Protein, Nucleic Acid, and Nanomaterial Engineering for Biosensors and Monitoring
Abstract
1. Introduction
2. Protein Engineering for Biosensors
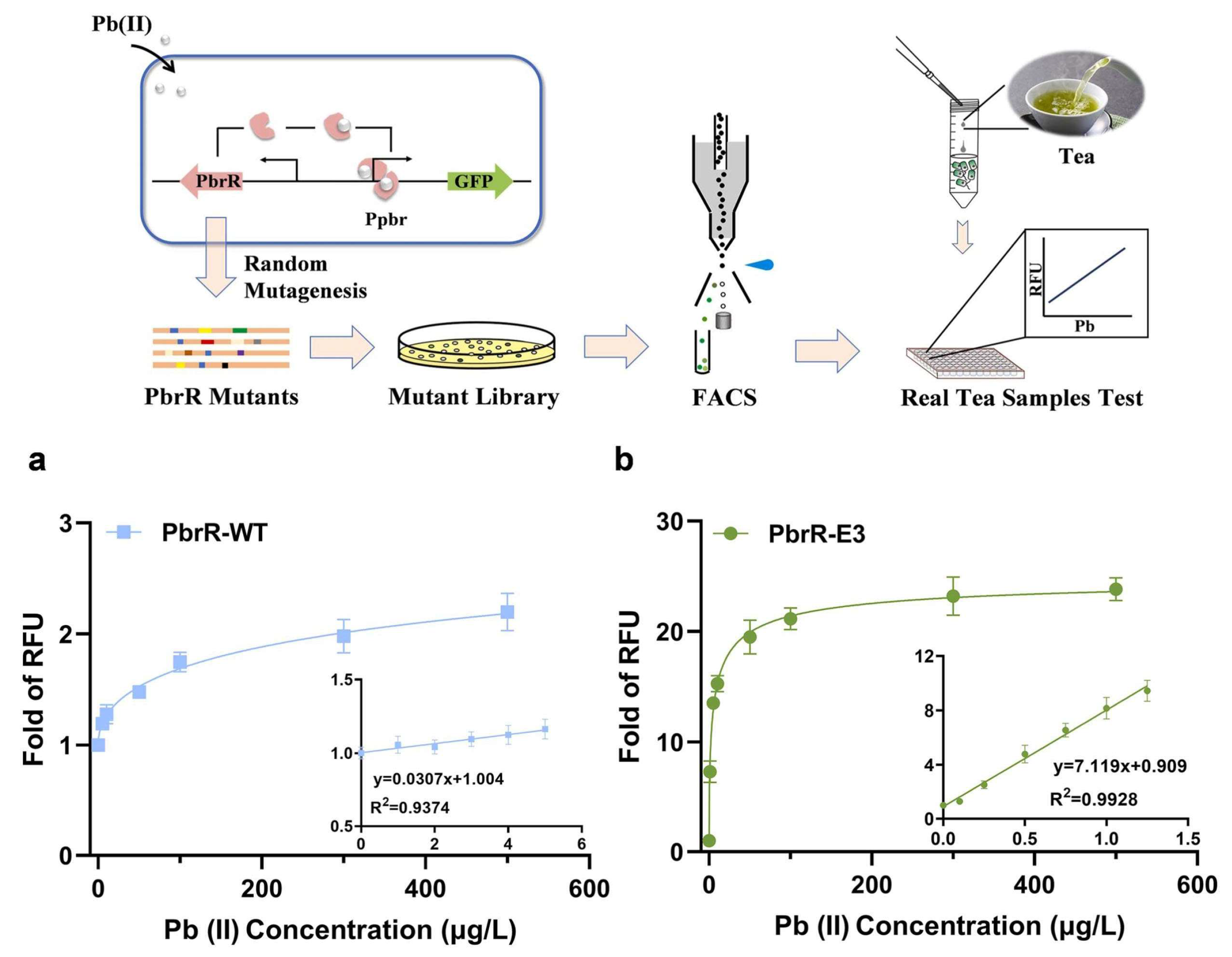
2.1. Directed Evolution of Proteins for Biosensors

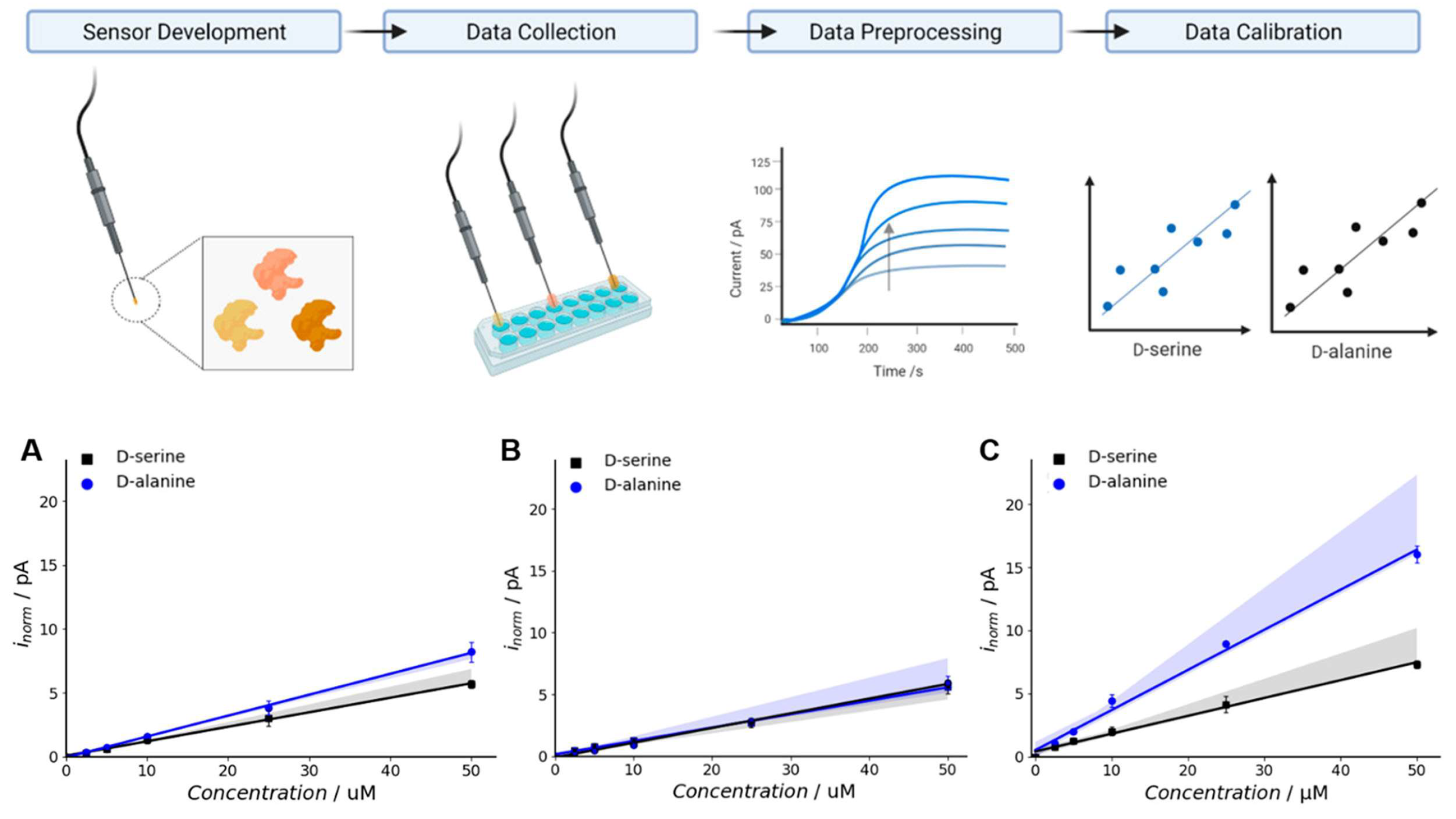
2.2. Semi-Rational Design of Proteins for Biosensors
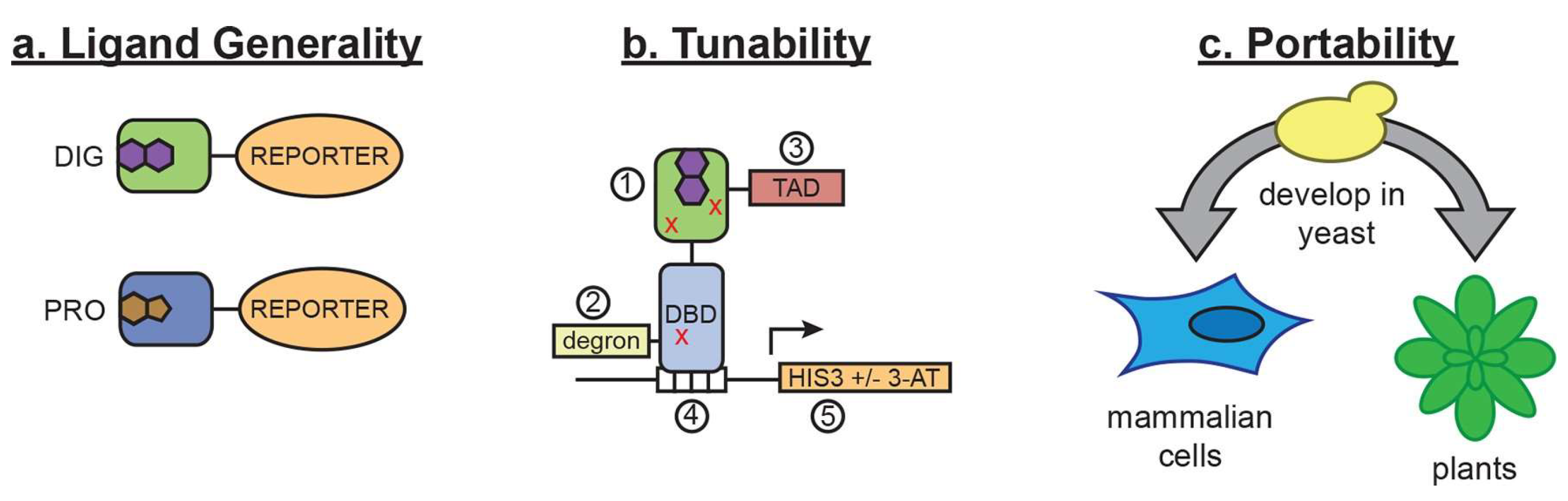
2.3. Rational Design of Proteins for Biosensors
3. Nucleic Acid Engineering for Biosensors
| Biosensor | Detection | LOD | Linear Range | Sample | References |
| Protein engineering for biosensors | |||||
| TP2-Rho P53 peptide biosensor | R248Q mutant of protein p53 | Nanomolar concentrations of p53 mutant in lung cancer cell extract | NR | Lung cancer cells | [1] |
| Pb whole-cell biosensor | Detection of Pb | 0.045 µg/L | NR | Food products, water samples | [2] |
| Bacterial Cd biosensor (CadR) | Detection of Cd | 0.45 µg/L | NR | Soil and water | [7] |
| Bacterial biosensor for detecting lead toxicity | Detection of Pb | 0.012 µM Pb | 0.012–3.125 µM Pb | Artificially polluted water | [8] |
| Bacterial biosensor for C1 molecules | Detection of formate, formaldehyde, and methanol | NR | 1.0–250 mM formate 1.0–50 µM formaldehyde 5–400 mM methanol | Cell culture | [9] |
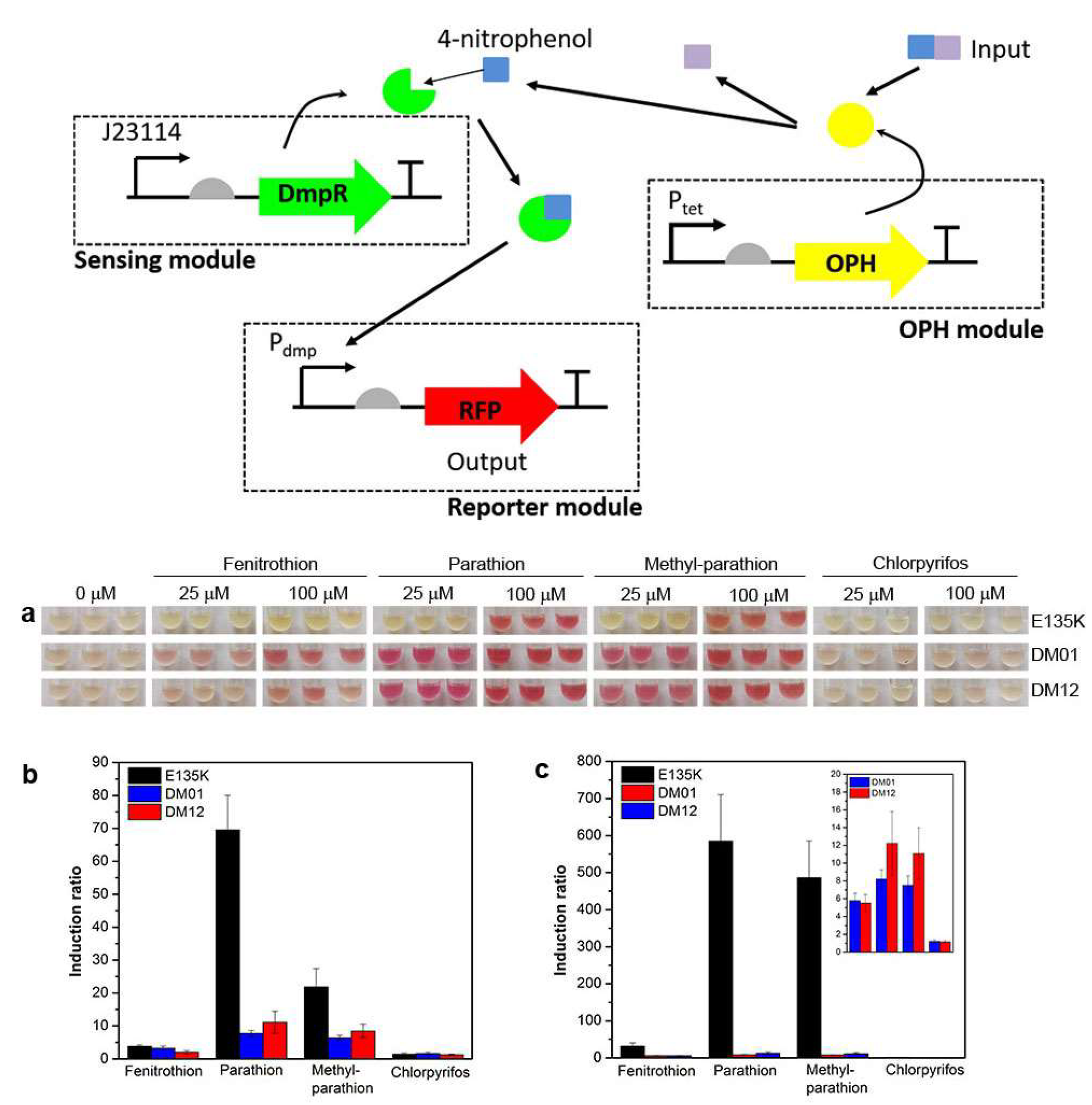
| Bacterial biosensor | Detection of organophosphorus compounds | 10 µM parathion | NR | Artificial samples | [25] |
| Nucleic acid engineering for biosensors | |||||
| Riboswitch biosensor | Detection of sesquiterpene | NR | 10–100 mg/mL amorpha-4,11—diene | Samples for high throughput screening and for metabolic engineering | [29] |
| Nucleotide aptamer | Detection of levofloxacin | 100 µM | NR | Artificial samples | [33] |
| Nucleotide aptamer | Detection of ciprofloxacin | The maximum residue limit (MRL) | NR | Food samples | [34] |
4. Nanomaterials Engineering for Biosensors
| Electrode Nanomaterial | Analyte | Linear Range | LOD | Real Matrices | Reference |
|---|---|---|---|---|---|
| Graphitic carbon nitride/zirconium dioxide/multiwalled carbon nanotubes | Metalloproteinase-9 | 50 to 1250 pg/mL | 10.51 pg/mL | Human serum and saliva | [40] |
| Gold/manganese (IV) oxide/multiwalled carbon nanotubes | Homocysteine | 5–125 μM/L | 0.6173 μM/L | Serum homocysteine | [41] |
| Graphene-oxide-doped gold nanoparticles conjugated with polythiophene | H. pylori | 10–900 nM/L | 0.0080 μM/L with impedance; 0.0067 μM/L for SWV method | / | [46] |
| Polypyrrole nanotubes and carbon nanotubes nanocomposite | H. pylori outer membrane protein (HopQ) | 5 pg/mL 1.063 ng/mL | 2.06 pg/mL | Spiked drinking water | [47] |
| Self-supported PtPdMnCoFe | Neuron-specific enolase (NSE) | 0.1 pg/mL–200 ng/mL | 0.0036 pg/mL | Human serum samples | [49] |
| Dendritic quinary PtRhMoCoFe | cTnI | 0.0001–200 ng/mL | 0.0095 pg/mL | Serum samples | [50] |
| MOF@Pt@MOF nanozyme | Exosomal miRNA | 1 fM/L–1 nM/L | 0.29 fM/L | MCF-7 cells, MCF-10 A cells, and serums of a breast cancer patient and a healthy person were tested | [53] |
