Smart Nucleic Acid Hydrogel-Based Biosensors: From Molecular Recognition and Responsive Mechanisms to Applications
Abstract
1. Introduction
2. Molecular Recognition Elements in SNAHs
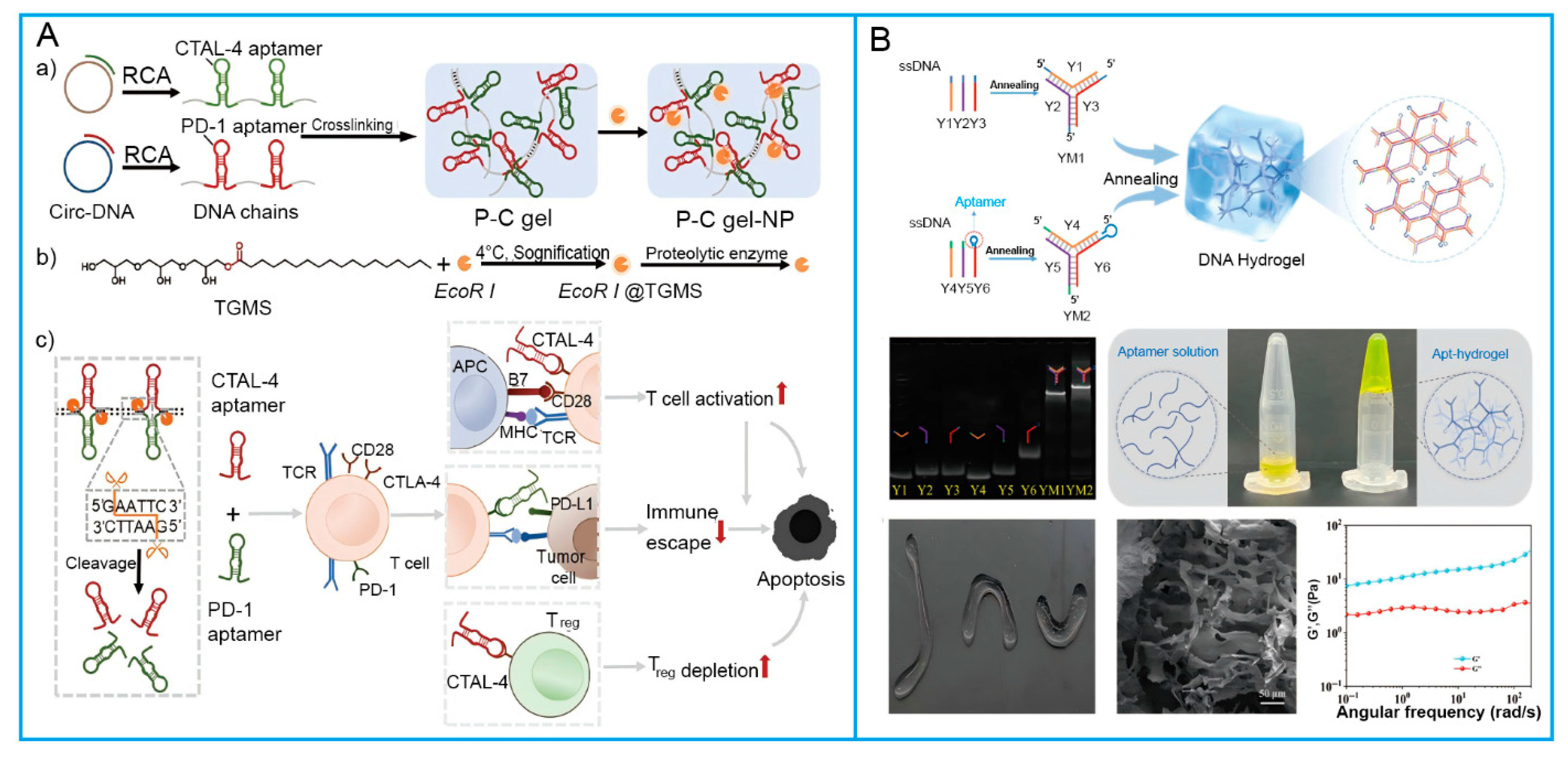
2.1. Aptamer
2.2. DNAzymes
2.3. Antibodies
2.4. Peptides
2.5. MIPs
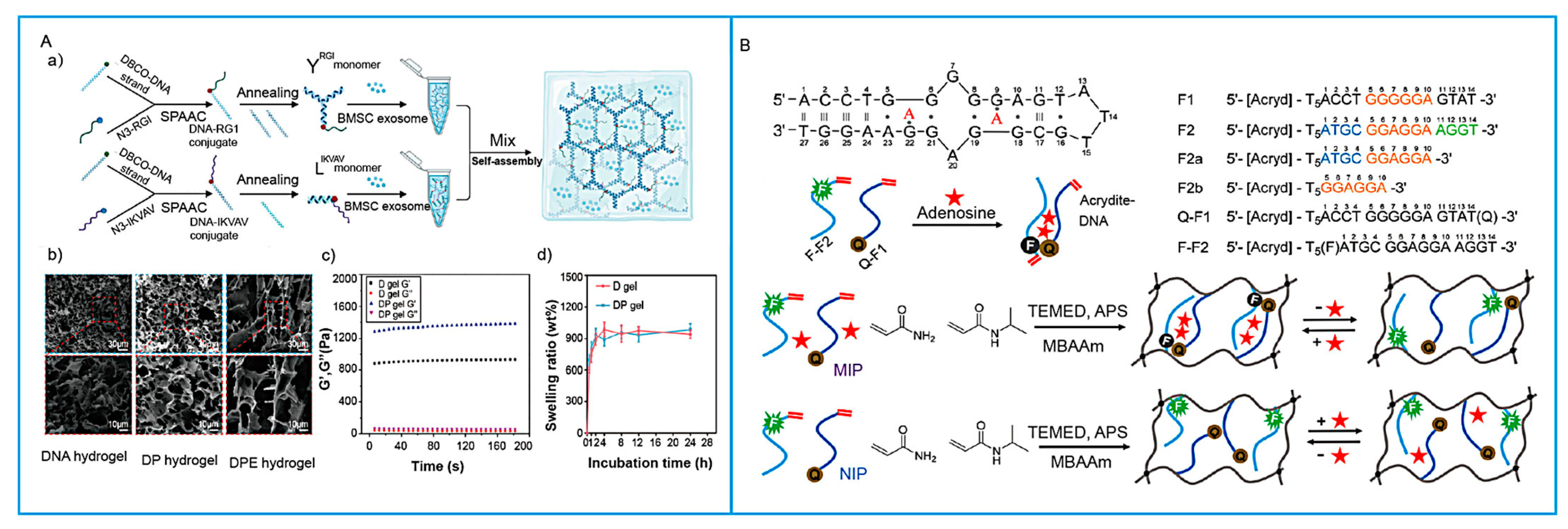
3. Stimulus Response Mechanisms and Signal Transduction in SNAHs
3.1. Temperature-Responsive SNAHs
3.2. Light-Responsive SNAHs
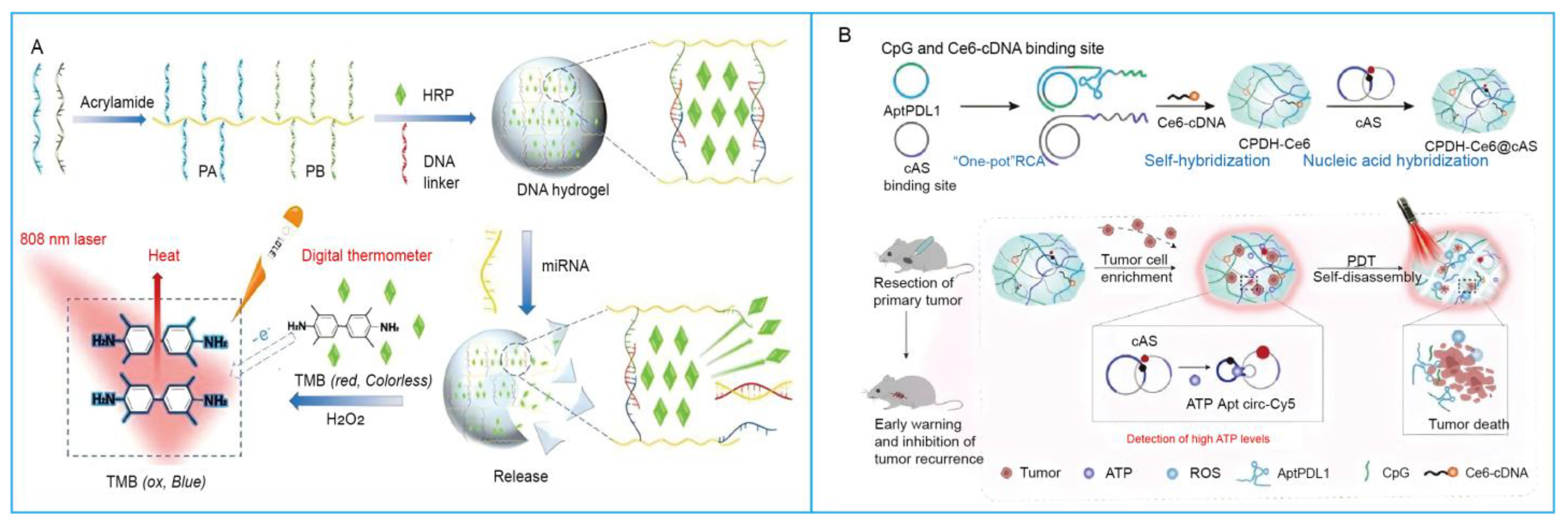
3.3. Magnetic-Responsive SNAHs
3.4. pH-/Ion-Responsive SNAHs
3.5. Nucleic Acid-Responsive SNAHs
3.6. Protein-Responsive SNAHs
3.7. Small Molecule-Responsive SNAHs
3.8. Pathogen-Responsive SNAHs
4. Application of SNAHs in Biosensors
4.1. Environmental Monitoring
4.2. Food Safety
4.3. Disease Diagnosis
5. Conclusions and Prospects
- Expansion of recognition targets and development of next-generation intelligent responsive modules. Future advancements of SNAHs should prioritize the fundamental optimization of sequence stability, structural precision, and selective recognition before enabling more advanced smart sensing functions. Correspondingly, the next research phase should focus on enhancing AI- or machine learning-assisted nucleic acid sequence engineering to improve binding affinity, prediction accuracy, and structural robustness. Furthermore, the rational integration of non-natural functional groups (e.g., hydrophobic moieties, redox-active groups, photocrosslinkers, and metal-coordination sites, etc.) should be guided by well-documented mechanisms derived from existing SNAH systems, rather than being proposed as a generic strategy.
- Advancement of multiplexed detection and multimodal signal output strategies. To address the growing demand for parallel, high-content analysis, future SNAH systems should focus on integrating multiple recognition modules with well-defined logic-gating circuits to achieve accurate multiplexed decision-making. This step represents the second tier of development, which is only feasible after resolving foundational issues related to stability and specificity. Regarding external stimuli, the systems reviewed in this work suggest several priority directions. Optical stimuli offer high spatiotemporal precision suitable for in vitro and wearable monitoring; magnetic fields provide deep-tissue penetration and remote actuation ideal for in vivo sensing; electrical stimulation is highly compatible with flexible electronics. Importantly, machine learning-guided optimization of hydrogel composition, porosity, and signal-transduction pathways could offer automated strategies for improving multiplex sensing fidelity.
- Promotion of theranostic integration, microfluidic convergence, and wearable applications. Given their biocompatibility and programmability, SNAHs offer substantial advantages for future translational use. Beyond conventional drug delivery, hybrid organic–inorganic or multi-stimuli composite hydrogels (e.g., SNAH-MOF, SNAH-metal nanoparticle systems) could enable high-performance theranostic platforms with amplified sensing and therapeutic functions. A particularly important direction is the integration of SNAHs with microfluidic systems, flexible electronics, and wearable platforms. Microfluidics can provide precise sample manipulation and automated cascaded analysis, while flexible or epidermal electronics allow for continuous, on-body sensing with real-time feedback. Furthermore, embedding SNAHs in microneedle patches, strain-responsive hydrogel circuits, or self-powered sensing devices may enable next-generation personalized health monitoring with closed-loop intervention capability.
