Multiplexed CRISPR Assay for Amplification-Free Detection of miRNAs
Abstract
1. Introduction
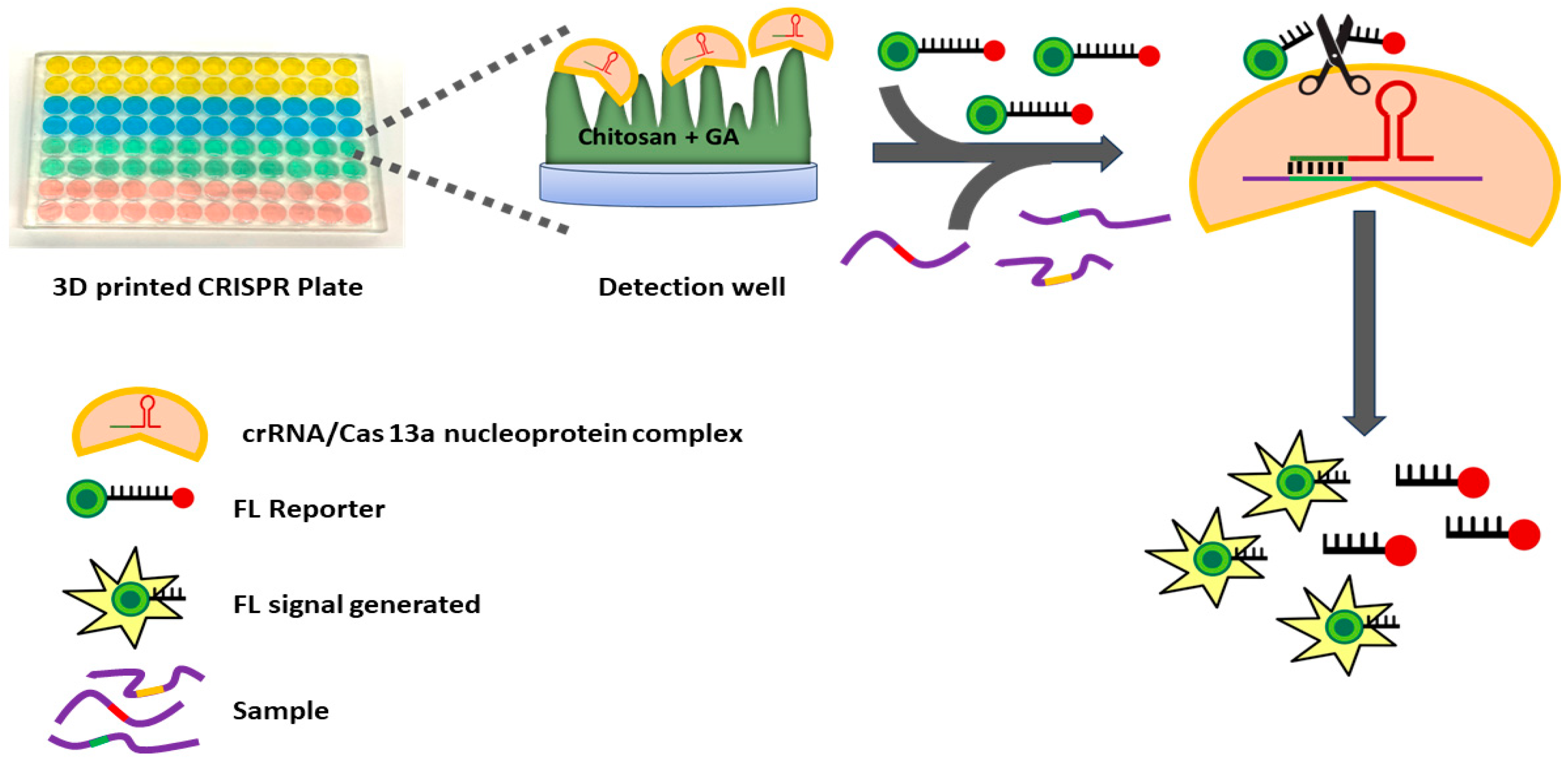
2. Materials and Methods
3. Results
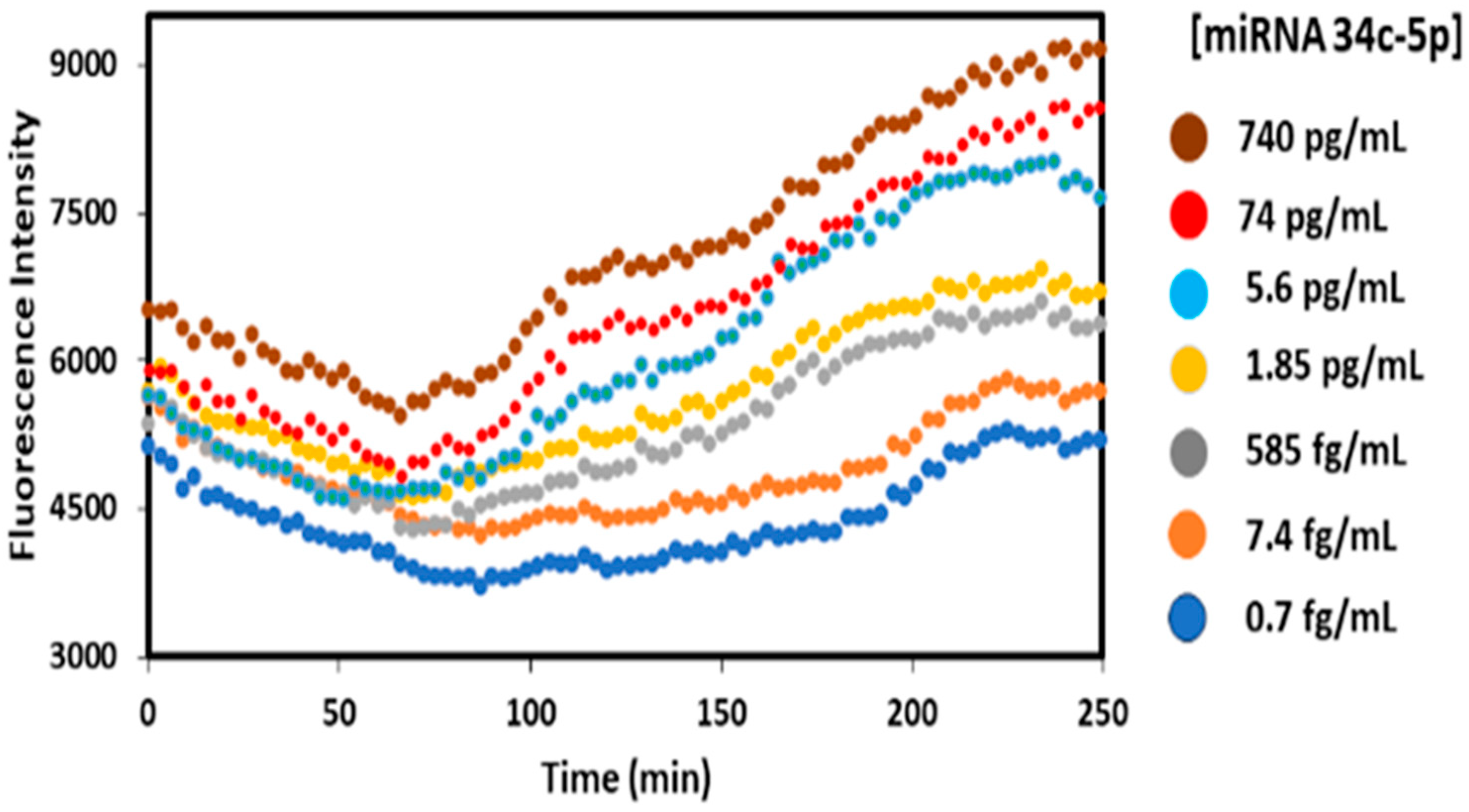
3.1. Assay Optimization
3.2. Calibration and Validation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rusling, J.F.; Kumar, C.V.; Gutkind, J.S.; Patel, V. Measurement of Biomarker Proteins for Point-of-Care Early Detection and Monitoring of Cancer. Analyst 2010, 135, 2496–2511. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Imran, M.; Ahsan, H. Biomarkers as Biomedical Bioindicators: Approaches and Techniques for the Detection, Analysis, and Validation of Novel Biomarkers of Diseases. Pharmaceutics 2023, 15, 1630. [Google Scholar] [CrossRef]
- Song, J.G.; Barol, K.C.; Kim, G.-L.; Park, J.-W.; Seo, S.-H.; Kim, D.-H.; Jung, D.H.; Ifekpolugo, N.L.; Han, H.-K. Quantitative analysis of therapeutic proteins in biological fluids: Recent advancement in analytical techniques. Drug Deliv. 2023, 30, 2183816. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Costelloe, C.M.; Rohren, E.M.; Madewell, J.E.; Hamaoka, T.; Theriault, R.L.; Yu, T.K.; Lewis, V.O.; Ma, J.; Stafford, R.J.; Tari, A.M.; et al. Imaging Bone Metastasis in Breast Cancer: Techniques and recommendations for diagnosis. Lancet 2009, 10, 606–614. [Google Scholar] [CrossRef]
- Tucaszewski, B.; Nazar, J.; Goch, M. Diagnostic Methods for Detection of bone metastasis. Contemp. Oncol. 2017, 21, 98–103. [Google Scholar] [CrossRef]
- Self, W.K.; Holzman, D.M. Emerging diagnostics and therapeutics for Alzheimer disease. Nat. Med. 2023, 29, 2187–2199. [Google Scholar] [CrossRef]
- Jiang, L.; Lin, X.; Chen, F.; Qin, X.; Yan, Y.; Ren, L.; Yu, H.; Chang, L.; Wang, Y. Current research status of tumor cell biomarker detection. Microsyst. Nanoeng. 2023, 9, 123. [Google Scholar] [CrossRef]
- Rusling, J.F.; Forster, R.J. Biosensors Designed for Clinical Applications. Biomedicines 2021, 9, 702. [Google Scholar] [CrossRef]