| 2D conductive MOF nanosheets/gold nanoparticles | H2O2 | 50 nM/L–16.4 mM/L | 5.6 nM/L | Human colon cells | [54] |
| Fe(II)-MOF nanozyme | Glucose | 100–600 μmol/L | 17.6 μmol/L | Artificial sweat samples | [55] |
5. Biosensors for Rare Diseases Monitoring
5.1. Biosensors for the Detection of Aβ42, a Biomarker for Alzheimer’s Disease Diagnosis
5.2. Biosensors for the Monitoring of Hepatitis B
5.3. Biosensors for the Monitoring of Human Papillomavirus
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pellerano, M.; Naud-Martin, D.; Mahuteau-Betzer, F.; Morille, M.; Morris, M.C. Fluorescent Biosensor for Detection of the R248Q Aggregation-Prone Mutant of p53. ChemBioChem 2019, 20, 605–613. [Google Scholar] [CrossRef]
- Shen, L.; Chen, Y.W.; Pan, J.J.; Yu, X.; Zhang, Y.B.; Guo, B.X.; Wang, J.Q.; Liu, Y.; Xiao, X.; Chen, S.P.; et al. Development of a highly sensitive PbrR-based biosensor via directed evolution and its application for lead detection. J. Hazard. Mater. 2025, 488, 137489. [Google Scholar] [CrossRef]
- Feng, J.; Jester, B.W.; Tinberg, C.E.; Mandell, D.J.; Antunes, M.S.; Chari, R.; Morey, K.J.; Rios, X.; Medford, J.I.; Church, G.M.; et al. A general strategy to construct small molecule biosensors in eukaryotes. eLife 2015, 4, e10606. [Google Scholar] [CrossRef]
- Paulmurugan, R.; Afjei, R.; Sekar, T.V.; Babikir, H.A.; Massoud, T.F. A protein folding molecular imaging biosensor monitors the effects of drugs that restore mutant p53 structure and its downstream function in glioblastoma cells. Oncotarget 2018, 9, 21495–21511. [Google Scholar] [CrossRef][Green Version]
- Taylor, N.D.; Garruss, A.S.; Moretti, R.; Chan, S.; Arbing, M.A.; Cascio, D.; Rogers, J.K.; Isaacs, F.J.; Kosuri, S.; Baker, D.; et al. Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods 2016, 13, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Gohil, K.; Wu, S.Y.; Takahashi-Yamashiro, K.; Shen, Y.; Campbell, R.E. Biosensor Optimization Using a Forster Resonance Energy Transfer Pair Based on mScarlet Red Fluorescent Protein and an mScarlet-Derived Green Fluorescent Protein. ACS Sens. 2023, 8, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhu, K.; Shen, L.; Ma, J.; Bao, L.; Chen, D.; Wei, L.; Wei, N.; Liu, B.; Wu, Y.; et al. Evolved Biosensor with High Sensitivity and Specificity for Measuring Cadmium in Actual Environmental Samples. Environ. Sci. Technol. 2022, 56, 10062–10071. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, Z.L.; Zhu, D.L.; Hu, S.Y.; Li, H.; Hui, C.Y. Anthocyanin biosynthetic pathway switched by metalloregulator PbrR to enable a biosensor for the detection of lead toxicity. Front. Microbiol. 2022, 13, 975421. [Google Scholar] [CrossRef]
- Lee, J.Y.; Sung, B.H.; Oh, S.H.; Kwon, K.K.; Lee, H.; Kim, H.; Lee, D.H.; Yeom, S.J.; Lee, S.G. C1 Compound Biosensors: Design, Functional Study, and Applications. Int. J. Mol. Sci. 2019, 20, 2253. [Google Scholar] [CrossRef]