- Enhancement of matrix stability, device standardization, and pathways toward scalable industrial translation. Prior to the translation of SNAH-based biosensors into practical or commercial applications, several foundational challenges must be effectively addressed. These include nonspecific adsorption, susceptibility to nucleases, mechanical fragility, and environmental interference—all of which directly affect device reproducibility and lifespan. Therefore, priority should be given to chemical modification (e.g., backbone stabilization, protective coatings, zwitterionic surfaces), mechanical reinforcement, and stabilization strategies validated in the current literature. To enable cross-laboratory reproducibility, standardized fabrication protocols, quality-control metrics, and benchmark testing systems should be established. Moreover, coupling SNAHs with low-cost mass-production techniques such as microfluidic extrusion, injection molding, or 3D printing represents an important pathway toward industrialization.
Funding
Conflicts of Interest
Abbreviations
| 3D | three-dimensional |
| AFM1 | aflatoxin M1 |
| AFP | alpha-fetoprotein |
| AFs | aflatoxins |
| AgNCs | silver nanoclusters |
| AMPs | antimicrobial peptides |
| APC | antigen-presenting cell |
| APS | ammonium persulfate |
| ATP | adenosine triphosphate |
| AuNPs | gold nanoparticles |
| AuNRs | gold nanorods |
| CAP | chloramphenicol |
| cDNA | complementary DNA |
| COR | coralyne |
| CTLA-4 | cytotoxic T lymphocyte antigen 4 |
| DGT | diffusive gradients |
| DOX | doxorubicin |
| EDC/NHS | 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide/N-Hydroxysuccinimide |
| ELISA | enzyme-linked immunosorbent assay |
| EpCAM | epithelial cell adhesion molecule |
| FB1 | fumonisin B1 |
| FLD-MS | fluorescence detection–mass spectrometry |
| GC | gas chromatography |
| HPLC | high-performance liquid chromatography |
| HRCA | hyperbranched rolling circle amplification |
| HRP | horseradish peroxidase |
| ITO | Indium-tin oxide |
| LC | liquid chromatography |
| LCST | lower critical solution temperature |
| LOD | limit of detection |
| LPS | lipopolysaccharides |
| MAF | malathion aptamer |
| MBAAm | methylene bis(acrylamide) |
| MB-Dox | doxorubicin-methylene blue conjugates |
| MC-LR | Microcystin-LR |
| MEL | melamine |
| microRNA | miRNA |
| MIPs | molecular imprinted polymers |
| MNAzymes | multicomponent nucleic acid enzymes |
| MNPs | magnetic nanoparticles |
| MOFs | metal–organic frameworks |
| MUC1 | mucin 1 protein |
| NFC | near-field communication |
| NIR | near-infrared |
| nM | nanomolar |
| OTA | ochratoxin A |
| PCR | polymerase chain reaction |
| PD-1 | cell death protein 1 |
| pM | picomolar |
| pNIPAM | poly(N-isopropylacrylamide) |
| QD | quantum dots |
| RAC | ractopamine |
| RCA | rolling circle amplification |
| RhB | rhodamine B |
| ROS | reactive oxygen species |
| SELEX | Systematic Evolution of Ligands by Exponential Enrichment |
| SEM | scanning electron microscopy |
| SERS | surface-enhanced Raman scattering |
| SNAHs | smart nucleic acid hydrogels |
| SPR | surface plasmon resonance |
| ssDNA | single-stranded DNA |
| T cell | T lymphocytes |
| TBO | toluidine blue O |
| TEMED | N,N,N′,N′-tetramethylethylenediamine |
| tFNAs | tetrahedral framework nucleic acids |
| TGMS | Triglycerol monostearate |
| TMB | 3,3′,5,5′-tetramethylbenzidine |
| TNF-α | tumor necrosis factor-α |
| UO22+ | uranyl ion |
| V.P. | Vibrio parahaemolyticus |
| ZEN | zearalenone |
| μM | micromolar |
References
- Gao, X.; Bayinqiaoge, B.; Li, M.; Chandrawati, R.; Li, X.; Sun, L.; Wang, C.H.; Zhang, C.; Tang, S.-Y. Stimuli-Responsive Smart Materials Enabled High-Performance Biosensors for Liquid Biopsies. J. Nanobiotechnol. 2025, 23, 477. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Yang, H.; Cao, J.; Zhang, J.; He, Q.; Deng, R. Isothermal Nucleic Acid Amplification for Food Safety Analysis. TrAC Trends Anal. Chem. 2022, 153, 116641. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, J.; Pan, H.; Ma, X.; Jiang, L.; Zhu, Q.; Wu, H.; Wang, Z. Development and Preliminary Application of a Triplex Real-Time Quantitative PCR Assay for the Simultaneous Detection of Entamoeba Histolytica, Giardia Lamblia, and Cryptosporidium Parvum. Front. Microbiol. 2022, 13, 888529. [Google Scholar] [CrossRef]
- Xia, X.; He, Q.; Dong, Y.; Deng, R.; Li, J. Aptamer-Based Homogeneous Analysis for Food Control. Curr. Anal. Chem. 2020, 16, 4–13. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Zhu, L.; Chen, K.; Li, X.; Xu, W. Smart Nucleic Acid Hydrogels with High Stimuli-Responsiveness in Biomedical Fields. Int. J. Mol. Sci. 2022, 23, 1068. [Google Scholar] [CrossRef]
- Yao, C.; Zhang, R.; Tang, J.; Yang, D. Rolling Circle Amplification (RCA)-Based DNA Hydrogel. Nat. Protoc. 2021, 16, 5460–5483. [Google Scholar] [CrossRef]
- Žuržul, N.; Stokke, B.T. DNA Aptamer Functionalized Hydrogels for Interferometric Fiber-Optic Based Continuous Monitoring of Potassium Ions. Biosensors 2021, 11, 266. [Google Scholar] [CrossRef]
- Lim, S.; Kuang, Y.; Ardoña, H.A.M. Evolution of Supramolecular Systems towards Next-Generation Biosensors. Front. Chem. 2021, 9, 723111. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Shang, Y.; Wang, Z.; Wang, J.; Wu, Q.; Shen, Y.; Ding, Y. Programmable DNA Hydrogels for Biosensing and Point-of-Care Test. Coord. Chem. Rev. 2024, 518, 216084. [Google Scholar] [CrossRef]
- Xu, L.; Zhu, L.; Zhang, Y.; Zhou, C.; Ni, K.; Hu, B.; Cao, J.; Xu, W. Programmable and Multifunctional Nucleic Acid Hydrogels for Accelerating Chronic Wound Healing. Chem. Eng. J. 2025, 520, 166107. [Google Scholar] [CrossRef]
- Röthlisberger, P.; Hollenstein, M. Aptamer Chemistry. Adv. Drug Delivery Rev. 2018, 134, 3–21. [Google Scholar]
- McConnell, E.M.; Cozma, I.; Mou, Q.; Brennan, J.D.; Lu, Y.; Li, Y. Biosensing with DNAzymes. Chem. Soc. Rev. 2021, 50, 8954–8994. [Google Scholar] [CrossRef]
- Trier, N.; Hansen, P.; Houen, G. Peptides, Antibodies, Peptide Antibodies and More. Int. J. Mol. Sci. 2019, 20, 6289. [Google Scholar] [CrossRef]
- Huang, Y.; Pan, J.; Liu, Y.; Wang, M.; Deng, S.; Xia, Z. A SPE Method with Two MIPs in Two Steps for Improving the Selectivity of MIPs. Anal. Chem. 2019, 91, 8436–8442. [Google Scholar] [CrossRef]
- Mahinroosta, M.; Jomeh Farsangi, Z.; Allahverdi, A.; Shakoori, Z. Hydrogels as Intelligent Materials: A Brief Review of Synthesis, Properties and Applications. Mater. Today Chem. 2018, 8, 42–55. [Google Scholar] [CrossRef]
- Li, F.; Tang, J.; Geng, J.; Luo, D.; Yang, D. Polymeric DNA Hydrogel: Design, Synthesis and Applications. Prog. Polym. Sci. 2019, 98, 101163. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Ayachit, N.H.; Aminabhavi, T.M. Biosensors and Microfluidic Biosensors: From Fabrication to Application. Biosensors 2022, 12, 543. [Google Scholar] [CrossRef]
- McConnell, E.M.; Nguyen, J.; Li, Y. Aptamer-Based Biosensors for Environmental Monitoring. Front. Chem. 2020, 8, 434. [Google Scholar] [CrossRef]
- Kotsiri, Z.; Vidic, J.; Vantarakis, A. Applications of Biosensors for Bacteria and Virus Detection in Food and Water–a Systematic Review. J. Environ. Sci. 2022, 111, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Justino, C.I.L.; Duarte, A.C.; Rocha-Santos, T.A.P. Critical Overview on the Application of Sensors and Biosensors for Clinical Analysis. TrAC Trends Anal. Chem. 2016, 85, 36–60. [Google Scholar] [CrossRef]
- Li, J.; Mo, L.; Lu, C.-H.; Fu, T.; Yang, H.-H.; Tan, W. Functional Nucleic Acid-Based Hydrogels for Bioanalytical and Biomedical Applications. Chem. Soc. Rev. 2016, 45, 1410–1431. [Google Scholar] [CrossRef]
- Marrazza, G. Aptamer Sensors. Biosensors 2017, 7, 5. [Google Scholar] [CrossRef]
- Wen, C.; Wang, Y.; Wang, Y. Aptamers in Drug Delivery Development. Mater. Today 2025, 88, 855–870. [Google Scholar] [CrossRef]
