Recent Uses of Paper Microfluidics in Isothermal Nucleic Acid Amplification Tests
Abstract
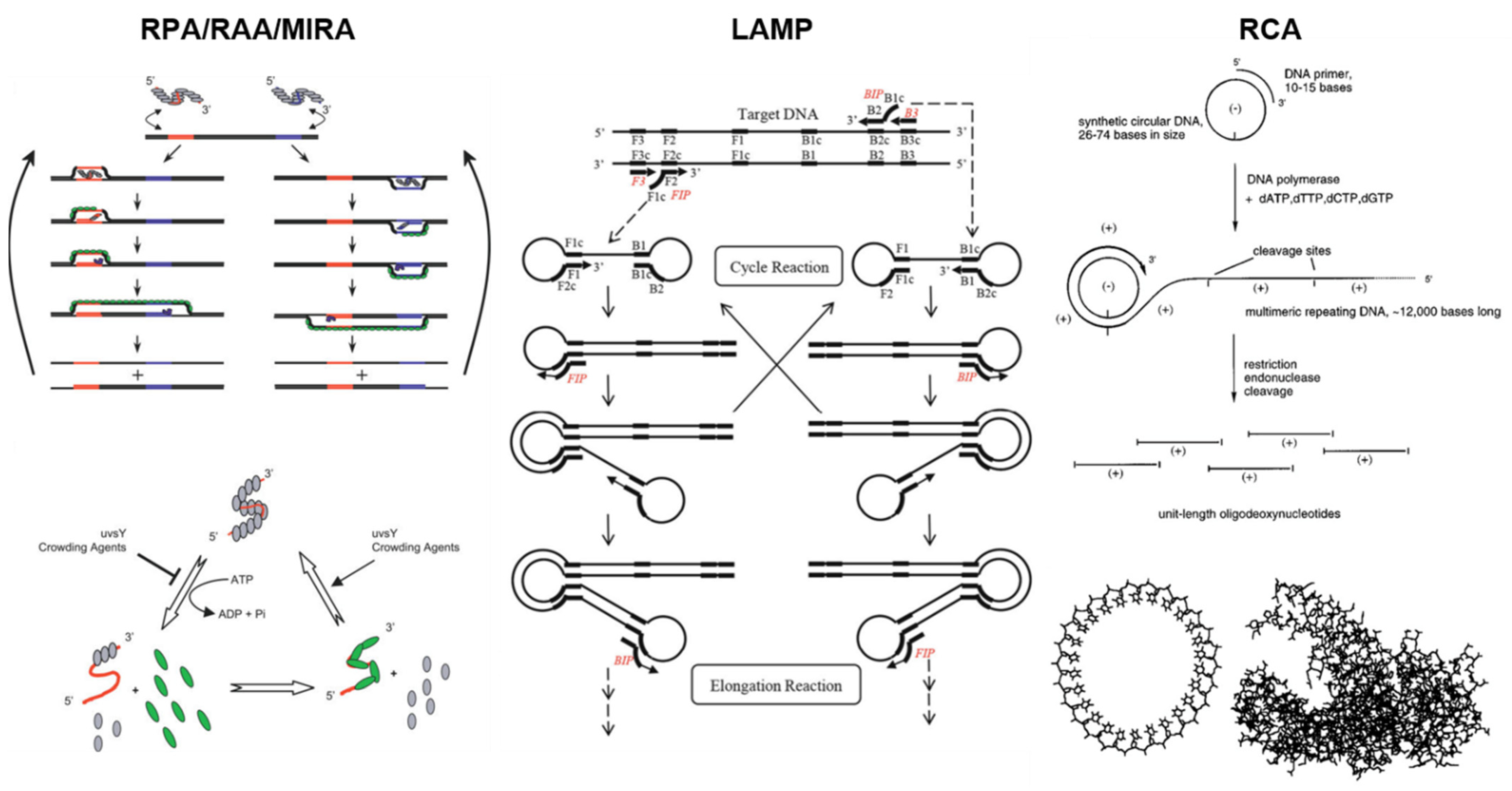
:1. Introduction
2. RPA with Paper Microfluidics
3. RAA and MIRA with Paper Microfluidics

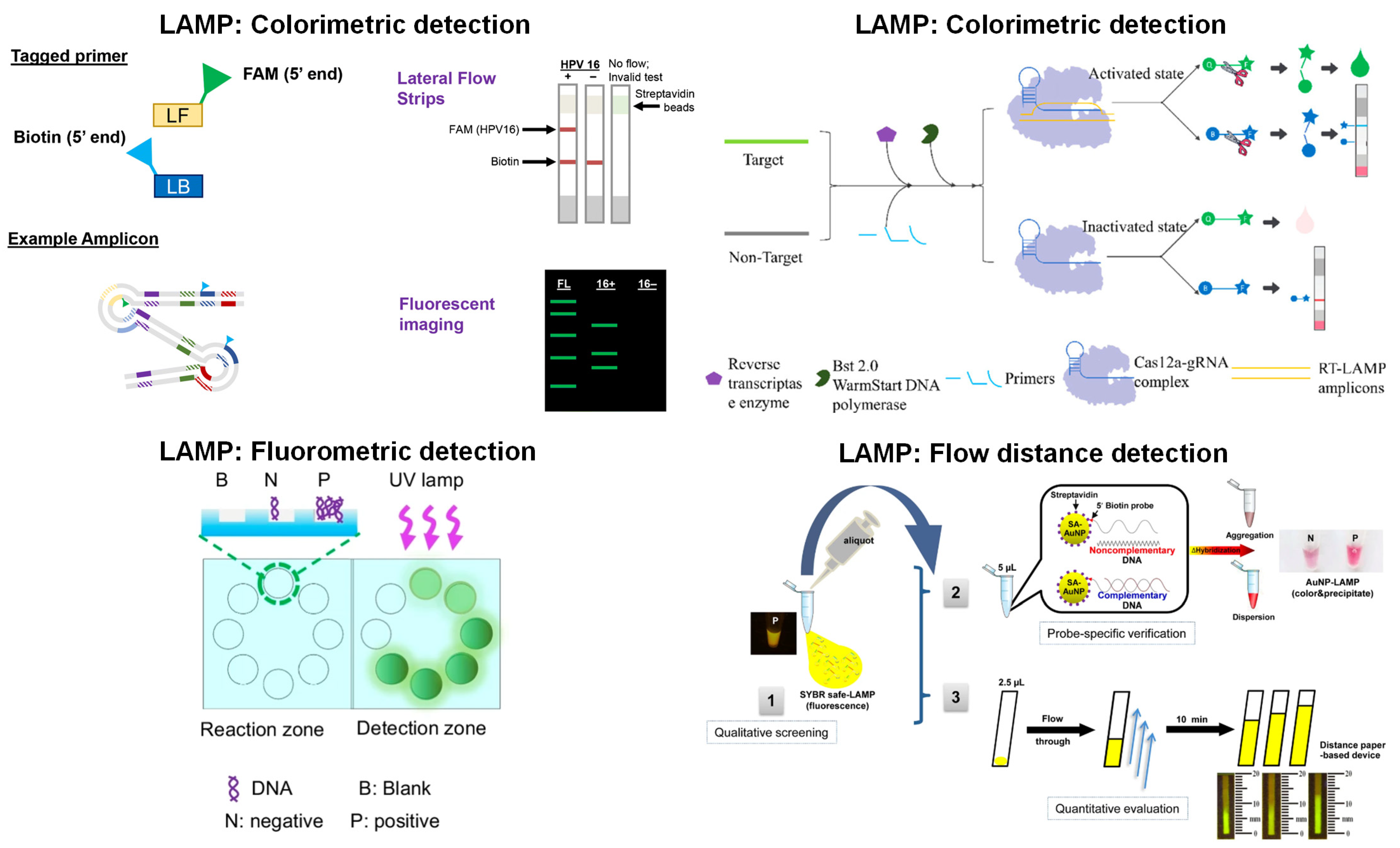
4. LAMP with Paper Microfluidics
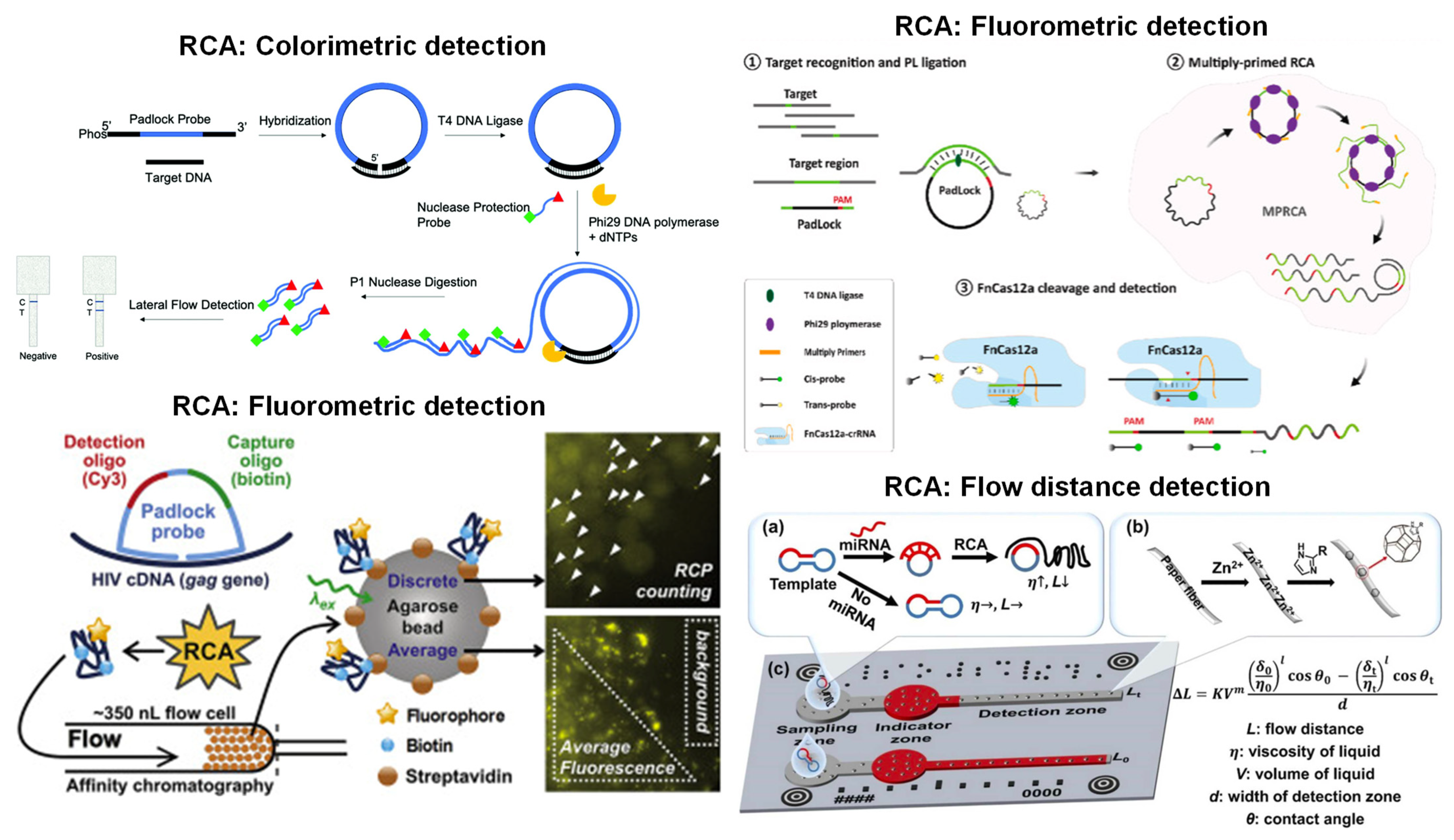
5. RCA with Paper Microfluidics
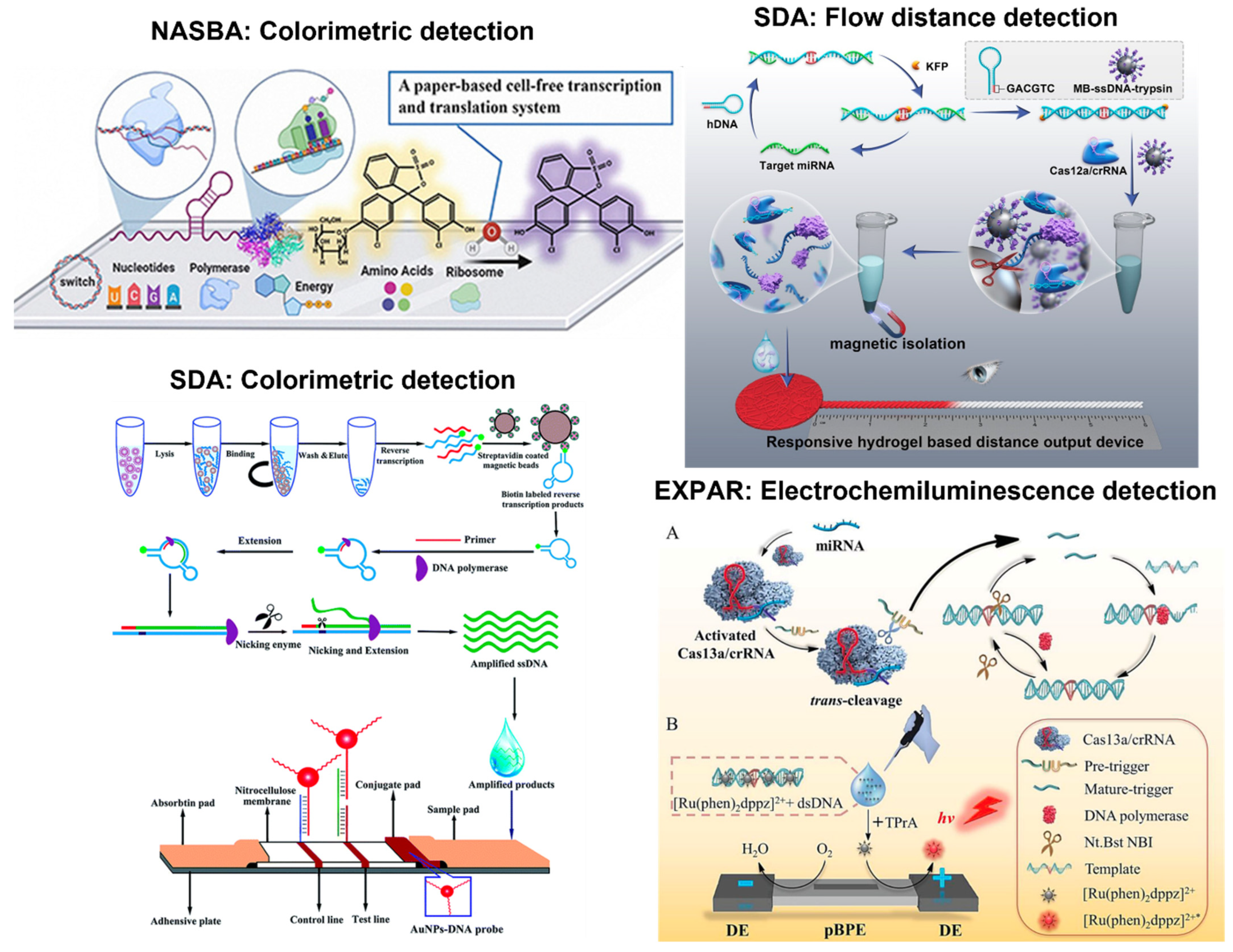
6. Other Isothermal Amplification Tests with Paper Microfluidics
7. Discussion
8. Future Directions and Fully Integrated Systems
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Geng, L.; Deng, D.D.; Wubben, M.J.; Jenkins, J.N.; McCarty, J.C., Jr.; Abdurakhmonov, I. A High-Throughput Standard PCR-Based Genotyping Method for Determining Transgene Zygosity in Segregating Plant Populations. Front. Plant Sci. 2017, 8, 1252. [Google Scholar] [CrossRef]
- Ahn, H.; Batule, B.S.; Seok, Y.; Kim, M.-G. Single-Step Recombinase Polymerase Amplification Assay Based on a Paper Chip for Simultaneous Detection of Multiple Foodborne Pathogens. Anal. Chem. 2018, 90, 10211–10216. [Google Scholar] [CrossRef]
- D’Agata, R.; Breveglieri, G.; Zanoli, L.M.; Borgatti, M.; Spoto, G.; Gambari, R. Direct Detection of Point Mutations in Nonamplified Human Genomic DNA. Anal. Chem. 2011, 83, 8711–8717. [Google Scholar] [CrossRef]
- Mullis, K.B.; Faloona, F.A. [21] Specific Synthesis of DNA in Vitro via a Polymerase-Catalyzed Chain Reaction. Meth. Enzymol. 1987, 155, 335–350. [Google Scholar] [CrossRef]
- Louie, M.; Louie, L.; Simor, A.E. The Role of DNA Amplification Technology in the Diagnosis of Infectious Diseases. Can. Med. Assoc. J. 2000, 163, 301–309. [Google Scholar] [CrossRef]
- Varlamov, D.A.; Blagodatskikh, K.A.; Smirnova, E.V.; Kramarov, V.M.; Ignatov, K.B. Combinations of PCR and Isothermal Amplification Techniques Are Suitable for Fast and Sensitive Detection of SARS-CoV-2 Viral RNA. Front. Bioeng. Biotechnol. 2020, 8, 604793. [Google Scholar] [CrossRef]
- Katzman, B.M.; Wockenfus, A.M.; Kelley, B.R.; Karon, B.S.; Donato, L.J. Evaluation of the Visby Medical COVID-19 Point of Care Nucleic Acid Amplification Test. Clin. Biochem. 2023, 117, 1–3. [Google Scholar] [CrossRef]
- Liu, H.Y.; Hopping, G.C.; Vaidyanathan, U.; Ronquillo, Y.C.; Hoopes, P.C.; Moshirfar, M. Polymerase Chain Reaction and Its Application in the Diagnosis of Infectious Keratitis. Med Hypothesis Discov. Innov. Ophthalmol. 2019, 8, 152–155. [Google Scholar]
- Chen, Y.; Qian, C.; Liu, C.; Shen, H.; Wang, Z.; Ping, J.; Wu, J.; Chen, H. Nucleic Acid Amplification Free Biosensors for Pathogen Detection. Biosens. Bioelectron. 2020, 153, 112049. [Google Scholar] [CrossRef]