- Ebrahimi Fana, S.; Fazaeli, A.; Aminian, M. Directed evolution of cholesterol oxidase with improved thermostability using error-prone PCR. Biotechnol. Lett. 2023, 45, 1159–1167. [Google Scholar] [CrossRef]
- Ding, N.; Zhou, S.; Yuan, Z.; Zhang, X.; Chen, J.; Deng, Y. Fine-tuning biosensor dynamic range based on rational design of cross-ribosome-binding sites in bacteria. bioRxiv 2020, 2020.01.27.922302. [Google Scholar]
- Moussa, S.; Murtas, G.; Pollegioni, L.; Mauzeroll, J. Enhancing Electrochemical Biosensor Selectivity with Engineered D-Amino Acid Oxidase Enzymes for D-Serine and D-Alanine Quantification. ACS Appl. Bio Mater. 2021, 4, 5598–5604. [Google Scholar] [CrossRef]
- Nadler, D.C.; Morgan, S.A.; Flamholz, A.; Kortright, K.E.; Savage, D.F. Rapid construction of metabolite biosensors using domain-insertion profiling. Nat. Commun. 2016, 7, 12266. [Google Scholar] [CrossRef]
- Zhou, Y.K.; Yuan, Y.M.; Wu, Y.A.; Li, L.; Jameel, A.; Xing, X.H.; Zhang, C. Encoding Genetic Circuits with DNA Barcodes Paves the Way for Machine Learning-Assisted Metabolite Biosensor Response Curve Profiling in Yeast. ACS Synth. Biol. 2022, 11, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.Q.; Zhang, Z.W.; Gu, Q.Y.; Yu, X.B. Semi-rational design for enhancing thermostability of acetylcholinesterase and sensitivity analysis of acephate. Sci. Total Environ. 2024, 934, 173282. [Google Scholar] [CrossRef]
- Balaz, A.M.; Stevanovic, J.; Ostafe, R.; Blazic, M.; Durdic, K.I.; Fischer, R.; Prodanovic, R. Semi-rational design of cellobiose dehydrogenase for increased stability in the presence of peroxide. Mol. Divers. 2020, 24, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.X.; Zhai, H.T.; Hou, J. Biosensors design in yeast and applications in metabolic engineering. Fems Yeast Res. 2019, 19, foz082. [Google Scholar] [CrossRef]
- Pedotti, M.; Ferrero, V.E.V.; Lettieri, T.; Colpo, P.; Follonier, S.; Calzolai, L.; Varani, L. Rationally Modified Estrogen Receptor Protein as a Bio-Recognition Element for the Detection of EDC Pollutants: Strategies and Opportunities. Int. J. Environ. Res. Public Health 2015, 12, 2612–2621. [Google Scholar] [CrossRef]
- Rogers, J.K.; Guzman, C.D.; Taylor, N.D.; Raman, S.; Anderson, K.; Church, G.M. Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res. 2015, 43, 7648–7660. [Google Scholar] [CrossRef]
- Hiraka, K.; Kojima, K.; Tsugawa, W.; Asano, R.; Ikebukuro, K.; Sode, K. Rational engineering of L-lactate oxidase for the mediator modification to achieve quasi-direct electron transfer type lactate sensor. Biosens. Bioelectron. 2020, 151, 111974. [Google Scholar] [CrossRef]
- Golynskiy, M.V.; Koay, M.S.; Vinkenborg, J.L.; Merkx, M. Engineering Protein Switches: Sensors, Regulators, and Spare Parts for Biology and Biotechnology. ChemBiochem 2011, 12, 353–361. [Google Scholar] [CrossRef]