- Wang, J.; Huo, J.; Huang, H.; Yao, C.; Yang, D. Construction of Nucleic Acid Aptamer-Functionalized DNA Hydrogels and Their Application in Biomedicine. Mater. Today 2025, 88, 552–566. [Google Scholar] [CrossRef]
- Yao, C.; Tang, H.; Wu, W.; Tang, J.; Guo, W.; Luo, D.; Yang, D. Double Rolling Circle Amplification Generates Physically Cross-Linked DNA Network for Stem Cell Fishing. J. Am. Chem. Soc. 2020, 142, 3422–3429. [Google Scholar] [CrossRef] [PubMed]
- Um, S.H.; Lee, J.B.; Park, N.; Kwon, S.Y.; Umbach, C.C.; Luo, D. Enzyme-Catalysed Assembly of DNA Hydrogel. Nat. Mater. 2006, 5, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Zheng, W.; Duan, M.; Su, T.; Wang, Z.; Wu, S.; Duan, N. Construction of Aptamer-Functionalized DNA Hydrogels for Effective Inhibition of Shiga Toxin II Toxicity. J. Agric. Food Chem. 2024, 72, 23533–23543. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, Y.-R.; Wu, J.; Lv, J.; Chen, Y.; Lin, M.; Liu, R.; Yin, H.; Wu, Z.-S. Programmable DNA Dendrimer from a Single Symmetric Y-Shaped Structural Unit for Targeted Tumor Therapy with One Hundred Inhibition Efficiency. Adv. Funct. Mater. 2025, 35, 2421245. [Google Scholar] [CrossRef]
- Ahn, S.Y.; Kim, J.; Vellampatti, S.; Oh, S.; Lim, Y.T.; Park, S.H.; Luo, D.; Chung, J.; Um, S.H. Protein-Encoding Free-Standing RNA Hydrogel for Sub-Compartmentalized Translation. Adv. Mater. 2022, 34, 2110424. [Google Scholar] [CrossRef]
- Huang, Z.; Kangovi, G.N.; Wen, W.; Lee, S.; Niu, L. An RNA Aptamer Capable of Forming a Hydrogel by Self-Assembly. Biomacromolecules 2017, 18, 2056–2063. [Google Scholar] [CrossRef]
- Zhang, R.; Lv, Z.; Chang, L.; Wang, J.; Tang, J.; Wang, Z.; Li, S.; Guo, J.; Yao, C.; Yang, D. A Responsive DNA Hydrogel Containing Poly-Aptamers as Dual-Target Inhibitors for Localized Cancer Immunotherapy. Adv. Funct. Mater. 2024, 34, 2401563. [Google Scholar] [CrossRef]
- Mao, X.; Simon, A.J.; Pei, H.; Shi, J.; Li, J.; Huang, Q.; Plaxco, K.W.; Fan, C. Activity Modulation and Allosteric Control of a Scaffolded DNAzyme Using a Dynamic DNA Nanostructure. Chem. Sci. 2016, 7, 1200–1204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, L.; Tian, J.; Zhu, L.; Ma, X.; He, X.; Huang, K.; Ren, F.; Xu, W. Smart and Functionalized Development of Nucleic Acid-Based Hydrogels: Assembly Strategies, Recent Advances, and Challenges. Adv. Sci. 2021, 8, 2100216. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Fan, H.; Satyavolu, N.S.R.; Wang, W.; Lake, R.; Jiang, J.-H.; Lu, Y. Imaging Endogenous Metal Ions in Living Cells Using a DNAzyme–Catalytic Hairpin Assembly Probe. Angew. Chem. Int. Ed. 2017, 56, 8721–8725. [Google Scholar] [CrossRef]
- Khajouei, S.; Ravan, H.; Ebrahimi, A. DNA Hydrogel-Empowered Biosensing. Adv. Colloid Interface Sci. 2020, 275, 102060. [Google Scholar] [CrossRef]
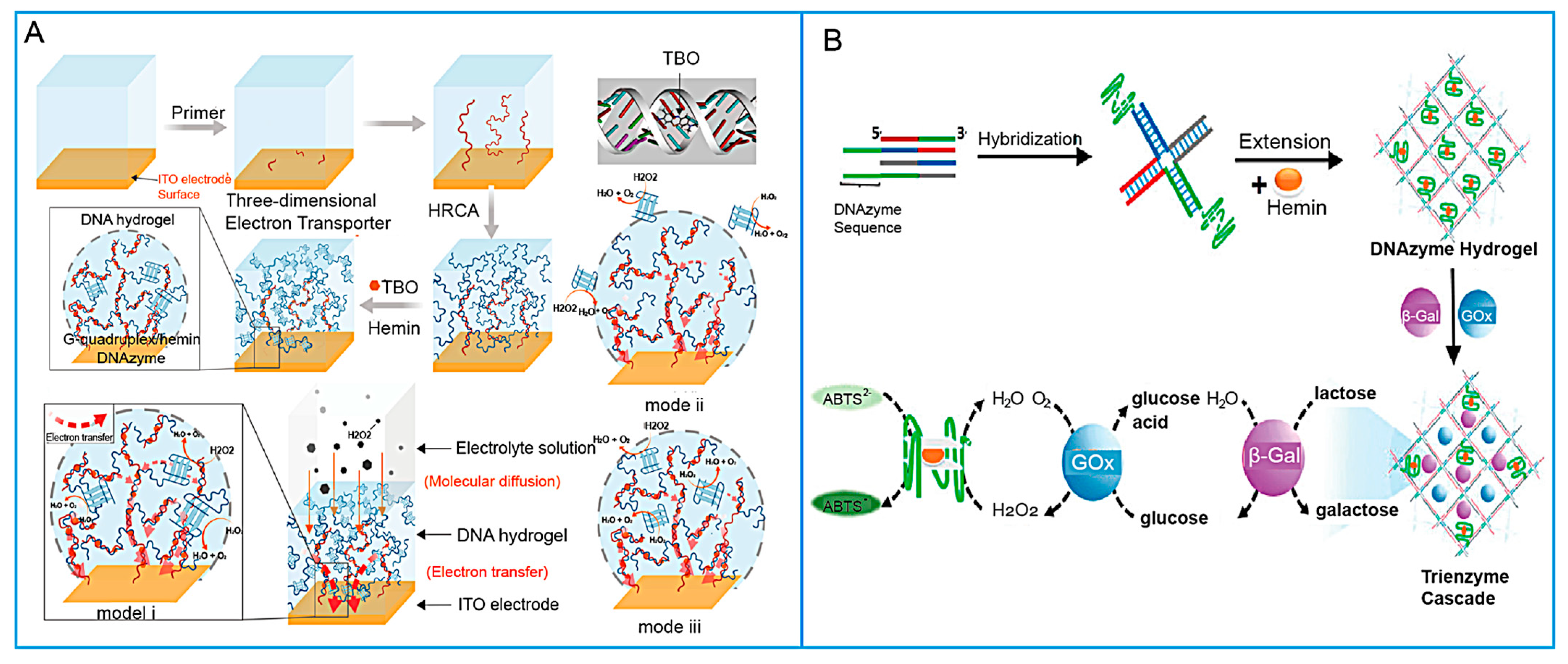
- Mao, X.; Mao, D.; Chen, T.; Jalalah, M.; Al-Assiri, M.S.; Harraz, F.A.; Zhu, X.; Li, G. DNA Hydrogel-Based Three-Dimensional Electron Transporter and Its Application in Electrochemical Biosensing. ACS Appl. Mater. Interfaces 2020, 12, 36851–36859. [Google Scholar] [CrossRef]
- Mao, X.; Pan, S.; Zhou, D.; He, X.; Zhang, Y. Fabrication of DNAzyme-Functionalized Hydrogel and Its Application for Visible Detection of Circulating Tumor DNA. Sens. Actuators B Chem. 2019, 285, 385–390. [Google Scholar] [CrossRef]
- Jiang, C.; Li, Y.; Wang, H.; Chen, D.; Wen, Y. A Portable Visual Capillary Sensor Based on Functional DNA Crosslinked Hydrogel for Point-of-Care Detection of Lead Ion. Sens. Actuators B Chem. 2020, 307, 127625. [Google Scholar] [CrossRef]
- Chai, H.; Yan, C.; Guo, J.; Lei, F.; Miao, P. Electrochemical Analysis of Ca2+ Based on DNAzyme Catalyzed Degradation of DNA Hydrogel. Electrochem. Commun. 2024, 165, 107755. [Google Scholar] [CrossRef]
- Hou, M.; Yin, X.; Jiang, J.; He, J. DNAzyme-Triggered Sol–Gel–Sol Transition of a Hydrogel Allows Target Cell Enrichment. ACS Appl. Mater. Interfaces 2021, 13, 15031–15039. [Google Scholar] [CrossRef]
- Xiang, B.; He, K.; Zhu, R.; Liu, Z.; Zeng, S.; Huang, Y.; Nie, Z.; Yao, S. Self-Assembled DNA Hydrogel Based on Enzymatically Polymerized DNA for Protein Encapsulation and Enzyme/DNAzyme Hybrid Cascade Reaction. ACS Appl. Mater. Interfaces 2016, 8, 22801–22807. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wang, X.; Luo, S.; Zhang, P.; Li, N.; Chen, C.; Li, J.; Shi, H.; Dong, H.; Huang, R.-P. Constructions, Purifications and Applications of DNA-Antibody Conjugates: A Review. ACS Omega 2024, 9, 47951–47963. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Linko, V. Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem. Int. Ed. 2020, 59, 15818–15833. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wang, K.; Luo, S.; Li, F.; Zuo, X.; Fan, C.; Li, Q. Programmable DNA Hydrogels as Artificial Extracellular Matrix. Small 2022, 18, 2107640. [Google Scholar] [CrossRef]
- Zhu, Y.; Shi, R.; Lu, W.; Shi, S.; Chen, Y. Framework Nucleic Acids as Promising Reactive Oxygen Species Scavengers for Anti-Inflammatory Therapy. Nanoscale 2024, 16, 7363–7377. [Google Scholar] [CrossRef]
- Dovgan, I.; Koniev, O.; Kolodych, S.; Wagner, A. Antibody–Oligonucleotide Conjugates as Therapeutic, Imaging, and Detection Agents. Bioconjugate Chem. 2019, 30, 2483–2501. [Google Scholar] [CrossRef]
- Dugal-Tessier, J.; Thirumalairajan, S.; Jain, N. Antibody-Oligonucleotide Conjugates: A Twist to Antibody-Drug Conjugates. J. Clin. Med. 2021, 10, 838. [Google Scholar] [CrossRef]
- Watson, E.E.; Winssinger, N. Synthesis of Protein-Oligonucleotide Conjugates. Biomolecules 2022, 12, 1523. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, S.; Chen, N.; Yang, C.; Wang, Y. Programmable Display of DNA–Protein Chimeras for Controlling Cell–Hydrogel Interactions via Reversible Intermolecular Hybridization. Biomacromolecules 2013, 14, 1174–1180. [Google Scholar] [CrossRef]
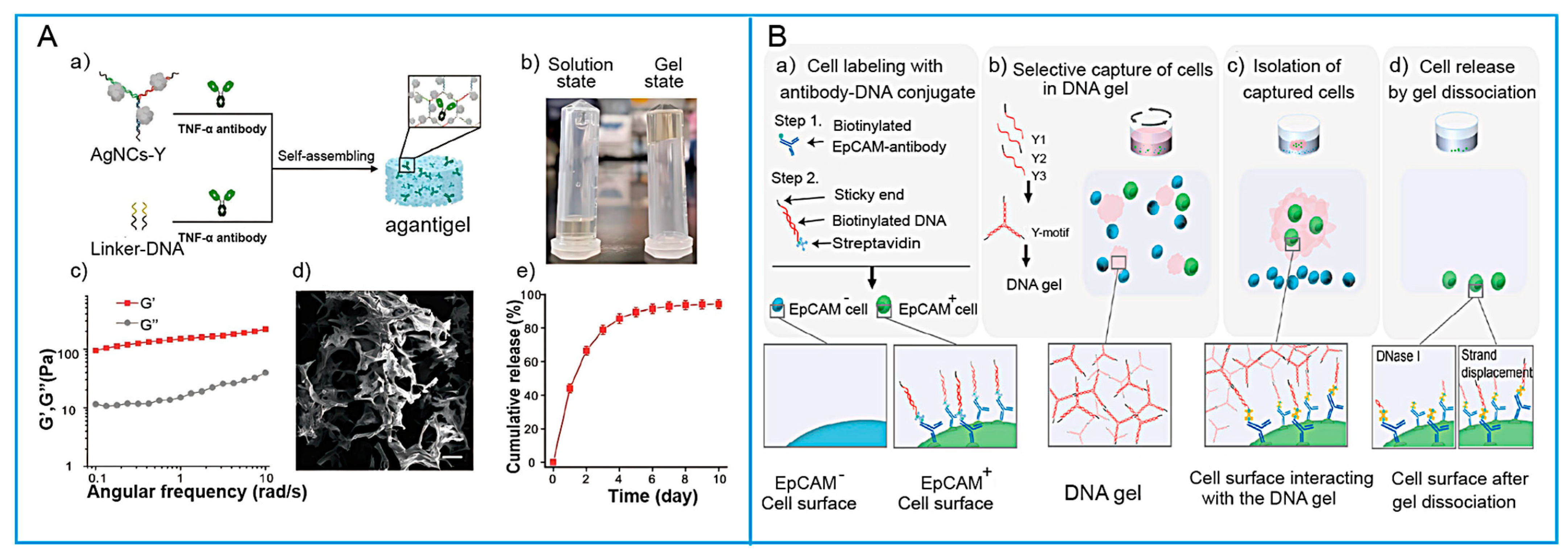
- Peng, L.; Li, W.; Peng, G.; Wei, D.; Gou, L.; Zhou, Y.; Zhou, Y.; Chen, X.; Wu, L.; Zhang, W.; et al. Antibacterial and DNA-Based Hydrogels In Situ Block TNF-α to Promote Diabetic Alveolar Bone Rebuilding. Macromol. Rapid Commun. 2024, 45, 2300559. [Google Scholar] [CrossRef]
- Bourdon, L.; Cochard, A.; Tauran, Y.; Sugitani, Y.; Sato, Y.; Takinoue, M.; Fujita, H.; Fujii, T.; Kim, S.H.; Genot, A.J. Antibody-Functionalized DNA Hydrogels Recognize and Isolate Living Tumor Cells. Adv. Mater. Interfaces 2025, e00619. [Google Scholar] [CrossRef]
- Adhikari, S.; Ghosh, S.; Sarathi Addy, P. Advancing Biomarker Detection with Peptide-Integrated Fluorescent Probes. Chem.-Asian J. 2025, 20, e00211. [Google Scholar] [CrossRef] [PubMed]
- Hoyos-Nogués, M.; Gil, F.J.; Mas-Moruno, C. Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies. Molecules 2018, 23, 1683. [Google Scholar] [CrossRef]
- Liu, X.; Marrakchi, M.; Xu, D.; Dong, H.; Andreescu, S. Biosensors Based on Modularly Designed Synthetic Peptides for Recognition, Detection and Live/Dead Differentiation of Pathogenic Bacteria. Biosens. Bioelectron. 2016, 80, 9–16. [Google Scholar] [CrossRef]
- Ahn, S.; Kim, N.; Choi, Y.; Kim, J.; Hwang, H.; Kim, C.; Lee, H.-Y.; Kim, S.; Kim, J.S.; Lee, H.H.; et al. Peptide-Decorated Microneedles for the Detection of Microplastics. Biosensors 2024, 14, 140. [Google Scholar] [CrossRef]
- Chen, M.; Song, Z.; Yang, X.; Song, Z.; Luo, X. Antifouling Peptides Combined with Recognizing DNA Probes for Ultralow Fouling Electrochemical Detection of Cancer Biomarkers in Human Bodily Fluids. Biosens. Bioelectron. 2022, 206, 114162. [Google Scholar] [CrossRef]
- He, X.; Sun, N.; Jia, H.; Hou, M.; Tan, Z.; Lu, X. Antifouling Electrochemical Biosensor Based on Conductive Hydrogel of DNA Scaffold for Ultrasensitive Detection of ATP. ACS Appl. Mater. Interfaces 2022, 14, 40624–40632. [Google Scholar] [CrossRef]
- Li, C.; Chen, P.; Shao, Y.; Zhou, X.; Wu, Y.; Yang, Z.; Li, Z.; Weil, T.; Liu, D. A Writable Polypeptide–DNA Hydrogel with Rationally Designed Multi-modification Sites. Small 2015, 11, 1138–1143. [Google Scholar] [CrossRef]