- Sharafeldin, M.; Kadimisetty, K.; Bhalero, K.R.; Chen, T.; Rusling, J.F. 3D-printed Immunosensor arrays for cancer diagnostics. Sensors 2020, 20, 4514. [Google Scholar] [CrossRef]
- Dhanapala, L.; Krause, C.E.; Jones, A.L.; Rusling, J.F. Printed electrodes in microfluidic arrays for cancer biomarker protein detection. Biosensors 2020, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Dhanapala, L.; Kankanamage, R.N.T.; Kumar, C.V.; Rusling, J.F. Multiplexed Immunosensors and Immunoarrays. Anal. Chem. 2020, 92, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Dhanapala, L.; Baldo, T.A.; Sharafeldin, M.; Krause, C.; Shen, M.; Faria, R.C.; Dey, D.; Moghaddam, S.; Watson, R.W.; et al. Prostate Cancer Diagnosis in the Clinic Using an 8-Protein Biomarker Panel. Anal. Chem. 2021, 93, 1059–1067. [Google Scholar] [CrossRef]
- Dhanapala, L.; Jones, A.L.; Czarnecki, P.; Rusling, J.F. Sub-Zeptomole Detection of Biomarker Proteins Using a Microfluidic Immunoarray with Nanostructured Sensors. Anal. Chem. 2020, 92, 8021–8025. [Google Scholar] [CrossRef]
- de Puig, H.; Lee, R.A.; Najjar, D.; Tan, X.; Soenksen, L.R.; Angenent-Mari, N.M.; Donghia, N.M.; Weckman, N.E.; Ory, A.; Ng, C.F.; et al. Minimally Instrumented SHERLOCK (miSHERLOCK) for CRISPR-Based Point-of-Care Diagnosis of SARS-CoV-2 and Emerging Variants. Sci. Adv. 2021, 7, eabh2944. [Google Scholar] [CrossRef]
- Krishnan, S.; Mani, V.; Wasalathanthri, D.; Kumar, C.V.; Rusling, J.F. Attomolar Detection of a Cancer Biomarker Protein in Serum by Surface Plasmon Resonance Using Superparamagnetic Particle Labels. Angew. Chem. Int. Ed. 2011, 50, 1175–1178. [Google Scholar] [CrossRef]
- Das, S.; Devireddy, R.; Gartia, M.R. Surface plasmon resonance (SPR) sensor for cancer biomarker detection. Biosensors 2023, 13, 396. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020, 11, 3475. [Google Scholar] [CrossRef]