- Hellweg, L.; Edenhofer, A.; Barck, L.; Huppertz, M.C.; Frei, M.S.; Tarnawski, M.; Bergner, A.; Koch, B.; Johnsson, K.; Hiblot, J. A general method for the development of multicolor biosensors with large dynamic ranges. Nat. Chem. Biol. 2023, 19, 1147–1157. [Google Scholar] [CrossRef]
- Kimura, Y.; Kawai-Noma, S.; Saito, K.; Umeno, D. Directed Evolution of the Stringency of the LuxR Quorum Sensor without OFF-State Selection. Acs Synth. Biol. 2020, 9, 567–575. [Google Scholar] [CrossRef]
- Du, H.X.; Liang, Y.Y.; Li, J.N.; Yuan, X.Y.; Tao, F.L.; Dong, C.J.; Shen, Z.K.; Sui, G.C.; Wang, P.C. Directed Evolution of 4-Hydroxyphenylpyruvate Biosensors Based on a Dual Selection System. Int. J. Mol. Sci. 2024, 25, 1533. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.Q.; Ching, C.B. Development of Colorimetric-Based Whole-Cell Biosensor for Organophosphorus Compounds by Engineering Transcription Regulator DmpR. Acs Synth. Biol. 2016, 5, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Z.; Su, R.T.; Fu, Y.; Wang, Y.P.; Chang, L.Q. Recent Progress in DNA Biosensors for Detecting Biomarkers in Living Cells. ACS Biomater. Sci. Eng. 2024, 10, 5595–5608. [Google Scholar] [CrossRef] [PubMed]
- Stephen, B.J.; Suchanti, S.; Jain, D.; Dhaliwal, H.; Sharma, V.; Kaur, R.; Mishra, R.; Singh, A. DNA biosensor based detection for neglected tropical disease: Moving towards smart diagnosis. Sens. Rev. 2022, 42, 517–525. [Google Scholar] [CrossRef]
- Machado, L.F.M.; Currin, A.; Dixon, N. Directed evolution of the PcaV allosteric transcription factor to generate a biosensor for aromatic aldehydes. J. Biol. Eng. 2019, 13, 91. [Google Scholar] [CrossRef]
- Huo, Y.Y.; Zhang, S.D.; Bi, H.R.; Wang, K.; Fang, Y.M.; Wang, M.; Tan, T.W. Development of a specific biosensor for sesquiterpene based on SELEX and directed evolution platforms. Talanta 2025, 283, 127186. [Google Scholar] [CrossRef]
- Townshend, B.; Kaplan, M.; Smolke, C.D. Highly multiplexed selection of RNA aptamers against a small molecule library. PLoS ONE 2022, 17, e0273381. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Nikolaus, N.; Strehlitz, B. Capture-SELEX: Selection of DNA Aptamers for Aminoglycoside Antibiotics. J. Anal. Methods Chem. 2012, 2012, 415697. [Google Scholar] [CrossRef]
- Ao, Y.Q.; Duan, A.Q.; Chen, B.F.; Yu, X.M.; Wu, Y.Y.; Zhang, X.J.; Li, S.S. Integration of an Expression Platform in the SELEX Cycle to Select DNA Aptamer Binding to a Disease Biomarker. ACS Omega 2022, 7, 10804–10811. [Google Scholar] [CrossRef] [PubMed]
- Kramat, J.; Kraus, L.; Gunawan, V.J.; Smyej, E.; Froehlich, P.; Weber, T.E.; Spiehl, D.; Koeppl, H.; Blaeser, A.; Suess, B. Sensing Levofloxacin with an RNA Aptamer as a Bioreceptor. Biosensors 2024, 14, 56. [Google Scholar] [CrossRef]
- Jaeger, J.; Groher, F.; Stamm, J.; Spiehl, D.; Braun, J.; Dörsam, E.; Suess, B. Characterization and Inkjet Printing of an RNA Aptamer for Paper-Based Biosensing of Ciprofloxacin. Biosensors 2019, 9, 7. [Google Scholar] [CrossRef]