- Wei, Z.; Li, X.; Chen, Y.; Han, Z.; Li, Y.; Gan, L.; Yang, Y.; Chen, Y.; Zhang, F.; Ye, X.; et al. Programmable DNA-Peptide Conjugated Hydrogel via Click Chemistry for Sequential Modulation of Peripheral Nerve Regeneration. Adv. Funct. Mater. 2025, 35, 2419915. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, X.; Zhang, Y.; Xie, M.; Wang, F.; Qin, J.; Ye, H.; Zhang, H.; Zhang, C.; Hong, J. A Nucleic Acid-Based LYTAC plus Platform to Simultaneously Mediate Disease-Driven Protein Downregulation. Adv. Sci. 2024, 11, 2306248. [Google Scholar] [CrossRef]
- Yin, W.; Chen, X.; Bai, L.; Li, Y.; Chen, W.; Jiang, Y.; He, Y.; Yang, Y.; Lin, Y.; Tian, T.; et al. BBPs-Functionalized Tetrahedral Framework Nucleic Acid Hydrogel Scaffold Captures Endogenous BMP-2 to Promote Bone Regeneration. Biomaterials 2025, 319, 123194. [Google Scholar] [CrossRef]
- Obuobi, S.; Tay, H.K.-L.; Tram, N.D.T.; Selvarajan, V.; Khara, J.S.; Wang, Y.; Ee, P.L.R. Facile and Efficient Encapsulation of Antimicrobial Peptides via Crosslinked DNA Nanostructures and Their Application in Wound Therapy. J. Control. Release 2019, 313, 120–130. [Google Scholar] [CrossRef]
- BelBruno, J.J. Molecularly Imprinted Polymers. Chem. Rev. 2019, 119, 94–119. [Google Scholar] [CrossRef]
- Haupt, K.; Medina Rangel, P.X.; Bui, B.T.S. Molecularly Imprinted Polymers: Antibody Mimics for Bioimaging and Therapy. Chem. Rev. 2020, 120, 9554–9582. [Google Scholar] [CrossRef]
- Ogiso, M.; Minoura, N.; Shinbo, T.; Shimizu, T. Detection of a Specific DNA Sequence by Electrophoresis through a Molecularly Imprinted Polymer. Biomaterials 2006, 27, 4177–4182. [Google Scholar] [CrossRef]
- Byrne, M.E.; Salian, V. Molecular Imprinting within Hydrogels II: Progress and Analysis of the Field. Int. J. Pharm. 2008, 364, 188–212. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Lyu, Y.; Liu, Z. Recent Advances of Molecular Imprinting Technology for the Separation and Recognition of Complex Biological Sample Systems. Chin. J. Chromatogr. 2024, 42, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Lusina, A.; Cegłowski, M. Molecularly Imprinted Polymers as State-of-the-Art Drug Carriers in Hydrogel Transdermal Drug Delivery Applications. Polymers 2022, 14, 640. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, J. Molecular Imprinting with Functional DNA. Small 2019, 15, 1805246. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Z.; Liu, B.; Liu, J. Incorporation of Boronic Acid into Aptamer-Based Molecularly Imprinted Hydrogels for Highly Specific Recognition of Adenosine. ACS Appl. Bio Mater. 2020, 3, 2568–2576. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, J. Molecularly Imprinted Polymers with DNA Aptamer Fragments as Macromonomers. ACS Appl. Mater. Interfaces 2016, 8, 6371–6378. [Google Scholar] [CrossRef]
- Song, M.; Zhang, J.; Shen, K.; Hu, Y.; Shen, W.; Tang, S.; Lee, H.K. Application of Smart-Responsive Hydrogels in Nucleic Acid and Nucleic Acid-Based Target Sensing: A Review. Biosens. Bioelectron. 2025, 267, 116803. [Google Scholar] [CrossRef]
- Shahbazi, M.-A.; Bauleth-Ramos, T.; Santos, H.A. DNA Hydrogel Assemblies: Bridging Synthesis Principles to Biomedical Applications. Adv. Ther. 2018, 1, 1800042. [Google Scholar] [CrossRef]
- Uzumcu, A.T.; Guney, O.; Okay, O. Nanocomposite DNA Hydrogels with Temperature Sensitivity. Polymer 2016, 100, 169–178. [Google Scholar] [CrossRef]
- Chen, J.; Ouyang, X.; Yu, C.; Xiang, J. Functionalized pNIPAM-DNA Hydrogel Colorimetric Platform for Visual Detection of Low-Mass Soluble β-Amyloid Oligomers. Anal. Chem. 2025, 97, 10417–10423. [Google Scholar] [CrossRef] [PubMed]
- Yata, T.; Takahashi, Y.; Tan, M.; Nakatsuji, H.; Ohtsuki, S.; Murakami, T.; Imahori, H.; Umeki, Y.; Shiomi, T.; Takakura, Y.; et al. DNA Nanotechnology-Based Composite-Type Gold Nanoparticle-Immunostimulatory DNA Hydrogel for Tumor Photothermal Immunotherapy. Biomaterials 2017, 146, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Wang, Y.; Gong, B.; Xiao, Y.; Chen, Y.; Wang, S.; Li, S.; Huang, F.; Shen, Y.; Xie, A. Reduced Graphene Oxide/Amaranth Extract/AuNPs Composite Hydrogel on Tumor Cells as Integrated Platform for Localized and Multiple Synergistic Therapy. ACS Appl. Mater. Interfaces 2015, 7, 11246–11256. [Google Scholar] [CrossRef]
- Wang, C.; Liu, X.; Wulf, V.; Vázquez-González, M.; Fadeev, M.; Willner, I. DNA-Based Hydrogels Loaded with Au Nanoparticles or Au Nanorods: Thermoresponsive Plasmonic Matrices for Shape-Memory, Self-Healing, Controlled Release, and Mechanical Applications. ACS Nano 2019, 13, 3424–3433. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Z.; Liu, F.; Zhang, X.; Huang, J.; Wang, G. Programmable Plasmonic Hydrogel Thermometers Actuated by DNA Breathing. Adv. Mater. Technol. 2024, 9, 2400243. [Google Scholar] [CrossRef]
- Lu, S.; Shen, J.; Fan, C.; Li, Q.; Yang, X. DNA Assembly-Based Stimuli-Responsive Systems. Adv. Sci. 2021, 8, 2100328. [Google Scholar] [CrossRef]
- Stricker, L.; Fritz, E.-C.; Peterlechner, M.; Doltsinis, N.L.; Ravoo, B.J. Arylazopyrazoles as Light-Responsive Molecular Switches in Cyclodextrin-Based Supramolecular Systems. J. Am. Chem. Soc. 2016, 138, 4547–4554. [Google Scholar] [CrossRef]
- Cangialosi, A.; Yoon, C.; Liu, J.; Huang, Q.; Guo, J.; Nguyen, T.D.; Gracias, D.H.; Schulman, R. DNA Sequence–Directed Shape Change of Photopatterned Hydrogels via High-Degree Swelling. Science 2017, 357, 1126–1130. [Google Scholar] [CrossRef]
- Peng, L.; You, M.; Yuan, Q.; Wu, C.; Han, D.; Chen, Y.; Zhong, Z.; Xue, J.; Tan, W. Macroscopic Volume Change of Dynamic Hydrogels Induced by Reversible DNA Hybridization. J. Am. Chem. Soc. 2012, 134, 12302–12307. [Google Scholar] [CrossRef]
- Yang, M.; Dong, Y.; Li, C. Light-Dissipative and Reprogrammable DNA Hydrogels Enabled by Merocyanine Photoacids. Chem. Mater. 2023, 35, 9978–9987. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Fadeev, M.; Li, Z.; Wulf, V.; Tian, H.; Willner, I. Chemical and Photochemical DNA “Gears” Reversibly Control Stiffness, Shape-Memory, Self-Healing and Controlled Release Properties of Polyacrylamide Hydrogels. Chem. Sci. 2019, 10, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Pi, W.; Cheng, S.; Gu, Z.; Zhang, K.; Min, T.; Zhang, W.; Du, H.; Zhang, P.; Wen, Y. Multifunctional DNA Hydrogels with Hydrocolloid-Cotton Structure for Regeneration of Diabetic Infectious Wounds. Adv. Funct. Mater. 2021, 31, 2106167. [Google Scholar] [CrossRef]
- Pandey, P.K.; Ulla, H.; Satyanarayan, M.N.; Rawat, K.; Gaur, A.; Gawali, S.; Hassan, P.A.; Bohidar, H.B. Fluorescent MoS2 Quantum Dot–DNA Nanocomposite Hydrogels for Organic Light-Emitting Diodes. ACS Appl. Nano Mater. 2020, 3, 1289–1297. [Google Scholar] [CrossRef]
- Wang, D.; Liu, J.; Duan, J.; Yi, H.; Liu, J.; Song, H.; Zhang, Z.; Shi, J.; Zhang, K. Enrichment and Sensing Tumor Cells by Embedded Immunomodulatory DNA Hydrogel to Inhibit Postoperative Tumor Recurrence. Nat. Commun. 2023, 14, 4511. [Google Scholar] [CrossRef]
- Liu, B.; Sun, J.; Zhu, J.; Li, B.; Ma, C.; Gu, X.; Liu, K.; Zhang, H.; Wang, F.; Su, J.; et al. Injectable and NIR-Responsive DNA–Inorganic Hybrid Hydrogels with Outstanding Photothermal Therapy. Adv. Mater. 2020, 32, 2004460. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, M.; Chen, Z.; Cui, J.; Yang, L.; Lu, Z.; Qi, F.; Wang, H. Photothermal Detection of MicroRNA Using a Horseradish Peroxidase-Encapsulated DNA Hydrogel with a Portable Thermometer. Front. Bioeng. Biotechnol. 2021, 9, 799370. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Chen, C.; Cheng, Y. Magnetic-Responsive Hydrogels: From Strategic Design to Biomedical Applications. J. Control. Release 2021, 335, 541–556. [Google Scholar] [CrossRef]