- Shen, Y.; Yang, W.; Liu, J.; Zhang, Y. Minimally invasive approaches for the early detection of endometrial cancer. Mol. Cancer 2023, 22, 53. [Google Scholar] [CrossRef]
- Hiniduma, K.; Bhalerao, K.S.; De Silva, P.I.T.; Chen, T.; Rusling, J.F. Design and Fabrication of a 3D-Printed Microfluidic Immunoarray for Ultrasensitive Multiplexed Protein Detection. Micromachines 2023, 14, 2187. [Google Scholar] [CrossRef]
- Ghorbani, A.; Hadifar, S.; Salari, R.; Izadpanah, K.; Burmistrz, M.; Afsharifar, A.; Eskandari, M.H.; Niazi, A.; Denes, C.E.; Neely, G.G. A short overview of CRISPR-Cas technology and its application in viral disease control. Transgen. Res. 2021, 30, 221–238. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/CAS, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef]
- Lau, A.; Ren, C.; Lee, L.P. Critical review on where CRISPR meets molecular diagnostics. Prog. Biomed. Eng. 2020, 3, 012001. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Prot. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Green, C.M.; Spangler, J.; Susumu, K.; Stenger, D.A.; Medintz, I.L.; Díaz, S.A. Quantum Dot-Based Molecular Beacons for Quantitative Detection of Nucleic Acids with CRISPR/Cas(N) Nucleases. ACS Nano 2022, 16, 20693–20704. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Bai, X.; Zhang, X.; Yuan, B.; Lin, L.; Guo, Y.; Cui, Y.; Liu, J.; Cui, H.; Ren, X.; et al. Development and application of DETECTR-based rapid detection for pathogenic Bacillus anthracis. Anal. Chim. Acta 2023, 1247, 340891. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xia, Q.; Wu, J.; Lin, Y.; Ju, H. A sensitive electrochemical method for rapid detection of dengue virus by CRISPR/Cas13a-assisted catalytic hairpin assembly. Anal. Chim. Acta 2021, 1187, 339131. [Google Scholar] [CrossRef]