- Player, R.; Verratti, K.; Staab, A.; Bradburne, C.; Grady, S.; Goodwin, B.; Sozhamannan, S. Comparison of the Performance of an Amplicon Sequencing Assay Based on Oxford Nanopore Technology to Real-Time PCR Assays for Detecting Bacterial Biodefense Pathogens. BMC Genom. 2020, 21, 166. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J. Advances in Isothermal Amplification: Novel Strategies Inspired by Biological Processes. Biosens. Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Asiello, P.J.; Baeumner, A.J. Miniaturized Isothermal Nucleic Acid Amplification, a Review. Lab Chip 2011, 11, 1420–1430. [Google Scholar] [CrossRef]
- Gillespie, T. The Capillary Rise of a Liquid in a Vertical Strip of Filter Paper. J. Colloid Sci. 1959, 14, 123–130. [Google Scholar] [CrossRef]
- Li, X.; Ballerini, D.R.; Shen, W. A Perspective on Paper-Based Microfluidics: Current Status and Future Trends. Biomicrofluidics 2012, 6, 11301–1130113. [Google Scholar] [CrossRef]
- Mason, M.G.; Botella, J.R. Rapid (30-Second), Equipment-Free Purification of Nucleic Acids Using Easy-to-Make Dipsticks. Nat. Protoc. 2020, 15, 3663–3677. [Google Scholar] [CrossRef]
- Seok, Y.; Joung, H.-A.; Byun, J.-Y.; Jeon, H.-S.; Shin, S.J.; Kim, S.; Shin, Y.-B.; Han, H.S.; Kim, M.-G. A Paper-Based Device for Performing Loop-Mediated Isothermal Amplification with Real-Time Simultaneous Detection of Multiple DNA Targets. Theranostics 2017, 7, 2220–2230. [Google Scholar] [CrossRef]
- Wang, A.G.; Dong, T.; Mansour, H.; Matamoros, G.; Sanchez, A.L.; Li, F. Paper-Based DNA Reader for Visualized Quantification of Soil-Transmitted Helminth Infections. ACS Sens. 2018, 3, 205–210. [Google Scholar] [CrossRef]
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M. Three-Dimensional Microfluidic Devices Fabricated in Layered Paper and Tape. Proc. Natl. Acad. Sci. USA 2008, 105, 19606–19611. [Google Scholar] [CrossRef]
- Jahanshahi-Anbuhi, S.; Henry, A.; Leung, V.; Sicard, C.; Pennings, K.; Pelton, R.; Brennan, J.D.; Filipe, C.D.M. Paper-Based Microfluidics with an Erodible Polymeric Bridge Giving Controlled Release and Timed Flow Shutoff. Lab Chip 2014, 14, 229–236. [Google Scholar] [CrossRef]
- Schonhorn, J.E.; Fernandes, S.C.; Rajaratnam, A.; Deraney, R.N.; Rolland, J.P.; Mace, C.R. A Device Architecture for Three-Dimensional, Patterned Paper Immunoassays. Lab Chip 2014, 14, 4653–4658. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Korvink, J.G.; Mager, D.; Land, K. Correction: The Potential of Paper-Based Diagnostics to Meet the ASSURED Criteria. RSC Adv. 2018, 8, 37841. [Google Scholar] [CrossRef] [PubMed]