- Shi, J.; Shi, Z.; Dong, Y.; Wu, F.; Liu, D. Responsive DNA-Based Supramolecular Hydrogels. ACS Appl. Bio Mater. 2020, 3, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; He, W.; Shen, H.; Zhou, Z.; Li, M.; Su, P.; Yang, Y. Self-Assembly of a Magnetic DNA Hydrogel as a New Biomaterial for Enzyme Encapsulation with Enhanced Activity and Stability. Chem. Commun. 2019, 55, 2449–2452. [Google Scholar] [CrossRef]
- Suhail, M.; Alamgir; Wahab, A.; Eggers, T.; Ahmad, Z.; Shehzad, K.; Iqbal, M.Z. Magnetically Responsive Hydrogel Systems: Fundamental Features, Emerging Applications, and Future Horizons. Coord. Chem. Rev. 2025, 543, 216916. [Google Scholar] [CrossRef]
- Ma, X.; Yang, Z.; Wang, Y.; Zhang, G.; Shao, Y.; Jia, H.; Cao, T.; Wang, R.; Liu, D. Remote Controlling DNA Hydrogel by Magnetic Field. ACS Appl. Mater. Interfaces 2017, 9, 1995–2000. [Google Scholar] [CrossRef]
- Tang, J.; Yao, C.; Gu, Z.; Jung, S.; Luo, D.; Yang, D. Super-Soft and Super-Elastic DNA Robot with Magnetically Driven Navigational Locomotion for Cell Delivery in Confined Space. Angew. Chem. Int. Ed. 2020, 59, 2490–2495. [Google Scholar] [CrossRef]
- Yao, C.; Yuan, Y.; Yang, D. Magnetic DNA Nanogels for Targeting Delivery and Multistimuli-Triggered Release of Anticancer Drugs. ACS Appl. Bio Mater. 2018, 1, 2012–2020. [Google Scholar] [CrossRef]
- Wang, D.; Duan, J.; Liu, J.; Yi, H.; Zhang, Z.; Song, H.; Li, Y.; Zhang, K. Stimuli-Responsive Self-Degradable DNA Hydrogels: Design, Synthesis, and Applications. Adv. Healthcare Mater. 2023, 12, 2203031. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, R.; Yang, S.; Li, S.; Gao, Z. Design and Application of Stimuli-Responsive DNA Hydrogels: A Review. Mater. Today Bio. 2022, 16, 100430. [Google Scholar] [CrossRef] [PubMed]
- Day, H.A.; Pavlou, P.; Waller, Z.A.E. I-Motif DNA: Structure, Stability and Targeting with Ligands. Bioorg. Med. Chem. 2014, 22, 4407–4418. [Google Scholar] [CrossRef]
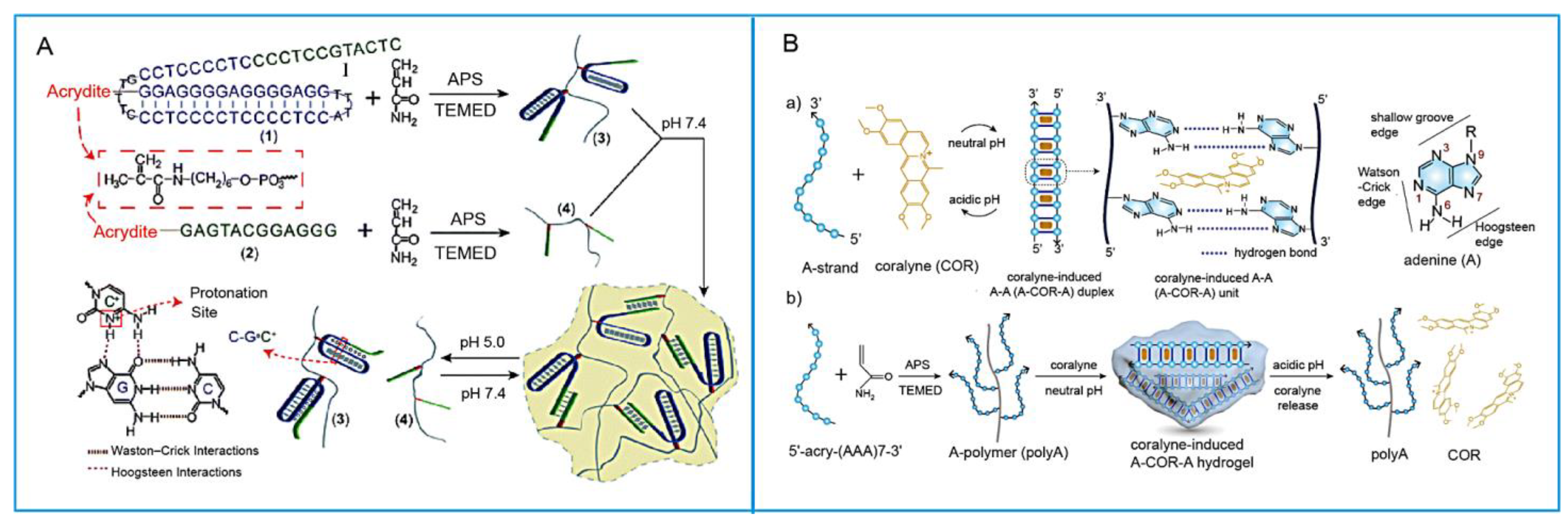
- Huang, Z.; Liu, B.; Liu, J. Parallel Polyadenine Duplex Formation at Low pH Facilitates DNA Conjugation onto Gold Nanoparticles. Langmuir 2016, 32, 11986–11992. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Gao, S.; Lu, H.; Ying, J.Y. Acid-Resistant and Physiological pH-Responsive DNA Hydrogel Composed of a-Motif and i-Motif toward Oral Insulin Delivery. J. Am. Chem. Soc. 2022, 144, 5461–5470. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lu, C.-H.; Guo, W.; Aleman-Garcia, M.A.; Ren, J.; Willner, I. A Shape Memory Acrylamide/DNA Hydrogel Exhibiting Switchable Dual pH-Responsiveness. Adv. Funct. Mater. 2015, 25, 6867–6874. [Google Scholar] [CrossRef]
- Ren, J.; Hu, Y.; Lu, C.-H.; Guo, W.; Aleman-Garcia, M.A.; Ricci, F.; Willner, I. pH-Responsive and Switchable Triplex-Based DNA Hydrogels. Chem. Sci. 2015, 6, 4190–4195. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Ong, C.Y.J.; Wong, J.Y.; Ke, Y.; Lim, J.Y.C.; Dong, Z.; Long, Y.; Hu, Y. Programming the Assembly of Oligo-Adenine with Coralyne into a pH-Responsive DNA Hydrogel. ACS Appl. Mater. Interfaces 2024, 16, 15394–15404. [Google Scholar] [CrossRef]
- Wang, X.; Chen, C.; Waterhouse, G.I.N.; Qiao, X.; Xu, Z. Ultra-Sensitive Detection of Streptomycin in Foods Using a Novel SERS Switch Sensor Fabricated by AuNRs Array and DNA Hydrogel Embedded with DNAzyme. Food Chem. 2022, 393, 133413. [Google Scholar] [CrossRef]
- Pi, K.; Liu, J.; Van Cappellen, P. A DNA-Based Biosensor for Aqueous Hg(II): Performance under Variable pH, Temperature and Competing Ligand Composition. J. Hazard. Mater. 2020, 385, 121572. [Google Scholar] [CrossRef]
- Yoshida, K.; Hayashi, T.; Takinoue, M.; Onoe, H. Repeatable Detection of Ag+ Ions Using a DNA Aptamer-Linked Hydrogel Biochemical Sensor Integrated with Microfluidic Heating System. Sci. Rep. 2022, 12, 9692. [Google Scholar] [CrossRef]
- Cai, X.; Luo, Y.; Zhang, W.; Du, D.; Lin, Y. pH-Sensitive ZnO Quantum Dots–Doxorubicin Nanoparticles for Lung Cancer Targeted Drug Delivery. ACS Appl. Mater. Interfaces 2016, 8, 22442–22450. [Google Scholar] [CrossRef]
- Cai, X.; Luo, Y.; Yan, H.; Du, D.; Lin, Y. pH-Responsive ZnO Nanocluster for Lung Cancer Chemotherapy. ACS Appl. Mater. Interfaces 2017, 9, 5739–5747. [Google Scholar] [CrossRef]
- Yao, S.; Xiang, L.; Wang, L.; Gong, H.; Chen, F.; Cai, C. pH-Responsive DNA Hydrogels with Ratiometric Fluorescence for Accurate Detection of miRNA-21. Anal. Chim. Acta 2022, 1207, 339795. [Google Scholar] [CrossRef] [PubMed]
- Simmel, F.C.; Yurke, B.; Singh, H.R. Principles and Applications of Nucleic Acid Strand Displacement Reactions. Chem. Rev. 2019, 119, 6326–6369. [Google Scholar] [CrossRef] [PubMed]
- Oishi, M.; Nakatani, K. Dynamically Programmed Switchable DNA Hydrogels Based on a DNA Circuit Mechanism. Small 2019, 15, 1900490. [Google Scholar] [CrossRef]
- Lin, D.C.; Yurke, B.; Langrana, N.A. Mechanical Properties of a Reversible, DNA-Crosslinked Polyacrylamide Hydrogel. J. Biomech. Eng. 2004, 126, 104–110. [Google Scholar] [CrossRef]
- Ho, M.S.M.; Lim, A.Z.T.; Ke, Y.; Loh, W.W.; Zheng, X.T.; Yang, L.; Dong, Z.; Wang, F.; Lim, J.Y.C.; Hu, Y. Cascade DNA Structural Transitions Enable Stimuli-Responsive Hydrogels. ACS Appl. Mater. Interfaces 2025, 17, 27116–27125. [Google Scholar] [CrossRef]
- Zhang, R.; Zhou, X.; Deng, H.; Yuan, R.; Yuan, Y. Efficient Multidriven Strand Displacement Reaction for Biosensing. Anal. Chem. 2024, 96, 16735–16742. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Wen, B.; Cheng, W.; Zhou, X.; Duan, X.; Zhao, M.; Xia, Q.; Ding, S. An Enzyme-Free and Label-Free Surface Plasmon Resonance Biosensor for Ultrasensitive Detection of Fusion Gene Based on DNA Self-Assembly Hydrogel with Streptavidin Encapsulation. Biosens. Bioelectron. 2018, 112, 120–126. [Google Scholar] [CrossRef]
- He, Y.; Yang, X.; Yuan, R.; Chai, Y. Switchable Target-Responsive 3D DNA Hydrogels As a Signal Amplification Strategy Combining with SERS Technique for Ultrasensitive Detection of miRNA 155. Anal. Chem. 2017, 89, 8538–8544. [Google Scholar] [CrossRef]
- Chang, W.-H.; Lee, Y.-F.; Liu, Y.-W.; Willner, I.; Liao, W.-C. Stimuli-Responsive Hydrogel Microcapsules for the Amplified Detection of microRNAs. Nanoscale 2021, 13, 16799–16808. [Google Scholar] [CrossRef]
- Eguchi, Y.; Kato, T.; Tanaka, T.; Maruyama, T. A DNA–Gold Nanoparticle Hybrid Hydrogel Network Prepared by Enzymatic Reaction. Chem. Commun. 2017, 53, 5802–5805. [Google Scholar] [CrossRef]
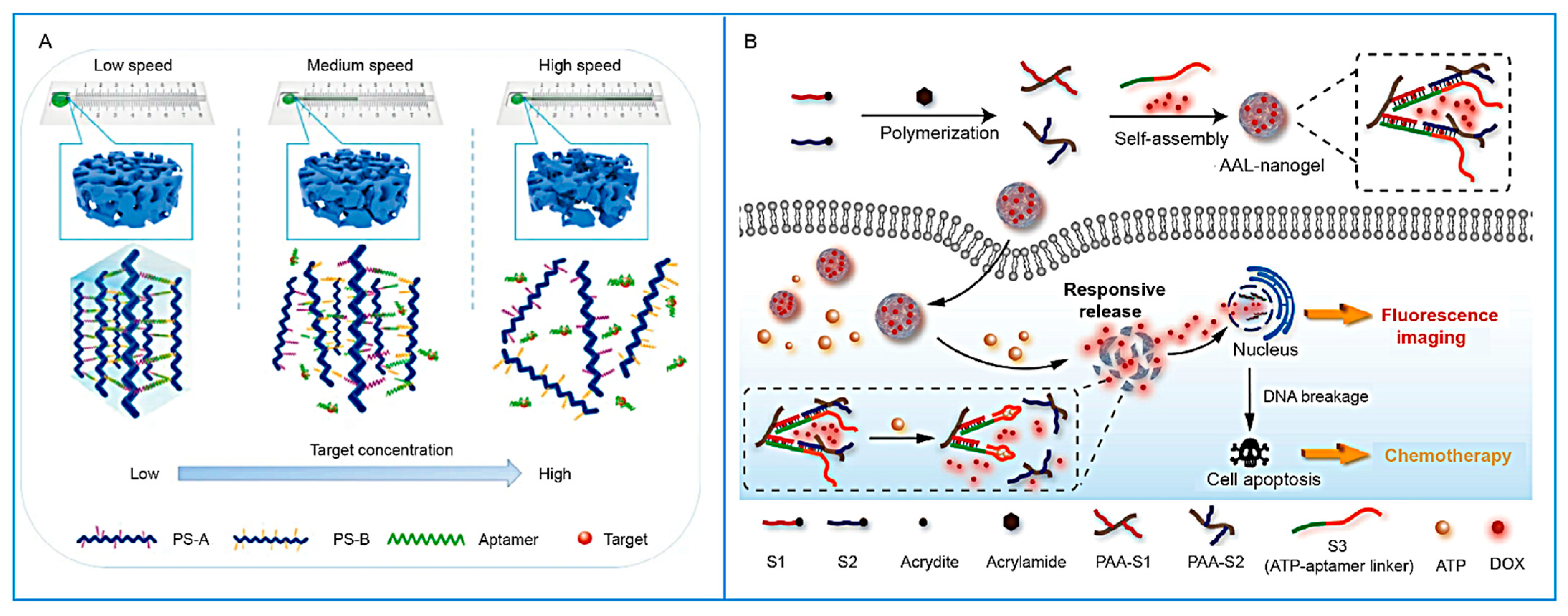
- Zhang, Z.; Liu, C.; Yang, C.; Wu, Y.; Yu, F.; Chen, Y.; Du, J. Aptamer-Patterned Hydrogel Films for Spatiotemporally Programmable Capture and Release of Multiple Proteins. ACS Appl. Mater. Interfaces 2018, 10, 8546–8554. [Google Scholar] [CrossRef]
- English, M.A.; Soenksen, L.R.; Gayet, R.V.; de Puig, H.; Angenent-Mari, N.M.; Mao, A.S.; Nguyen, P.Q.; Collins, J.J. Programmable CRISPR-Responsive Smart Materials. Science 2019, 365, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Du, J.; Li, Y.; Wu, J.; Yu, F.; Chen, Y. An Aptamer-Patterned Hydrogel for the Controlled Capture and Release of Proteins via Biorthogonal Click Chemistry and DNA Hybridization. J. Mater. Chem. B 2017, 5, 5974–5982. [Google Scholar] [CrossRef]
- He, L.; Chen, C.; Liu, Y.; Hai, H.; Li, J. Ultrasensitive Detection of CA125 Based on a Triple Signal Amplification Strategy with a Huge Number of Loaded Probes via Exonuclease Cyclic Cleavage, Rolling Cyclic Amplification and Strand Self-Growth. Analyst 2023, 148, 3217–3225. [Google Scholar] [CrossRef]
- Sun, K.; Chen, P.; Yan, S.; Yuan, W.; Wang, Y.; Li, X.; Dou, L.; Zhao, C.; Zhang, J.; Wang, Q.; et al. Ultrasensitive Nanopore Sensing of Mucin 1 and Circulating Tumor Cells in Whole Blood of Breast Cancer Patients by Analyte-Triggered Triplex-DNA Release. ACS Appl. Mater. Interfaces 2021, 13, 21030–21039. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Lv, H.; Sun, X.; Ding, C. Green-Emitting Carbon Dot Loaded Silica Nanoparticles Coated with DNA-Cross-Linked Hydrogels for Sensitive Carcinoembryonic Antigen Detection and Effective Targeted Cancer Therapy. Chem. Commun. 2019, 55, 15101–15104. [Google Scholar] [CrossRef]