- Holzinger, M.; Le Goff, A.; Cosnier, S. Synergetic Effects of Combined Nanomaterials for Biosensing Applications. Sensors 2017, 17, 1010. [Google Scholar] [CrossRef] [PubMed]
- El-Kady, M.M.; Ansari, I.; Arora, C.; Rai, N.; Soni, S.; Verma, D.K.; Singh, P.; Mahmoud, A.D. Nanomaterials: A comprehensive review of applications, toxicity, impact, and fate to environment. J. Mol. Liq. 2023, 370, 121046. [Google Scholar] [CrossRef]
- Mekuye, B.; Abera, B. Nanomaterials: An overview of synthesis, classification, characterization, and applications. Nano Sel. 2023, 4, 486–501. [Google Scholar] [CrossRef]
- Hwang, H.S.; Jeong, J.W.; Kim, Y.A.; Chang, M. Carbon Nanomaterials as Versatile Platforms for Biosensing Applications. Micromachines 2020, 11, 814. [Google Scholar] [CrossRef]
- Eivazzadeh-Keihan, R.; Noruzi, E.B.; Chidar, E.; Jafari, M.; Davoodi, F.; Kashtiaray, A.; Gorab, M.G.; Hashemi, S.M.; Javanshir, S.; Cohan, R.A.; et al. Applications of carbon-based conductive nanomaterials in biosensors. Chem. Eng. J. 2022, 442, 136183. [Google Scholar] [CrossRef]
- Ansari, M.A.; Mohd-Naim, N.F.; Ahmed, M.U. Electrochemical Nanoaptasensor Based on Graphitic Carbon Nitride/Zirconium Dioxide/Multiwalled Carbon Nanotubes for Matrix Metalloproteinase-9 in Human Serum and Saliva. ACS Appl. Bio Mater. 2024, 7, 1579–1587. [Google Scholar] [CrossRef]
- Shi, Z.H.; Zhao, H.L.; Zhou, F.F.; Mao, Y.; Gong, Z.; Lan, M.B. Sensitive Electrochemical Biosensor Using Gold and Manganese (IV) Oxide Nanomaterials on Multiwalled Carbon Nanotubes for the Determination of Homocysteine With a Portable Potentiostat. Anal. Lett. 2025, 58, 492–505. [Google Scholar] [CrossRef]
- du Plooy, J.; Kock, B.; Jahed, N.; Iwuoha, E.; Pokpas, K.; Hou, P.X.; He, M.S. Carbon Nanostructured Immunosensing of Anti-SARS-CoV-2 S-Protein Antibodies. Molecules 2023, 28, 8022. [Google Scholar] [CrossRef]
- Younis, M.R.; He, G.; Lin, J.; Huang, P. Recent Advances on Graphene Quantum Dots for Bioimaging Applications. Front. Chem. 2020, 8, 424. [Google Scholar] [CrossRef] [PubMed]
- Kansara, V.; Shukla, R.; Bahadur, P.; Tiwari, S.; Flora, S.J.S. Graphene quantum dots: Synthesis, optical properties and navigational applications against cancer. Mater. Today Commun. 2022, 31, 103359. [Google Scholar] [CrossRef]
- El-Said, W.A.; Abdelshakour, M.; Choi, J.H.; Choi, J.W. Application of Conducting Polymer Nanostructures to Electrochemical Biosensors. Molecules 2020, 25, 307. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.S.; Mehrdadi, N.; Bidhendi, G.N.; Pourmadadi, M.; Ahmadi, M.; Meknatkhah, S. Novel detection of H. pylori using ultrasensitive electrochemical aptasensor based on surface modified graphene oxide doped gold nanoparticles conjugated polythiophene(vol 200, 110279, 2024). Microchem. J. 2024, 204, 110572. [Google Scholar] [CrossRef]