- Leonard, P.; Hearty, S.; Brennan, J.; Dunne, L.; Quinn, J.; Chakraborty, T.; O’Kennedy, R. Advances in Biosensors for Detection of Pathogens in Food and Water. Enzyme Microb. Technol. 2003, 32, 3–13. [Google Scholar] [CrossRef]
- Sullivan, B.P. Paper-Based Nucleic Acid Amplification Testing at the Point-of-Care for HIV Viral Load Monitoring. Ph.D. Thesis, University of Washington, Seattle, WA, USA, 2022. [Google Scholar]
- Jia, Y.; Sun, H.; Tian, J.; Song, Q.; Zhang, W. Paper-Based Point-of-Care Testing of SARS-CoV-2. Front. Bioeng. Biotechnol. 2021, 9, 773304. [Google Scholar] [CrossRef]
- Magro, L.; Escadafal, C.; Garneret, P.; Jacquelin, B.; Kwasiborski, A.; Manuguerra, J.; Monti, B.; Sakuntabhai, A.; Vanhomwegen, J.; Lafayee, P.; et al. Paper Microfluidics for Nucleic Acid Amplification Testing (NAAT) of Infectious Diseases. Lab Chip 2017, 17, 2347–2371. [Google Scholar] [CrossRef]
- Kaur, N.; Toley, B.J. Paper-Based Nucleic Acid Amplification Tests for Point-of-Care Diagnostics. Analyst 2018, 143, 2213–2234. [Google Scholar] [CrossRef]
- Martinez, A.W.; Phillips, S.T.; Wiley, B.J.; Gupta, M.; Whitesides, G.M. FLASH: A Rapid Method for Prototyping Paper-Based Microfluidic Devices. Lab Chip 2008, 8, 2146. [Google Scholar] [CrossRef]
- Asghar, W.; Yuksekkaya, M.; Shafiee, H.; Zhang, M.; Ozen, M.O.; Inci, F.; Kocakulak, M.; Demirci, U. Engineering Long Shelf Life Multi-Layer Biologically Active Surfaces on Microfluidic Devices for Point of Care Applications. Sci. Rep. 2016, 6, 21163. [Google Scholar] [CrossRef]
- Pelton, R. Bioactive Paper Provides a Low-Cost Platform for Diagnostics. TrAC–Trends Anal. Chem. 2009, 28, 925–942. [Google Scholar] [CrossRef]
- Das, D.; Masetty, M.; Priye, A. Paper-Based Loop Mediated Isothermal Amplification (LAMP) Platforms: Integrating the Versatility of Paper Microfluidics with Accuracy of Nucleic Acid Amplification Tests. Chemosensors 2023, 11, 163. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-Mediated Isothermal Amplification (LAMP): Principle, Features, and Future Prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef]
- Liu, D.; Daubendiek, S.L.; Zillman, M.A.; Ryan, K.; Kool, E.T. Rolling Circle DNA Synthesis: Small Circular Oligonucleotides as Efficient Templates for DNA Polymerases. J. Am. Chem. Soc. 1996, 118, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Zhao, Y.; Shu, X.; Li, D.; Ao, Y.; Li, M.; Wang, S.; Cui, J.; An, X.; Zhan, X.; et al. Recombinase Polymerase Amplification with Lateral Flow Strip for Detecting Babesia Microti Infections. Parasitol. Int. 2021, 83, 102351. [Google Scholar] [CrossRef]