- Wang, C.; Sun, W.; Wright, G.; Wang, A.Z.; Gu, Z. Inflammation-Triggered Cancer Immunotherapy by Programmed Delivery of CpG and Anti-PD1 Antibody. Adv. Mater. 2016, 28, 8912–8920. [Google Scholar] [CrossRef]
- Gao, X.; Li, X.; Sun, X.; Zhang, J.; Zhao, Y.; Liu, X.; Li, F. DNA Tetrahedra-Cross-Linked Hydrogel Functionalized Paper for Onsite Analysis of DNA Methyltransferase Activity Using a Personal Glucose Meter. Anal. Chem. 2020, 92, 4592–4599. [Google Scholar] [CrossRef] [PubMed]
- Gayet, R.V.; de Puig, H.; English, M.A.; Soenksen, L.R.; Nguyen, P.Q.; Mao, A.S.; Angenent-Mari, N.M.; Collins, J.J. Creating CRISPR-Responsive Smart Materials for Diagnostics and Programmable Cargo Release. Nat. Protoc. 2020, 15, 3030–3063. [Google Scholar] [CrossRef]
- Zhao, J.; Gao, J.; Xue, W.; Di, Z.; Xing, H.; Lu, Y.; Li, L. Upconversion Luminescence-Activated DNA Nanodevice for ATP Sensing in Living Cells. J. Am. Chem. Soc. 2018, 140, 578–581. [Google Scholar] [CrossRef]
- Luan, Y.; Wang, N.; Li, C.; Guo, X.; Lu, A. Advances in the Application of Aptamer Biosensors to the Detection of Aminoglycoside Antibiotics. Antibiotics 2020, 9, 787. [Google Scholar] [CrossRef]
- Liu, D.; Jia, S.; Zhang, H.; Ma, Y.; Guan, Z.; Li, J.; Zhu, Z.; Ji, T.; Yang, C.J. Integrating Target-Responsive Hydrogel with Pressuremeter Readout Enables Simple, Sensitive, User-Friendly, Quantitative Point-of-Care Testing. ACS Appl. Mater. Interfaces 2017, 9, 22252–22258. [Google Scholar] [CrossRef]
- Yan, C.; Hua, Y.; Guo, J.; Miao, P. Programmable DNA Hydrogels Construction with Functional Regulations for Biosensing Applications. TrAC Trends Anal. Chem. 2024, 173, 117628. [Google Scholar] [CrossRef]
- Chen, Q.; Tian, R.; Liu, G.; Wen, Y.; Bian, X.; Luan, D.; Wang, H.; Lai, K.; Yan, J. Fishing Unfunctionalized SERS Tags with DNA Hydrogel Network Generated by Ligation-Rolling Circle Amplification for Simple and Ultrasensitive Detection of Kanamycin. Biosens. Bioelectron. 2022, 207, 114187. [Google Scholar] [CrossRef]
- Hu, M.; Yue, F.; Dong, J.; Tao, C.; Bai, M.; Liu, M.; Zhai, S.; Chen, S.; Liu, W.; Qi, G.; et al. Screening of Broad-Spectrum Aptamer and Development of Electrochemical Aptasensor for Simultaneous Detection of Penicillin Antibiotics in Milk. Talanta 2024, 269, 125508. [Google Scholar] [CrossRef]
- Sun, Z.; Han, L.; Yin, Y.; Mou, Y.; Tian, Y.; Zhang, W.; Chen, D.; Wu, Y.; Sun, X.; Guo, Y.; et al. Screening Broad-Spectrum Aptamers for Cephalosporin Antibiotics Using Real Samples and Development of a DNA Walker-Driven Dual-Mode Aptasensor. Biosens. Bioelectron. 2025, 280, 117446. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Y.; Jiao, X.; Li, T.; Lv, Z.; Yang, C.J.; Zhang, X.; Wen, Y. Control of Capillary Behavior through Target-Responsive Hydrogel Permeability Alteration for Sensitive Visual Quantitative Detection. Nat. Commun. 2019, 10, 1036. [Google Scholar] [CrossRef]
- Liu, H.; Cao, T.; Xu, Y.; Dong, Y.; Liu, D. Tuning the Mechanical Properties of a DNA Hydrogel in Three Phases Based on ATP Aptamer. Int. J. Mol. Sci. 2018, 19, 1633. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, Y.; Lu, C. Self-Assembled ATP-Responsive DNA Nanohydrogel for Specifically Activated Fluorescence Imaging and Chemotherapy in Cancer Cells. Anal. Chem. 2022, 94, 10221–10226. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.C.; Goh, S.S.; Loh, X.J. Bottom-up Engineering of Responsive Hydrogel Materials for Molecular Detection and Biosensing. ACS Mater. Lett. 2020, 2, 918–950. [Google Scholar] [CrossRef]
- Yu, J.; Xiao, S.; Yu, Z.; Hui, Y.; Li, T.; Wu, D.; Bi, W.; Gan, N.; Jia, Z. On-Site and Dual-Mode Detection of Live Vibrio Parahaemolyticus in Waters: A Universal Pathogen Sensing Platform Based on a Smart Hydrogel Aptasensor Imbedded with Gold Nanoclusters. Sens. Actuators B Chem. 2022, 366, 131947. [Google Scholar] [CrossRef]
- Nam, J.; Jang, W.S.; Kim, J.; Lee, H.; Lim, C.S. Lamb Wave-Based Molecular Diagnosis Using DNA Hydrogel Formation by Rolling Circle Amplification (RCA) Process. Biosens. Bioelectron. 2019, 142, 111496. [Google Scholar] [CrossRef] [PubMed]
- Na, W.; Nam, D.; Lee, H.; Shin, S. Rapid Molecular Diagnosis of Infectious Viruses in Microfluidics Using DNA Hydrogel Formation. Biosens. Bioelectron. 2018, 108, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Abbas, N.; Shin, S. A Rapid Diagnosis of SARS-CoV-2 Using DNA Hydrogel Formation on Microfluidic Pores. Biosens. Bioelectron. 2021, 177, 113005. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Kang, Q.; Cui, S.; Cao, J.; Lin, T.; Ma, C.; Xiao, Z.; Du, T.; Wang, N.; Du, X.; et al. Target-Driven Functionalized DNA Hydrogel Capillary Sensor for SARS-CoV-2 Dual-Mode Detection. Talanta 2025, 285, 127342. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, G.; Tan, Q.; Gao, M.; Chen, G.; Huang, X.; Xu, X.; Li, L.; Wang, J.; Zhang, Y.; et al. Polysaccharide-Based Biopolymer Hydrogels for Heavy Metal Detection and Adsorption. J. Adv. Res. 2023, 44, 53–70. [Google Scholar] [CrossRef]
- Malik, L.A.; Bashir, A.; Qureashi, A.; Pandith, A.H. Detection and Removal of Heavy Metal Ions: A Review. Environ. Chem. Lett. 2019, 17, 1495–1521. [Google Scholar] [CrossRef]
- Lambropoulou, D.A.; Albanis, T.A. Methods of Sample Preparation for Determination of Pesticide Residues in Food Matrices by Chromatography–Mass Spectrometry-Based Techniques: A Review. Anal. Bioanal. Chem. 2007, 389, 1663–1683. [Google Scholar] [CrossRef]
- Zhou, T.; Xiao, X.; Li, G. Microwave Accelerated Selective Soxhlet Extraction for the Determination of Organophosphorus and Carbamate Pesticides in Ginseng with Gas Chromatography/Mass Spectrometry. Anal. Chem. 2012, 84, 5816–5822. [Google Scholar] [CrossRef]
- Yang, N.; You, D.; Wang, J.; Ge, L. Progress in DNA-Based Hydrogels for Biosensing. Mater. Technol. 2022, 37, 798–813. [Google Scholar] [CrossRef]
- Ono, A.; Togashi, H. Highly Selective Oligonucleotide-Based Sensor for Mercury(II) in Aqueous Solutions. Angew. Chem. Int. Ed. 2004, 43, 4300–4302. [Google Scholar] [CrossRef] [PubMed]
- Dave, N.; Chan, M.Y.; Huang, P.-J.J.; Smith, B.D.; Liu, J. Regenerable DNA-Functionalized Hydrogels for Ultrasensitive, Instrument-Free Mercury(II) Detection and Removal in Water. J. Am. Chem. Soc. 2010, 132, 12668–12673. [Google Scholar] [CrossRef] [PubMed]
- Pi, K.; Liu, J.; Van Cappellen, P. Direct Measurement of Aqueous Mercury(II): Combining DNA-Based Sensing with Diffusive Gradients in Thin Films. Environ. Sci. Technol. 2020, 54, 13680–13689. [Google Scholar] [CrossRef]
- Chu, J.; Chen, C.; Li, X.; Yu, L.; Li, W.; Cheng, M.; Tang, W.; Xiong, Z. A Responsive Pure DNA Hydrogel for Label-Free Detection of Lead Ion. Anal. Chim. Acta 2021, 1157, 338400. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhou, X.; Liu, W.; Liu, Y.; Wang, X. Flexible DNA Hydrogel SERS Active Biofilms for Conformal Ultrasensitive Detection of Uranyl Ions from Aquatic Products. Langmuir 2020, 36, 2930–2936. [Google Scholar] [CrossRef]
- Liu, C.; Gou, S.; Bi, Y.; Gao, Q.; Sun, J.; Hu, S.; Guo, W. Smart DNA-Gold Nanoparticle Hybrid Hydrogel Film Based Portable, Cost-Effective and Storable Biosensing System for the Colorimetric Detection of Lead (II) and Uranyl Ions. Biosens. Bioelectron. 2022, 210, 114290. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, R.; Wang, H.; Wang, C.; Zhang, X.; Yang, Y.; Pang, W.; Ren, S.; Yang, J.; Yang, C.; et al. A Colorimetric/Electrochemical Microfluidic Biosensor Using Target-Triggered DNA Hydrogels for Organophosphorus Detection. Biosens. Bioelectron. 2024, 263, 116558. [Google Scholar] [CrossRef]
- Zandieh, M.; Luo, X.; Zhao, Y.; Feng, C.; Liu, J. Selection of Plastic-Binding DNA Aptamers for Microplastics Detection. Angew. Chem. Int. Ed. 2025, 64, e202421438. [Google Scholar] [CrossRef]