- Jaradat, H.; Hryniewicz, B.M.; Pasti, I.A.; Valerlo, T.L.; Al -Hamry, A.; Marchesi, L.F.; Vidotti, M.; Kanoun, O. Detection of H. pylori outer membrane protein (HopQ) biomarker using electrochemical impedimetric immunosensor with polypyrrole nanotubes and carbon nanotubes nanocomposite on screen-printed carbon electrode. Biosens. Bioelectron. 2024, 249, 115937. [Google Scholar] [CrossRef]
- Yu, T.T.; Xu, H.T.; Jin, Z.Y.; Zhang, Y.Y.; Qiu, H.J. Noble metal-free high-entropy oxide/Co-N-C bifunctional electrocatalyst enables highly reversible and durable Zn-air batteries. Appl. Surf. Sci. 2023, 610, 155624. [Google Scholar] [CrossRef]
- Lv, C.L.; Tang, C.; Zhou, H.Y.; Wang, A.J.; Feng, J.J.; Cheang, T.Y. Self-supported PtPdMnCoFe high-entropy alloy with nanochain-like internetworks for ultrasensitive electrochemical immunoassay of biomarker. Sens. Actuators B-Chem. 2024, 401, 135041. [Google Scholar] [CrossRef]
- Tang, C.; Lv, C.L.; Chen, P.F.; Wang, A.J.; Feng, J.J.; Cheang, T.Y.; Xia, H.M. Dendritic quinary PtRhMoCoFe high-entropy alloy as a robust immunosensing nanoplatform for ultrasensitive detection of biomarker. Bioelectrochemistry 2024, 157, 108639. [Google Scholar] [CrossRef]
- Kurup, C.P.; Ahmed, M.U. Nanozymes towards Personalized Diagnostics: A Recent Progress in Biosensing. Biosensors 2023, 13, 461. [Google Scholar] [CrossRef] [PubMed]
- Keum, C.; Park, S.; Kim, H.; Kim, H.; Lee, K.H.; Jeong, Y. Modular conductive MOF-gated field-effect biosensor for sensitive discrimination on the small molecular scale. Chem. Eng. J. 2023, 456, 141079. [Google Scholar] [CrossRef]
- Li, X.Y.; Li, X.M.; Li, D.D.; Zhao, M.; Wu, H.P.; Shen, B.; Liu, P.; Ding, S.J. Electrochemical biosensor for ultrasensitive exosomal miRNA analysis by cascade primer exchange reaction and MOF@Pt@MOF nanozyme. Biosens. Bioelectron. 2020, 168, 112554. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Xu, Y.; Wang, Z.P.; Liao, K.; Zhang, Y.; Sun, Y.M. Dual nanozyme based on ultrathin 2D conductive MOF nanosheets intergraded with gold nanoparticles for electrochemical biosensing of HO in cancer cells. Talanta 2022, 249, 123612. [Google Scholar] [CrossRef]
- Koukouviti, E.; Plessas, A.K.; Pagkali, V.; Economou, A.; Papaefstathiou, G.S.; Kokkinos, C. 3D-printed electrochemical glucose device with integrated Fe(II)-MOF nanozyme. Microchim. Acta 2023, 190, 274. [Google Scholar] [CrossRef]
- Yu, Y.Y.; Sun, X.Y.; Tang, D.Q.; Li, C.L.; Zhang, L.; Nie, D.X.; Yin, X.X.; Shi, G.Y. Gelsolin bound β-amyloid peptides: Electrochemical evaluation of levels of soluble peptide associated with Alzheimer’s disease. Biosens. Bioelectron. 2015, 68, 115–121. [Google Scholar] [CrossRef]
- Diba, F.S.; Kim, S.; Lee, H.J. Electrochemical immunoassay for amyloid-beta 1-42 peptide in biological fluids interfacing with a gold nanoparticle modified carbon surface. Catal. Today 2017, 295, 41–47. [Google Scholar] [CrossRef]