- Petrucci, S.; Costa, C.; Broyles, D.; Kaur, A.; Dikici, E.; Daunert, S.; Deo, S.K. Monitoring Pathogenic Viable E. coli O157:H7 in Food Matrices Based on the Detection of RNA Using Isothermal Amplification and a Paper-Based Platform. Anal. Chem. 2022, 94, 2485–2492. [Google Scholar] [CrossRef]
- Chondrogiannis, G.; Réu, P.; Hamedi, M.M. Paper-based Bacterial Lysis Enables Sample-to-answer Home-based DNA Testing. Adv. Mater. Technol. 2023, 8, 2201004. [Google Scholar] [CrossRef]
- Song, Y.; Huang, P.; Yu, M.; Li, Y.; Jin, H.; Qiu, J.; Li, Y.; Gao, Y.; Zhang, H.; Wang, H. Rapid and Visual Detection of SARS-CoV-2 RNA Based on Reverse Transcription-Recombinase Polymerase Amplification with Closed Vertical Flow Visualization Strip Assay. Microbiol. Spectr. 2023, 11, e0296622. [Google Scholar] [CrossRef]
- Zhang, S.; Xie, H.; Liu, M.; Zheng, A.; Yan, H.; Duan, M.; Wei, X.; Hu, J.; Zhang, H.; Xia, X. Rapid Visual Detection of Streptococcus suis and Actinobacillus pleuropneumoniae through Duplex Recombinase Polymerase Amplification Combined with Lateral Flow Dipsticks. Kafkas Univ. Vet. Fak. Derg. 2022, 28, 601–611. [Google Scholar] [CrossRef]
- Farrera-Soler, L.; Gonse, A.; Kim, K.T.; Barluenga, S.; Winssinger, N. Combining Recombinase Polymerase Amplification and DNA-Templated Reaction for SARS-CoV-2 Sensing with Dual Fluorescence and Lateral Flow Assay Output. Biopolymers 2022, 113, e23485. [Google Scholar] [CrossRef]
- Ghosh, D.K.; Kokane, S.B.; Gowda, S. Development of a Reverse Transcription Recombinase Polymerase Based Isothermal Amplification Coupled with Lateral Flow Immunochromatographic Assay (CTV-RT-RPA-LFICA) for Rapid Detection of Citrus Tristeza Virus. Sci. Rep. 2020, 10, 20593. [Google Scholar] [CrossRef]
- Gao, X.; Liu, X.; Zhang, Y.; Wei, Y.; Wang, Y. Rapid and Visual Detection of Porcine Deltacoronavirus by Recombinase Polymerase Amplification Combined with a Lateral Flow Dipstick. BMC Vet. Res. 2020, 16, 130. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Y.; Chen, X.; Peng, C.; Wang, L.; Xu, X.; Wu, J.; Wei, W.; Xu, J. A Highly Integrated System with Rapid DNA Extraction, Recombinase Polymerase Amplification, and Lateral Flow Biosensor for on-Site Detection of Genetically Modified Crops. Anal. Chim. Acta 2020, 1109, 158–168. [Google Scholar] [CrossRef]
- Du, Y.; Liu, D.; Wang, M.; Guo, F.; Lin, J.S. Preparation of DNA Aptamer and Development of Lateral Flow Aptasensor Combining Recombinase Polymerase Amplification for Detection of Erythromycin. Biosens. Bioelectron. 2021, 181, 113157. [Google Scholar] [CrossRef]
- Zhang, Y.-M.; Zhang, Y.; Xie, K. Evaluation of CRISPR/Cas12a-Based DNA Detection for Fast Pathogen Diagnosis and GMO Test in Rice. Mol. Breed. 2020, 40, 11. [Google Scholar] [CrossRef]