- Dukhovny, A.; Lamkiewicz, K.; Chen, Q.; Fricke, M.; Jabrane-Ferrat, N.; Marz, M.; Jung, J.U.; Sklan, E.H. A CRISPR Activation Screen Identifies Genes That Protect against Zika Virus Infection. J. Virol. 2019, 93, e00211-19. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and Visual Detection of SARS-CoV-2 Using All-in-One Dual CRISPR-Cas12a Assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]
- Jiang, Y.; Hu, M.; Liu, A.-A.; Lin, Y.; Liu, L.; Yu, B.; Zhou, X.; Pang, D.-W. Detection of SARS-COV-2 by CRISPR/CAS12A-Enhanced Colorimetry. ACS Sens. 2021, 6, 1086–1093. [Google Scholar] [CrossRef]
- Palaz, F.; Kalkan, A.K.; Can, Ö.; Demir, A.N.; Tozluyurt, A.; Özcan, A.; Ozsoz, M. CRISPR-CAS13 system as a promising and versatile tool for cancer diagnosis, therapy, and research. ACS Synth. Biol. 2021, 10, 1245–1267. [Google Scholar] [CrossRef] [PubMed]
- Kadimisetty, K.; Malla, S.; Bhalerao, K.S.; Mosa, I.M.; Bhakta, S.; Lee, N.H.; Rusling, J.F. Automated 3D-Printed Microfluidic Array for Rapid Nanomaterial-enhanced (ECL) Detection of Multiple Proteins. Anal. Chem. 2018, 90, 7569–7577. [Google Scholar] [CrossRef]
- Corstjens, P.L.M.; Chen, Z.; Zuiderwijk, M.; Bau, H.H.; Abrams, W.R.; Malamud, D.; Niedbala, R.S.; Tanke, H.J. Rapid assay format for multiplex detection of humoral immune responses to infectious disease pathogens (HIV, HCV, and TB). Ann. N. Y. Acad. Sci. 2007, 1098, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Engin, E.D. The use of multiplexing technology in the immunodiagnosis of infectious agents. J. Immunoass. Immunochem. 2019, 40, 109–122. [Google Scholar] [CrossRef]
- Hao, L.; Zhao, R.T.; Welch, N.L.; Tan, E.K.W.; Zhong, Q.; Harzallah, N.S.; Ngambenjawong, C.; Ko, H.; Fleming, H.E.; Sabeti, P.C.; et al. CRISPR-Cas-amplified urinary biomarkers for multiplexed and portable cancer diagnostics. Nat. Nanotechnol. 2023, 18, 798–807. [Google Scholar] [CrossRef]