- Ding, M.L.; Shu, Q.; Zhang, N.; Yan, C.R.; Niu, H.Z.; Li, X.Q.; Guan, P.; Hu, X.L. Electrochemical Immunosensor for the Sensitive Detection of Alzheimer’s Biomarker Amyloid-β (1-42) Using the Heme-amyloid-β (1-42) Complex as the Signal Source. Electroanalysis 2022, 34, 263–274. [Google Scholar] [CrossRef]
- Hsu, C.H.; Gupta, A.K.; Purwidyantri, A.; Prabowo, B.A.; Chen, C.H.; Chuang, C.C.; Tian, Y.C.; Lu, Y.J.; Lai, C.S. Sensing Alzheimer’s Disease Utilizing Au Electrode by Controlling Nanorestructuring. Chemosensors 2022, 10, 94. [Google Scholar] [CrossRef]
- Le, H.T.N.; Park, J.; Cho, S. A Probeless Capacitive Biosensor for Direct Detection of Amyloid Beta 1-42 in Human Serum Based on an Interdigitated Chain-Shaped Electrode. Micromachines 2020, 11, 791. [Google Scholar] [CrossRef]
- Ribeiro, S.C.; Fernandes, R.; Moreira, F.T.C.; Sales, M.G.F. Potentiometric Biosensor Based on Artificial Antibodies for an Alzheimer Biomarker Detection. Appl. Sci. 2022, 12, 3625. [Google Scholar] [CrossRef]
- Chen, J.Y.; Weng, S.H.; Chen, Q.Q.; Liu, A.L.; Wang, F.Q.; Chen, J.; Yi, Q.; Liu, Q.C.; Lin, X.H. Development of an Electrochemical Sensing Technique for Rapid Genotyping of Hepatitis B Virus. Sensors 2014, 14, 5611–5621. [Google Scholar] [CrossRef]
- Teengam, P.; Tangkijvanich, P.; Chuaypen, N.; Chailapakul, O. An innovative wireless electrochemical card sensor for field-deployable diagnostics of Hepatitis B surface antigen. Sci. Rep. 2023, 13, 3523. [Google Scholar] [CrossRef]
- Fan, P.Y.; Ma, E.Y.; Liu, C.; Zhao, Y.; Wen, X.W.; Wang, L.; Li, L.; Qu, Q. A new electrochemical DNA biosensor based on the density control strategy of TiCNH MXene@Au nanocomposites for the detection of hepatitis B virus-DNA. Ionics 2024, 30, 541–552. [Google Scholar] [CrossRef]
- Li, X.; Scida, K.; Crooks, R.M. Detection of Hepatitis B Virus DNA with a Paper Electrochemical Sensor. Anal. Chem. 2015, 87, 9009–9015. [Google Scholar] [CrossRef] [PubMed]
- Pareek, S.; Jain, U.; Bharadwaj, M.; Saxena, K.; Roy, S.; Chauhan, N. An ultrasensitive electrochemical DNA biosensor for monitoring (HPV-16) using graphene oxide/Ag/Au nano-biohybrids. Anal. Biochem. 2023, 663, 115015. [Google Scholar] [CrossRef]
- Yang, Y.X.; Qing, Y.; Hao, X.D.; Fang, C.X.; Ouyang, P.; Li, H.Y.; Wang, Z.C.; Liao, Y.Z.; Fang, H.B.; Du, J. APTES-Modified Remote Self-Assembled DNA-Based Electrochemical Biosensor for Human Papillomavirus DNA Detection. Biosensors 2022, 12, 449. [Google Scholar] [CrossRef]
- Yang, Y.X.; Liao, Y.Z.; Qing, Y.; Li, H.Y.; Du, J. Electrochemical DNA Biosensors with Dual-Signal Amplification Strategy for Highly Sensitive HPV 16 Detection. Sensors 2023, 23, 7380. [Google Scholar] [CrossRef]
- Rasouli, E.; Basirun, W.J.; Johan, M.R.; Rezayi, M.; Mahmoudian, M.R.; Poenar, D.P. Electrochemical DNA-nano biosensor for the detection of cervical cancer-causing HPV-16 using ultrasmall Fe3O4-Au core-shell nanoparticles. Sens. Bio-Sens. Res. 2023, 40, 100562. [Google Scholar] [CrossRef]