- Lei, R.; Li, L.; Wu, P.; Fei, X.; Zhang, Y.; Wang, J.; Zhang, D.; Zhang, Q.; Yang, N.; Wang, X. RPA/CRISPR/Cas12a-Based On-Site and Rapid Nucleic Acid Detection of in the Environment. ACS Synth. Biol. 2022, 11, 1772–1781. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Qin, M.; Coleman, J.J.; Shang, W.; Hu, X. Rapid and Sensitive Detection of from Complex Samples Using CRISPR/Cas12a Technology Combined with RPA. Plant Dis. 2023, 107, 1664–1669. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, S.; Chu, C.; Yang, M.; Huo, D.; Hou, C. Dual Methylation-Sensitive Restriction Endonucleases Coupling with an RPA-Assisted CRISPR/Cas13a System (DESCS) for Highly Sensitive Analysis of DNA Methylation and Its Application for Point-of-Care Detection. ACS Sens. 2021, 6, 2419–2428. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Zhao, Z.; Lian, K.; Yin, L.; Wang, J.; Man, S.; Liu, G.; Ma, L. SERS-Based CRISPR/Cas Assay on Microfluidic Paper Analytical Devices for Supersensitive Detection of Pathogenic Bacteria in Foods. Biosens. Bioelectron. 2022, 207, 114167. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, B.P.; Chou, Y.-S.; Bender, A.T.; Martin, C.D.; Kaputa, Z.G.; March, H.; Song, M.; Posner, J.D. Quantitative Isothermal Amplification on Paper Membranes Using Amplification Nucleation Site Analysis. Lab Chip 2022, 22, 2352–2363. [Google Scholar] [CrossRef]
- Mei, J.; Wang, D.; Zhang, Y.; Wu, D.; Cui, J.; Gan, M.; Liu, P. Portable Paper-Based Nucleic Acid Enrichment for Field Testing. Adv. Sci. 2023, 10, e2205217. [Google Scholar] [CrossRef]
- Ge, R.; Dai, H.; Zhang, S.; Wei, J.; Jiao, T.; Chen, Q.; Chen, Q.; Chen, X. A Collection of RPA-Based Photoelectrochemical Assays for the Portable Detection of Multiple Pathogens. Anal. Chem. 2023, 95, 7379–7386. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kaundal, P.; Tiwari, R.K.; Lal, M.K.; Kumari, H.; Kumar, R.; Naga, K.C.; Kumar, A.; Singh, B.; Sagar, V.; et al. Development of Reverse Transcription Recombinase Polymerase Amplification (RT-RPA): A Methodology for Quick Diagnosis of Potato Leafroll Viral Disease in Potato. Int. J. Mol. Sci. 2023, 24, 2511. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kaundal, P.; Tiwari, R.K.; Siddappa, S.; Kumari, H.; Lal, M.K.; Naga, K.C.; Sharma, S.; Sagar, V.; Kumar, M. Establishment of a One-Step Reverse Transcription Recombinase Polymerase Amplification Assay for the Detection of Potato Virus S. J. Virol. Methods 2022, 307, 114568. [Google Scholar] [CrossRef]
- Nie, Z.; Lü, P.; Zhang, R.; Tu, Y.; Liu, Z.; Li, Y.; Tang, C.; Li, X.; Zhao, K.; Zhou, Q.; et al. A Simple and Rapid Method for Fish Sex Identification Based on Recombinase-Aided Amplification and Its Use in Cynoglossus semilaevis. Sci. Rep. 2021, 11, 10429. [Google Scholar] [CrossRef]