- Welch, N.L.; Zhu, M.; Hua, C.; Weller, J.; Mirhashemi, M.E.; Nguyen, T.G.; Mantena, S.; Bauer, M.R.; Shaw, B.M.; Ackerman, C.M.; et al. Multiplexed CRISPR-Based Microfluidic Platform for Clinical Testing of Respiratory Viruses and Identification of SARS-CoV-2 Variants. Nat. Med. 2022, 28, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Huang, Z.; Li, Z.; Hu, J.; Liu, R.; Lv, Y. CRISPR-Associated “Genetic Scissors” for Multiplexing Analysis. Trends Anal. Chem. 2024, 170, 117431. [Google Scholar] [CrossRef]
- Zhao, L.; Qiu, M.; Li, X.; Yang, J.; Li, J. CRISPR-Cas13a system: A novel tool for molecular diagnostics. Front. Microbiol. 2022, 13, 1060947. [Google Scholar] [CrossRef]
- Swarbrick, S.; Wragg, N.; Ghosh, S.; Stolzing, A. Systematic review of MIRNA as biomarkers in Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 6156–6167. [Google Scholar] [CrossRef]
- Cogswell, J.P.; Ward, J.; Taylor, I.A.; Waters, M.; Shi, Y.; Cannon, B.; Kelnar, K.; Kemppainen, J.; Brown, D.; Chen, C.; et al. Identification of miRNA Changes in Alzheimer’s Disease Brain and CSF Yields Putative Biomarkers and Insights into Disease Pathways. J. Alzheimer’s Dis. 2008, 14, 27–41. [Google Scholar] [CrossRef]
- Müller, M.; Kuiperij, H.B.; Claassen, J.A.; Küsters, B.; Verbeek, M.M. MicroRNAs in Alzheimer’s disease: Differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol. Aging 2014, 35, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Sharafeldin, M.; Chen, T.; Ozkaya, G.U.; Choudhary, D.; Molinolo, A.A.; Gutkind, J.S.; Rusling, J.F. Detecting Cancer Metastasis and Accompanying Protein Biomarkers at Single Cell Levels Using a 3D-Printed Microfluidic Immunoarray. Biosens. Bioelectron. 2021, 171, 112681. [Google Scholar] [CrossRef]
- Sharafeldin, M.; Kadimisetty, K.; Bhalerao, K.R.; Bist, I.; Jones, A.; Chen, T.; Lee, N.H.; Rusling, J.F. Accessible Telemedicine Diagnostics with ELISA in a 3D Printed Pipette Tip. Anal. Chem. 2019, 91, 7394–7402. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of Protein Using Bicinchoninic Acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Shah, V.P.; Midha, K.K.; Findlay, J.W.; Hill, H.M.; Hulse, J.D.; McGilveray, I.J.; McKay, G.; Miller, K.J.; Yacobi, A. Bioanalytical Method Validation—A Revisit with a Decade of Progress. Pharm. Res. 2000, 17, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Hao, Y.