- Chaibun, T.; Thanasapburachot, P.; Chatchawal, P.; Yin, L.S.; Jiaranuchart, S.; Jearanaikoon, P.; Promptmas, C.; Buajeeb, W.; Lertanantawong, B. A Multianalyte Electrochemical Genosensor for the Detection of High-Risk HPV Genotypes in Oral and Cervical Cancers. Biosensors 2022, 12, 290. [Google Scholar] [CrossRef]

| Field | Technique | Biosensor Type | Biomarkers | Advancements | Refs. |
|---|---|---|---|---|---|
| Protein Engineering | Directed evolution | Optical, Electrochemical | Mutant p53, Pb, Cd, C1 compounds, cholesterol, transcription factors | High-specificity biosensors via protein mutation and screening | [1,2,3,4,5,6,7,8,9,10] |
| Semi-rational design | Optical, Electrochemical | Glucarate, D-serine, D-alanine, metabolites, acephate | Enhanced enzyme selectivity/stability using structural data | [11,12,13,14,15,16,17] | |
| Rational design | Optical, Electrochemical | Estrogen, lactate, calcium, ATP, NAD+, HPP, pesticides | Targeted mutations for ligand binding, allosteric sensor design | [18,19,20,21,22,23,24,25] | |
| Nucleic Acid Engineering | SELEX and aptamer engineering | Fluorescent, Electrochemical | Aminoglycosides, levofloxacin | Improved aptamer affinity and specificity for antibiotics and proteins | [26,27,31,32,33] |
| Riboswitch development | Fluorescent | Amorpha-4,11-diene, aromatic aldehydes | Riboswitches for metabolite detection in synthetic biology | [28,29] | |
| RNA aptamer screening (DRIVER) | Fluorescent, Paper-based | Small molecules | High-throughput aptamer generation with multiplex selection | [30] | |
| Nanomaterials Engineering | Carbon nanomaterials (CNTs, graphene) | Electrochemical | MMP-9, H. pylori, SARS-CoV-2 IgG | Enhanced conductivity and immobilization for pathogen detection | [38,40,41,42,46,47] |
| MOF-based nanozymes | Electrochemical | Exosomal miRNA, H2O2, glucose | Ultrasensitive catalytic detection via MOF-Au composites | [52,53,54,55] | |
| High-entropy alloy nanomaterials | Electrochemical | NSE, cTnI | Multi-metallic platforms with ultralow LOD for disease markers | [48,49,50] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crnoglavac Popović, M.; Stanković, V.; Stanković, D.; Prodanović, R. Protein, Nucleic Acid, and Nanomaterial Engineering for Biosensors and Monitoring. Biosensors 2025, 15, 430. https://doi.org/10.3390/bios15070430
Crnoglavac Popović M, Stanković V, Stanković D, Prodanović R. Protein, Nucleic Acid, and Nanomaterial Engineering for Biosensors and Monitoring. Biosensors. 2025; 15(7):430. https://doi.org/10.3390/bios15070430
Chicago/Turabian StyleCrnoglavac Popović, Milica, Vesna Stanković, Dalibor Stanković, and Radivoje Prodanović. 2025. "Protein, Nucleic Acid, and Nanomaterial Engineering for Biosensors and Monitoring" Biosensors 15, no. 7: 430. https://doi.org/10.3390/bios15070430
APA StyleCrnoglavac Popović, M., Stanković, V., Stanković, D., & Prodanović, R. (2025). Protein, Nucleic Acid, and Nanomaterial Engineering for Biosensors and Monitoring. Biosensors, 15(7), 430. https://doi.org/10.3390/bios15070430