- Zheng, Y.-Z.; Chen, J.-T.; Li, J.; Wu, X.-J.; Wen, J.-Z.; Liu, X.-Z.; Lin, L.-Y.; Liang, X.-Y.; Huang, H.-Y.; Zha, G.-C.; et al. Reverse Transcription Recombinase-Aided Amplification Assay with Lateral Flow Dipstick Assay for Rapid Detection of 2019 Novel Coronavirus. Front. Cell. Infect. Microbiol. 2021, 11, 613304. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Li, S.; Zhao, H.; Yan, C.; Feng, Y.; Cui, J.; Jiang, T.; Yuan, J. Use of a Rapid Recombinase-Aided Amplification Assay for Mycoplasma Pneumoniae Detection. BMC Infect. Dis. 2020, 20, 79. [Google Scholar] [CrossRef]
- Li, D.; Zhao, J.; Lan, W.; Zhao, Y.; Sun, X. Effect of Food Matrix on Rapid Detection of Vibrio parahaemolyticus in Aquatic Products Based on toxR Gene. World J. Microbiol. Biotechnol. 2023, 39, 188. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Li, H.; Huang, W.; Wang, L.; Zhao, Y.; Wang, J.; Shao, H.; Tao, X.; Yong, B. Development of a New PCR Assay and a Recombinase-Aided Amplification Based Isothermal Amplification Coupled with Lateral Flow Dipstick Assay for Potato Late Blight Detection. Crop Prot. 2023, 168, 106235. [Google Scholar] [CrossRef]
- Guo, Z.; Xing, G.; Li, P.; Jin, Q.; Lu, Q.; Zhang, G. Development and Application of a Recombinase-Aided Amplification and Lateral Flow Assay for Rapid Detection of Pseudorabies Virus from Clinical Crude Samples. Int. J. Biol. Macromol. 2023, 224, 646–652. [Google Scholar] [CrossRef]
- Xia, W.; Chen, K.; Liu, W.; Yin, Y.; Yao, Q.; Ban, Y.; Pu, Y.; Zhan, X.; Bian, H.; Yu, S.; et al. Rapid and Visual Detection of Mycoplasma synoviae by Recombinase-Aided Amplification Assay Combined with a Lateral Flow Dipstick. Poult. Sci. 2022, 101, 101860. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, C.; Liu, F.; Wang, Y.; Li, R.; Chen, G. Recombinase-Aided Amplification Combined with Lateral Flow Dipstick for the Rapid Detection of Amphidinium carterae. J. Appl. Phycol. 2021, 34, 435–447. [Google Scholar] [CrossRef]
- He, Y.; Chen, W.; Fan, J.; Fan, S.; Ding, H.; Chen, J.; Yi, L. Recombinase-Aided Amplification Coupled with Lateral Flow Dipstick for Efficient and Accurate Detection of Porcine Parvovirus. Life 2021, 11, 762. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, C.; Zhang, P.; Yao, S.; Liu, J.; Zhai, X.; Zhang, T. Reverse Transcription Recombinase-Aided Amplification Assay Combined with a Lateral Flow Dipstick for Detection of Avian Infectious Bronchitis Virus. Poult. Sci. 2020, 99, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Lei, R.; Kuang, R.; Peng, X.; Jiao, Z.; Zhao, Z.; Cong, H.; Fan, Z.; Zhang, Y. Portable Rapid Detection of Maize Chlorotic Mottle Virus Using RT-RAA/CRISPR-Cas12a Based Lateral Flow Assay. Front. Plant Sci. 2023, 14, 1088544. [Google Scholar] [CrossRef]
