted, L. Effect of chicken meat environment on gene expression of Campylobacter jejuni and its relevance to survival in food. Int. J. Food Microbiol. 2011, 145, S111–S115. [Google Scholar] [CrossRef]
- González-Pérez, C.J.; Tanori-Cordova, J.; Aispuro-Hernández, E.; Vargas-Arispuro, I.; Martínez-Téllez, M. Morphometric parameters of foodborne related-pathogens estimated by transmission electron microscopy and their relation to optical density and colony forming units. J. Microbiol. Methods 2019, 165, 105691. [Google Scholar] [CrossRef]
- Clausen, P.T.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Maningat, C.C.; Jeradechachai, T.; Buttshaw, M.R. Textured wheat and pea proteins for meat alternative applications. Cereal Chem. 2022, 99, 37–66. [Google Scholar] [CrossRef]
- Hadi, J.; Brightwell, G. Safety of alternative proteins: Technological, environmental and regulatory aspects of cultured meat, plant-based meat, insect protein and single-cell protein. Foods 2021, 10, 1226. [Google Scholar] [CrossRef]
- Parvin, M.S.; Talukder, S.; Ali, Y.; Chowdhury, E.H.; Rahman, T.; Islam, T. Antimicrobial resistance pattern of Escherichia coli isolated from frozen chicken meat in Bangladesh. Pathogens 2020, 9, 420. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shaposhnikov, M.; Zhuang, S.; Tu, T.; Wang, H.; Wang, L. Growth and survival of common spoilage and pathogenic bacteria in ground beef and plant-based meat analogues. Food Res. Int. 2023, 164, 112408. [Google Scholar] [CrossRef] [PubMed]
- Suehr, Q.J.; Chen, F.; Anderson, N.M.; Keller, S.E. Effect of pH on survival of Escherichia coli O157, Escherichia coli O121, and Salmonella enterica during desiccation and short-term storage. J. Food Prot. 2020, 83, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-G.; Lin, T.-S.; Lin, W.-Y. Evaluating the growth of Listeria monocytogenes that has been inoculated into tofu containing background microflora. Food Control 2010, 21, 1764–1768. [Google Scholar] [CrossRef]
- de Freitas, C.G.; Santana, A.P.; da Silva, P.H.C.; Gonçalves, V.S.P.; de Aguiar Ferreira Barros, M.; Torres, F.A.G.; Murata, L.S.; Perecmanis, S. PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int. J. Food Microbiol. 2010, 139, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Mrema, N.; Mpuchane, S.; Gashe, B.A. Prevalence of Salmonella in raw minced meat, raw fresh sausages and raw burger patties from retail outlets in Gaborone, Botswana. Food Control 2006, 17, 207–212. [Google Scholar] [CrossRef]
- Osaili, T.M.; Al-Nabulsi, A.A.; Shaker, R.R.; Jaradat, Z.W.; Taha, M.; Al-Kherasha, M.; Meherat, M.; Holley, R. Prevalence of Salmonella serovars, Listeria monocytogenes, and Escherichia coli O157: H7 in Mediterranean ready-to-eat meat products in Jordan. J. Food Prot. 2014, 77, 106–111. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Cerda-Leal, F.; Contreras, A.; Valenzuela-Riffo, N.; Rodríguez, A.; Aguirre, J. Cronobacter sakazakii, Food alert, Microbiological parameters, Powdered infant formula, liquid dairy formula. Front. Microbiol. 2018, 9, 1708. [Google Scholar] [CrossRef]
- Polat Yemiş, G.; Delaquis, P. Natural compounds with antibacterial activity against Cronobacter spp. in powdered infant formula: A review. Front. Nutr. 2020, 7, 595964. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonaldo, F.; Avot, B.J.P.; De Cesare, A.; Aarestrup, F.M.; Otani, S. Foodborne Pathogen Dynamics in Meat and Meat Analogues Analysed Using Traditional Microbiology and Metagenomic Sequencing. Antibiotics 2024, 13, 16. https://doi.org/10.3390/antibiotics13010016
Bonaldo F, Avot BJP, De Cesare A, Aarestrup FM, Otani S. Foodborne Pathogen Dynamics in Meat and Meat Analogues Analysed Using Traditional Microbiology and Metagenomic Sequencing. Antibiotics. 2024; 13(1):16. https://doi.org/10.3390/antibiotics13010016
Chicago/Turabian StyleBonaldo, Francesco, Baptiste Jacques Philippe Avot, Alessandra De Cesare, Frank M. Aarestrup, and Saria Otani. 2024. "Foodborne Pathogen Dynamics in Meat and Meat Analogues Analysed Using Traditional Microbiology and Metagenomic Sequencing" Antibiotics 13, no. 1: 16. https://doi.org/10.3390/antibiotics13010016
APA StyleBonaldo, F., Avot, B. J. P., De Cesare, A., Aarestrup, F. M., & Otani, S. (2024). Foodborne Pathogen Dynamics in Meat and Meat Analogues Analysed Using Traditional Microbiology and Metagenomic Sequencing. Antibiotics, 13(1), 16. https://doi.org/10.3390/antibiotics13010016