-N.; Wang, X.-P.; Lu, W.-H.; Xie, L.-Y.; Niu, D. Bone marrow mesenchymal stem cell-derived exosomal miR-30e-5p ameliorates high-glucose induced renal proximal tubular cell pyroptosis by inhibiting ELAVL1. Ren. Fail. 2023, 45, 2177082. [Google Scholar] [CrossRef]
- Tu, Y.; Hu, Y. MiRNA-34c-5p protects against cerebral ischemia/reperfusion injury: Involvement of anti-apoptotic and anti-inflammatory activities. Metabol. Brain Dis. 2021, 36, 1341–1351. [Google Scholar] [CrossRef]
- Li, W.; Yang, S.; Chen, G.; He, S. MiR-200c-3p regulates pyroptosis by targeting SLC30A7 in diabetic retinopathy. Hum. Exp. Toxicol. 2022, 41, 096032712210995. [Google Scholar] [CrossRef]
- Zahra, A.; Shahid, A.; Shamim, A.; Khan, S.H.; Arshad, M.I. The SHERLOCK Platform: An Insight into Advances in Viral Disease Diagnosis. Mol. Biotechnol. 2022, 65, 699–714. [Google Scholar] [CrossRef]
- Yin, L.; Man, S.; Ye, S.; Liu, G.; Ma, L. CRISPR-Cas based virus detection: Recent advances and perspectives. Biosens. Bioelectron. 2021, 193, 113541. [Google Scholar] [CrossRef]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Hiniduma, K.; De Silva, P.I.T.; Canete, R.; Gunathillaka, H.; Clement, O.; Vora, P.; Shawky, S.M.; Rouge, J.L.; Mosa, I.; Breno, D.; et al. ECL-CRISPR Array for Multiplexed Detection of miRNAs. ACS Sens. 2025; submitted. [Google Scholar]


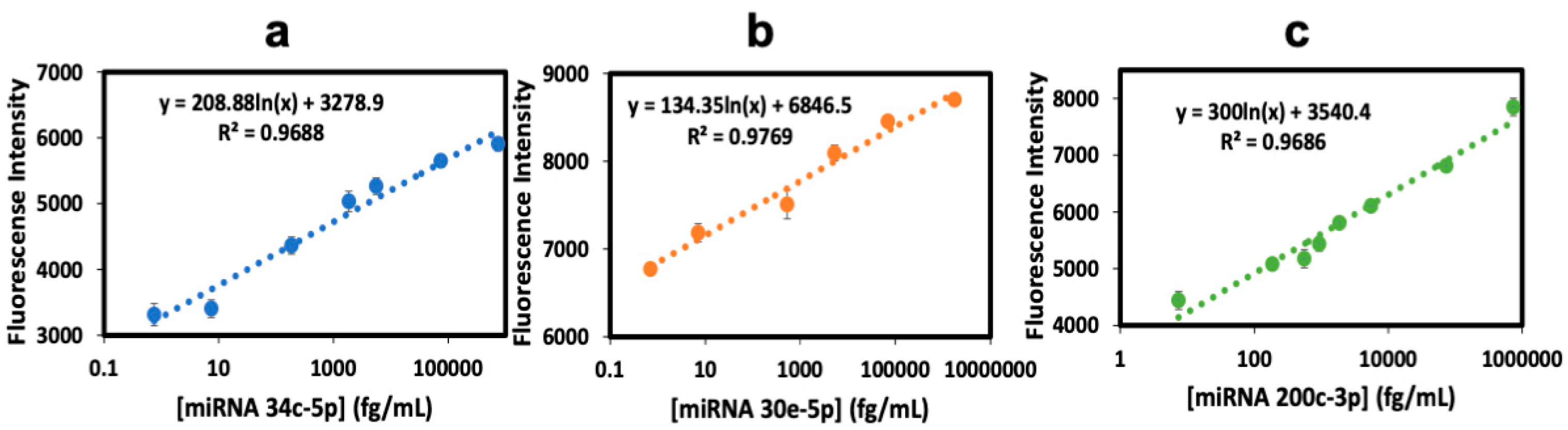
| Spiked Conc, (fg/mL) | miRNA 30e-5p | miRNA 200c-3p | miRNA 34c-5p | |||
|---|---|---|---|---|---|---|
| Found (fg/mL) | Recovery % | Found (fg/mL) | Recovery % | Found (fg/mL) | Recovery % | |
| 70 | 83 | 118 ± 12 | 60 | 86 ± 8 | 79 | 112 ± 11 |
| 850 | 808 | 95 ± 9 | 902 | 106 ± 10 | 703 | 83 ± 7 |
| 3500 | 4224 | 120 ± 14 | 4100 | 117 ± 13 | 3380 | 96 ± 7 |