- Ren, K.; Ding, S.; Shi, J.; Dong, J.; Du, F.; Tang, Z. Detection of Lead Contamination Using DNAzyme and Split Activator-Triggered CRISPR/Cas12a. Talanta 2025, 295, 128385. [Google Scholar] [CrossRef]
- He, W.; Wu, Z.; Liu, X.; Xu, H.; Jian, X.; Zhou, K.; Du, J.; Wang, J.; Zhang, D. MOF-Based Fluorescent Hydrogels for Rapid and Highly Selective Visual Detection of Heavy Metal Ions. Microchem. J. 2025, 217, 114962. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, X.; Tian, T.; Zhu, Z.; Lin, H.; Yang, C. Target-Responsive DNAzyme Hydrogel for Portable Colorimetric Detection of Lanthanide(III) Ions. Sci. China Chem. 2017, 60, 293–298. [Google Scholar] [CrossRef]
- Li, L.; Zhang, M.; Chen, W. Gold Nanoparticle-Based Colorimetric and Electrochemical Sensors for the Detection of Illegal Food Additives. J. Food Drug Anal. 2020, 28, 642–654. [Google Scholar] [CrossRef]
- Lu, Y.; Xia, Y.; Liu, G.; Pan, M.; Li, M.; Lee, N.A.; Wang, S. A Review of Methods for Detecting Melamine in Food Samples. Crit. Rev. Anal. Chem. 2017, 47, 51–66. [Google Scholar] [CrossRef]
- Chen, P.; Han, W.; Li, Y.; Gao, G.; Yang, H. Distance-Readout Paper-Based Microfluidic Chip with a DNA Hydrogel Valve for AFB1 Detection. Anal. Chem. 2025, 97, 5975–5981. [Google Scholar] [CrossRef]
- Lin, X.; Yan, H.; Zhao, L.; Duan, N.; Wang, Z.; Wu, S. Hydrogel-Integrated Sensors for Food Safety and Quality Monitoring: Fabrication Strategies and Emerging Applications. Crit. Rev. Food Sci. Nutr. 2024, 64, 6395–6414. [Google Scholar] [CrossRef]
- Kahn, J.S.; Hu, Y.; Willner, I. Stimuli-Responsive DNA-Based Hydrogels: From Basic Principles to Applications. Acc. Chem. Res. 2017, 50, 680–690. [Google Scholar] [CrossRef]
- Charusalaipong, P.; Gordon, M.-J.; Cantlay, L.; De Souza, N.; Horgan, G.W.; Bates, R.; Gratz, S.W. Frequent Dietary Multi-Mycotoxin Exposure in UK Children and Its Association with Dietary Intake. Toxins 2024, 16, 251. [Google Scholar] [CrossRef]
- Lin, J.; Deng, Y.; Lin, Y.; Luo, F.; Weng, Z.; Wang, J.; Lin, C.; Qiu, B.; Lin, Z. Advanced Dual-Signal Point-of-Care Testing Platform for Sensitive T-2 Toxin Detection: Integrating Copper-Based Conductive MOF with Target-Responsive DNA Hydrogel. Sens. Actuators B Chem. 2025, 438, 137802. [Google Scholar] [CrossRef]
- Sun, Y.; Qi, S.; Dong, X.; Qin, M.; Zhang, Y.; Wang, Z. Colorimetric Aptasensor Targeting Zearalenone Developed Based on the Hyaluronic Acid-DNA Hydrogel and Bimetallic MOFzyme. Biosens. Bioelectron. 2022, 212, 114366. [Google Scholar] [CrossRef]
- Sun, Y.; Qi, S.; Dong, X.; Qin, M.; Ding, N.; Zhang, Y.; Wang, Z. Colorimetric Aptasensor for Fumonisin B1 Detection Based on the DNA Tetrahedra-Functionalized Magnetic Beads and DNA Hydrogel-Coated Bimetallic MOFzyme. J. Hazard. Mater. 2023, 443, 130252. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Wang, W.; Shen, X.; Wang, S.; Li, Q.; An, F.; Wu, S. A Fluorescent DNA Hydrogel Aptasensor Based on the Self-Assembly of Rolling Circle Amplification Products for Sensitive Detection of Ochratoxin a. J. Agric. Food Chem. 2020, 68, 369–375. [Google Scholar] [CrossRef]
- Aran, G.C.; Bayraç, C. Simultaneous Dual-Sensing Platform Based on Aptamer-Functionalized DNA Hydrogels for Visual and Fluorescence Detection of Chloramphenicol and Aflatoxin M1. Bioconjugate Chem. 2023, 34, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Li, S.; Ye, X.; Ning, B.; Bai, J.; Peng, Y.; Li, L.; Han, T.; Zhou, H.; Gao, Z.; et al. Cu/Au/Pt Trimetallic Nanoparticles Coated with DNA Hydrogel as Target-Responsive and Signal-Amplification Material for Sensitive Detection of Microcystin-LR. Anal. Chim. Acta 2020, 1134, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Mann, H.; Khan, S.; Prasad, A.; Bayat, F.; Gu, J.; Jackson, K.; Li, Y.; Hosseinidoust, Z.; Didar, T.F.; Filipe, C.D.M. Bacteriophage-Activated DNAzyme Hydrogels Combined with Machine Learning Enable Point-of-Use Colorimetric Detection of Escherichia Coli. Adv. Mater. 2025, 37, 2411173. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, R.; Hou, Y.; Qin, Y.; Li, S.; Yang, S.; Gao, Z. DNA Hydrogels Combined with Microfluidic Chips for Melamine Detection. Anal. Chim. Acta 2022, 1228, 340312. [Google Scholar] [CrossRef]
- Bian, Y.; Zhou, Z.; Li, G.; Liu, S.; Li, S.; Gao, Z.; Kang, W. Bimetallic Nanozymes Laden DNA Hydrogel for Ultrasensitive Optical Detection of Ractopamine. Sens. Actuators B Chem. 2023, 380, 133402. [Google Scholar] [CrossRef]
- Chen, Y.; Qian, S.; Yu, X.; Wu, J.; Xu, J. Microfluidics: The Propellant of CRISPR-Based Nucleic Acid Detection. Trends Biotechnol. 2023, 41, 557–574. [Google Scholar] [CrossRef]
- Li, H.; Wu, Y.; Nunekpeku, X.; Yang, Y.; Zhao, J.; Sheng, W.; Wang, Y. On-Site AFB1 Detection in Food via Dual-Equipped NH2-ZIF-8@Cu2+ and Core-Shell UCNPs Embedded in SA-PAM Hydrogel. Sens. Actuators B Chem. 2025, 445, 138594. [Google Scholar] [CrossRef]
- Hu, Y.; Zhou, X.; Wang, L.; Gu, J.; Zuo, Y.; Zhao, L.; Lu, W.; Yu, Y. A Liposome-Based Aptasensor Integrated with Competitive Reaction Enabling Portable and Electrochemical Detection of Aβ Oligomer. Biosens. Bioelectron. 2023, 225, 115108. [Google Scholar] [CrossRef]
- Cheng, C.; Ganguly, S.; Li, P.; Tang, X. Detecting Hypoxia through the Non-Invasive and Simultaneous Monitoring of Sweat Lactate and Tissue Oxygenation. Biosensors 2024, 14, 584. [Google Scholar] [CrossRef]
- Lee, M.; Lee, M.; Kim, S.; Park, N. Stimuli-Responsive DNA Hydrogel Design Strategies for Biomedical Applications. Biosensors 2025, 15, 355. [Google Scholar] [CrossRef]
- Yamamoto, K.; Imamura, H.; Matsuyama, Y.; Kume, Y.; Ikeda, H.; Norman, G.L.; Shums, Z.; Aoki, T.; Hasegawa, K.; Beck, Y.; et al. AFP, AFP-L3, DCP, and GP73 as Markers for Monitoring Treatment Response and Recurrence and as Surrogate Markers of Clinicopathological Variables of HCC. J. Gastroenterol. 2010, 45, 1272–1282. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, Y.; Jiang, N.; Wang, J.; Yu, M.; Zhuang, X. Preparation of Aptamer Responsive DNA Functionalized Hydrogels for the Sensitive Detection of α-Fetoprotein Using SERS Method. Bioconjugate Chem. 2020, 31, 813–820. [Google Scholar] [CrossRef]
- Borum, R.M.; Moore, C.; Mantri, Y.; Xu, M.; Jokerst, J.V. Supramolecular Loading of DNA Hydrogels with Dye–Drug Conjugates for Real-Time Photoacoustic Monitoring of Chemotherapy. Adv. Sci. 2023, 10, 2204330. [Google Scholar] [CrossRef]
- Si, Y.; Xu, L.; Wang, N.; Zheng, J.; Yang, R.; Li, J. Target MicroRNA-Responsive DNA Hydrogel-Based Surface-Enhanced Raman Scattering Sensor Arrays for MicroRNA-Marked Cancer Screening. Anal. Chem. 2020, 92, 2649–2655. [Google Scholar] [CrossRef] [PubMed]
- Perala, R.S.; Chandrasekar, N.; Balaji, R.; Alexander, P.S.; Humaidi, N.Z.N.; Hwang, M.T. Corrigendum to “a Comprehensive Review on Graphene-Based Materials: From Synthesis to Contemporary Sensor Applications” [Mater. Sci. Eng. R 159 (2024) 100805]. Mater. Sci. Eng. R Rep. 2024, 160, 100816. [Google Scholar] [CrossRef]
- Xu, L.; Wang, R.; Kelso, L.C.; Ying, Y.; Li, Y. A Target-Responsive and Size-Dependent Hydrogel Aptasensor Embedded with QD Fluorescent Reporters for Rapid Detection of Avian Influenza Virus H5N1. Sens. Actuators B Chem. 2016, 234, 98–108. [Google Scholar] [CrossRef]
- Sun, X.; Ding, C.; Qin, M.; Li, J. Hydrogel-Based Biosensors for Bacterial Infections. Small 2024, 20, 2306960. [Google Scholar] [CrossRef]
- Li, X.; Lu, Y.; Hu, Y. A Wireless and Battery-Free DNA Hydrogel Biosensor for Wound Infection Monitoring. Matter 2022, 5, 2473–2475. [Google Scholar] [CrossRef]
- Li, S.; Dai, J.; Zhu, M.; Arroyo-Currás, N.; Li, H.; Wang, Y.; Wang, Q.; Lou, X.; Kippin, T.E.; Wang, S.; et al. Implantable Hydrogel-Protective DNA Aptamer-Based Sensor Supports Accurate, Continuous Electrochemical Analysis of Drugs at Multiple Sites in Living Rats. ACS Nano 2023, 17, 18525–18538. [Google Scholar] [CrossRef]