| 35,000 | 40,254 | 115 ± 11 | 40,790 | 116 ± 12 | 39,900 | 114 ± 7 |
| miRNA | % Cross-Reactivity with CRISPR RNA | ||
|---|---|---|---|
| 30e-5p | 34c-5p | 200c-3p | |
| 30e-5p | 3.3 | 3.3 | |
| 34c-5p | 4.1 | 4.7 | |
| 200c-3p | 2.5 | 3.6 | |
| Patient | 34c-5p (ng/mL) | 30e-5p (ng/mL) | 200c-3p (ng/mL) | ALZ Diagnosis |
|---|---|---|---|---|
| 1 | 0.000028 | 0.000023 | 0.000030 | Negative |
| 2 | 0.000041 | 0.000033 | 0.000044 | Negative |
| 3 | 0.655 | 0.0001834 | 0.965 | Positive |
| 4 | 2.16 | 3.11 | 1.02 | Positive |
| 5 | 2.74 | 6.52 | 1.21 | Positive |
| 6 | 3.74 | 21.2 | 1.69 | Positive |
| 7 | 9.12 | 86.4 | 3.11 | Positive |
| 8 | 24.2 | 349 | 3.01 | Positive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Silva, P.I.T.; Hiniduma, K.; Canete, R.; Bhalerao, K.S.; Shawky, S.M.; Gunathilaka, H.; Rouge, J.L.; Mosa, I.M.; Steffens, D.C.; Manning, K.; et al. Multiplexed CRISPR Assay for Amplification-Free Detection of miRNAs. Biosensors 2025, 15, 346. https://doi.org/10.3390/bios15060346
De Silva PIT, Hiniduma K, Canete R, Bhalerao KS, Shawky SM, Gunathilaka H, Rouge JL, Mosa IM, Steffens DC, Manning K, et al. Multiplexed CRISPR Assay for Amplification-Free Detection of miRNAs. Biosensors. 2025; 15(6):346. https://doi.org/10.3390/bios15060346
Chicago/Turabian StyleDe Silva, P. I. Thilini, Keshani Hiniduma, Rachelle Canete, Ketki S. Bhalerao, Sherif M. Shawky, Hansana Gunathilaka, Jessica L. Rouge, Islam M. Mosa, David C. Steffens, Kevin Manning, and et al. 2025. "Multiplexed CRISPR Assay for Amplification-Free Detection of miRNAs" Biosensors 15, no. 6: 346. https://doi.org/10.3390/bios15060346
APA StyleDe Silva, P. I. T., Hiniduma, K., Canete, R., Bhalerao, K. S., Shawky, S. M., Gunathilaka, H., Rouge, J. L., Mosa, I. M., Steffens, D. C., Manning, K., Diniz, B. S., & Rusling, J. F. (2025). Multiplexed CRISPR Assay for Amplification-Free Detection of miRNAs. Biosensors, 15(6), 346. https://doi.org/10.3390/bios15060346