- Hivare, P.; Gangrade, A.; Swarup, G.; Bhavsar, K.; Singh, A.; Gupta, R.; Thareja, P.; Gupta, S.; Bhatia, D. Peptide Functionalized DNA Hydrogel Enhances Neuroblastoma Cell Growth and Differentiation. Nanoscale 2022, 14, 8611–8620. [Google Scholar] [CrossRef] [PubMed]
- Salimian, R.; Nardin, C. Conjugated Polymers for Aptasensing Applications. Biomacromolecules 2023, 24, 3411–3437. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Mao, Y.; An, Y.; Tian, T.; Zhang, H.; Yan, J.; Zhu, Z.; Yang, C.J. Target-Responsive DNA Hydrogel for Non-Enzymatic and Visual Detection of Glucose. Analyst 2018, 143, 1679–1684. [Google Scholar] [CrossRef] [PubMed]
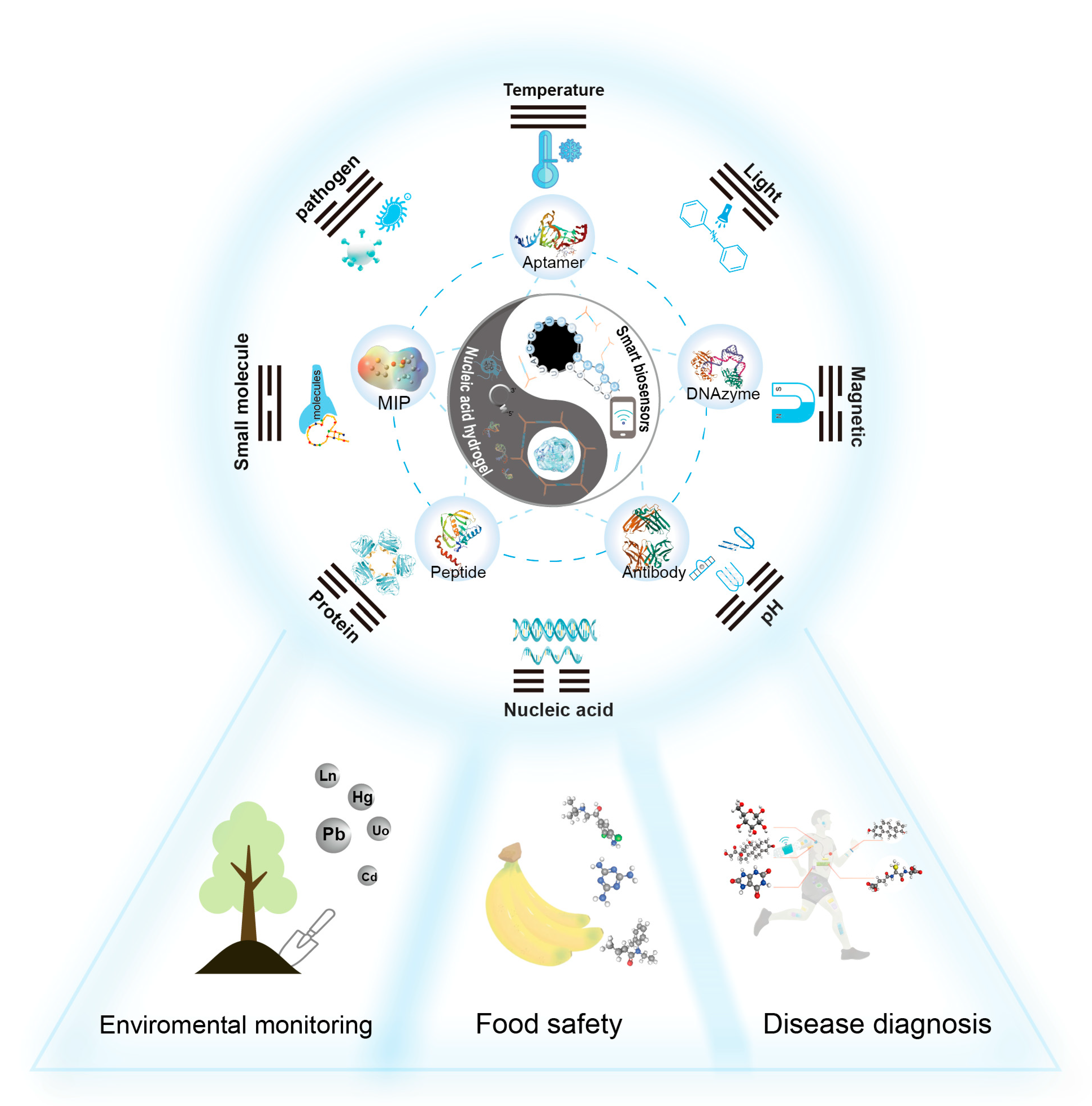
| Recognition Element | Principle | Characteristics |
|---|---|---|
| Aptamer | Single-stranded nucleic acids that fold into specific 3D conformations to bind targets via non-covalent interactions. | High affinity and selectivity (nM–pM) Chemical stability Facile modification Prone to nuclease degradation. |
| DNAzymes | Catalytic DNA sequences capable of mediating substrate cleavage or redox reactions, typically dependent on specific metal ions. | Catalytic and signal-amplifying Metal-ion selective Functions as a responsive crosslinker Activity modulated by ions and environment |
| Antibody | Protein receptors recognizing antigens through epitope-paratope complementarity. | Excellent affinity and biological relevance High cost Low stability under extreme pH/temperature |
| Peptides | Short amino acid chains interacting with targets via specific motifs or secondary structures. | Biocompatible Easily synthesized and modified Lower affinity and selectivity (μM range) |
| MIPs | Synthetic polymers with template-shaped cavities formed during polymerization. | Resistant to harsh conditions Scalable fabrication Limited by incomplete template removal and heterogeneous sites |
| Analyte | Recognition Element | Response Mechanism | Sensor Strategy | LOD | Reference |
|---|---|---|---|---|---|
| Pb2+ | DNAzyme | Cleavage of substrate strand induces hydrogel collapse, releasing nucleotide fragments. | Label-free pure DNA hydrogel biosensor | 7.7 nM | [154] |
| Pb2+ | DNAzyme | Enzyme-activated film degradation releases AuNPs for colorimetric readout. | DNAzyme–AuNP colorimetric sensor | 2.6 nM | [156] |
| Pb2+ | DNAzyme | Hydrogel mesh-size alteration modulates capillary flow velocity. | Capillary-based flow rate sensor | 10 nM | [38] |
| Hg2+ | T-rich DNA sequence | Specific T–Hg2+–T coordination complex formation, generating a fluorescent signal. | DNA-functionalized fluorescent hydrogel sensor | 0.05 nM | [153] |
| UO22+ | DNAzyme | Film disintegration via enzymatic cleavage releases AuNPs for colorimetric detection. | DNAzyme-mediated AuNP release-based colorimetric sensor | 10.3 nM | [156] |
| UO22+ | DNAzyme | Hydrogel breakdown releases Raman reporters, which are captured and enhanced by Ag-NPs@PAN membrane for SERS. | SERS-enhanced flexible hydrogel sensor | 0.838 pM | [155] |
| Ln3+ | DNAzyme | Hydrogel collapse releases AuNPs, resulting in a color change. | Colorimetric biosensor | 20 nM | [161] |
| Malathion | Aptamer | Competitive binding disrupts hydrogel network, releasing AuNPs (colorimetric) and reducing Fc-labeled aptamer (electrochemical). | Dual-mode colorimetric/electrochemical microfluidic chip sensor | 56 nM | [157] |
| Analyte | Recognition Element | Response Mechanism | Sensor Strategy | LOD | Reference |
|---|---|---|---|---|---|
| T-2 | Aptamer | Target-induced release of the nanozyme Cu3(HHTP)2 from a DNA hydrogel catalyzes TMB oxidation, enabling dual colorimetric and photothermal readout | Dual-modal colorimetric and photothermal sensor | 1.67 ng/mL | [168] |
| ZEN | Aptamer | ZEN-aptamer binding triggers hydrogel disintegration, exposing encapsulated MOFzyme that catalyzes substrate reaction | MOFzyme-based TMB colorimetric sensor | 0.8 pg/mL | [169] |
| FB1 | Aptamer | FB1 binding releases DNA strands to initiate a displacement reaction, leading to hydrogel dissolution and MOFzyme release | MOFzyme-based TMB colorimetric sensor | 0.38 pg/mL | [170] |
| AFB1 | Aptamer | AFB1 binding enhances electrostatic repulsion between probes, disrupting FRET effect and turning on CSUCNPs fluorescence | Turn-on fluorescent biosensor | 0.08 μg/kg | [178] |
| OTA | Aptamer | OTA binding releases primers to initiate RCA, generating long fluorescent DNA chains for signal amplification | Fluorescence signal-amplified biosensor | 0.01 ng/mL | [171] |
| MC-LR | Aptamer | Hydrogel dissociation releases encapsulated Cu/Au/Pt trimetallic nanoparticles (TNs) | Colorimetric sensor | 3.0 ng/L | [173] |
| E. coli | DNAzyme | DNAzyme cleavage degrades hydrogel network, releasing AuNPs for colorimetric response | DNAzyme-mediated AuNP release-based colorimetric sensor | 101 CFU mL−1 | [174] |
| RAC | Aptamer | Hydrogel collapse releases Au@Pd nanozymes that catalyze the H2O2-TMB reaction. | Ultrasensitive optical colorimetric sensor | 7.39 ng/L | [176] |
| MEL | Aptamer | Hydrogel dissociation releases entrapped AuNPs. | Colorimetric biosensor | 37 nM | [175] |
| Analyte | Recognition Element | Response Mechanism | Sensor Strategy | Reference |
|---|---|---|---|---|
| S. aureus DNase | DNA hydrogel | Enzymatic degradation of the hydrogel by DNase induces a change in dielectric properties | NFC module integrated with a smartphone for wireless readout | [189] |
| K+ | Aptamer | Target binding induces a concentration-dependent de-swelling of the aptamer-functionalized hydrogel | aptamer-hydrogel-based interferometric fiber sensor | [4] |
| MB-Dox | DNA-Crosslinked Hydrogel | Hydrogel degradation in response to endogenous nuclease activity releases MB-Dox, enhancing the photoacoustic signal | Enzyme-responsive disintegration of a DNA hydrogel matrix for turn-on photoacoustic imaging | [184] |
| Kanamycin | Aptamer | A hydrogel-coated aptamer sensor generates an electrochemical signal upon binding while providing antifouling capability | Electrochemical sensor based on a DNA aptamer with a protective hydrogel layer | [190] |
| AFB | Aptamer | Dissociation of the hydrogel network releases pre-embedded IgG, facilitating the formation of a sandwich complex with SERS probes and functionalized magnetic beads | Aptamer-responsive DNA hydrogel-based SERS biosensor | [183] |
| miRNA | MNAzymes | MNAzyme cleaves crosslinking substrates within the DNA hydrogel, leading to hydrogel dissolution and activation of the SERS signal | MNAzyme-responsive DNA hydrogel-based SERS biosensor | [185] |
| Virus | DNA template | Assembly of long DNA strands into a hydrogel network blocks the microfluidic pathways between beads | Microfluidic chip-integrated DNA hydrogel sensor | [143] |
| Glucose | Aptamer | Target binding triggers the release of pre-encapsulated gold nanoparticles (AuNPs) from the hydrogel | Colorimetric biosensor based on an aptamer-functionalized hydrogel | [193] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Zhu, L.; Wang, X.; Zhang, W.; He, X.; Zhang, Y.; Xu, W. Smart Nucleic Acid Hydrogel-Based Biosensors: From Molecular Recognition and Responsive Mechanisms to Applications. Biosensors 2025, 15, 799. https://doi.org/10.3390/bios15120799
Xu L, Zhu L, Wang X, Zhang W, He X, Zhang Y, Xu W. Smart Nucleic Acid Hydrogel-Based Biosensors: From Molecular Recognition and Responsive Mechanisms to Applications. Biosensors. 2025; 15(12):799. https://doi.org/10.3390/bios15120799
Chicago/Turabian StyleXu, Lu, Longjiao Zhu, Xiaoyu Wang, Wenqiang Zhang, Xiaoyun He, Yangzi Zhang, and Wentao Xu. 2025. "Smart Nucleic Acid Hydrogel-Based Biosensors: From Molecular Recognition and Responsive Mechanisms to Applications" Biosensors 15, no. 12: 799. https://doi.org/10.3390/bios15120799
APA StyleXu, L., Zhu, L., Wang, X., Zhang, W., He, X., Zhang, Y., & Xu, W. (2025). Smart Nucleic Acid Hydrogel-Based Biosensors: From Molecular Recognition and Responsive Mechanisms to Applications. Biosensors, 15(12), 799. https://doi.org/10.3390/bios15120799