- Qian, J.; Huang, D.; Ni, D.; Zhao, J.; Shi, Z.; Fang, M.; Xu, Z. A Portable CRISPR Cas12a Based Lateral Flow Platform for Sensitive Detection of Staphylococcus aureus with Double Insurance. Food Control 2022, 132, 108485. [Google Scholar] [CrossRef]
- Huang, D.; Ni, D.; Fang, M.; Shi, Z.; Xu, Z. Microfluidic Ruler-Readout and CRISPR Cas12a-Responded Hydrogel-Integrated Paper-Based Analytical Devices (μReaCH-PAD) for Visible Quantitative Point-of-Care Testing of Invasive Fungi. Anal. Chem. 2021, 93, 16965–16973. [Google Scholar] [CrossRef]
- Chen, H.; Sun, C.; Wang, Y.; Gao, X.; You, J.; Yu, W.; Sun, N.; Yang, Y.; Li, X. Rapid Detection of SARS-CoV-2 Using Duplex Reverse Transcription-Multienzyme Isothermal Rapid Amplification in a Point-of-Care Testing. Front. Cell. Infect. Microbiol. 2021, 11, 678703. [Google Scholar] [CrossRef]
- Ji, C.; Feng, Y.; Sun, R.; Gu, Q.; Zhang, Y.; Ma, J.; Pan, Z.; Yao, H. Development of a Multienzyme Isothermal Rapid Amplification and Lateral Flow Dipstick Combination Assay for Bovine Coronavirus Detection. Front. Vet. Sci. 2023, 9, 1059934. [Google Scholar] [CrossRef]
- Zhu, L.; Gong, F.; Liu, X.; Sun, X.; Yu, Y.; Shu, J.; Pan, Z. Integrating Filter Paper Extraction, Isothermal Amplification, and Lateral Flow Dipstick Methods to Detect in Milk within 15 Min. Front. Vet. Sci. 2023, 10, 1100246. [Google Scholar] [CrossRef]
- Liu, Q.; Chai, Z.; Cao, Y.; Yang, K.; Song, X.; Shao, Y.; Wang, Z.; Tu, J.; Qi, K. Rapid Visual Detection of Fowl Adenovirus Serotype 4 with Multienzyme Isothermal Rapid Amplification Combined with Lateral Flow Dipstick. SSRN 2022. [Google Scholar] [CrossRef]
- Hu, W.-W.; He, J.-W.; Guo, S.-L.; Li, J. Development and Evaluation of a Rapid and Sensitive Multienzyme Isothermal Rapid Amplification with a Lateral Flow Dipstick Assay for Detection of in Spiked Blood Specimens. Front. Cell. Infect. Microbiol. 2022, 12, 1010201. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.-L.; Lai, H.-Y.; Chong, N.-Y.; Liu, D.-F.; Zhang, Z.-Y.; Pang, B.; Yao, J. Simple and Feasible Detection of Hepatitis B Virus Combination of Multienzyme Isothermal Rapid Amplification and Lateral Flow Dipstick Strip. Front. Mol. Biosci. 2021, 8, 763079. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Niu, J.; Sun, M.; Li, Z.; Wang, X.; He, Y.; Qi, J. Rapid and Sensitive Detection of in Based on Multienzyme Isothermal Rapid Amplification. Int. J. Mol. Sci. 2023, 24, 7733. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.-L.; Zhong, Y.; Li, X.-N.; Yao, J.; Pan, Y.-Q. Simple and Feasible Detection of Hepatitis a Virus Using Reverse Transcription Multienzyme Isothermal Rapid Amplification and Lateral Flow Dipsticks without Standard PCR Laboratory. Artif. Cells Nanomed. Biotechnol. 2023, 51, 233–240. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Soroka, M.; Wasowicz, B.; Rymaszewska, A. Loop-Mediated Isothermal Amplification (LAMP): The Better Sibling of PCR? Cells 2021, 10, 1931. [Google Scholar] [CrossRef]
- Shirato, K. Detecting Amplicons of Loop-mediated Isothermal Amplification. Microbiol. Immunol. 2019, 63, 407. [Google Scholar] [CrossRef]
- Jawla, J.; Kumar, R.R.; Mendiratta, S.K.; Agarwal, R.K.; Singh, P.; Saxena, V.; Kumari, S.; Boby, N.; Kumar, D.; Rana, P. On-Site Paper-Based Loop-Mediated Isothermal Amplification Coupled Lateral Flow Assay for Pig Tissue Identification Targeting Mitochondrial CO I Gene. J. Food Compost. Anal. 2021, 102, 104036. [Google Scholar] [CrossRef]
- Jawla, J.; Kumar, R.R.; Mendiratta, S.K.; Agarwal, R.K.; Kumari, S.; Saxena, V.; Kumar, D.; Singh, P.; Boby, N.; Rana, P. Paper-Based Loop-Mediated Isothermal Amplification and Lateral Flow (LAMP-LF) Assay for Identification of Tissues of Cattle Origin. Anal. Chim. Acta 2021, 1150, 338220. [Google Scholar] [CrossRef]
- Landaverde, L.; Wong, W.; Hernandez, G.; Fan, A.; Klapperich, C. Method for the Elucidation of LAMP Products Captured on Lateral Flow Strips in a Point of Care Test for HPV 16. Anal. Bioanal. Chem. 2020, 412, 6199–6209. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Li, S.; Zhu, X.; Wang, X.; Huang, J.; Yang, X.; Tai, J. LAMP-CRISPR-Cas12-Based Diagnostic Platform for Detection of Mycobacterium tuberculosis Complex Using Real-Time Fluorescence or Lateral Flow Test. Microchim. Acta 2021, 188, 347. [Google Scholar] [CrossRef]
- Yee, E.H.; Sikes, H.D. Polymerization-Based Amplification for Target-Specific Colorimetric Detection of Amplified DNA on Cellulose. ACS Sens. 2020, 5, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Ruang-Areerate, T.; Saengsawang, N.; Ruang-Areerate, P.; Ratnarathorn, N.; Thita, T.; Leelayoova, S.; Siripattanapipong, S.; Choowongkomon, K.; Dungchai, W. Distance-Based Paper Device Using Combined SYBR Safe and Gold Nanoparticle Probe LAMP Assay to Detect Leishmania among Patients with HIV. Sci. Rep. 2022, 12, 14558. [Google Scholar] [CrossRef]
- Liu, M.; Zhao, Y.; Monshat, H.; Tang, Z.; Wu, Z.; Zhang, Q.; Lu, M. An IoT-Enabled Paper Sensor Platform for Real-Time Analysis of Isothermal Nucleic Acid Amplification Tests. Biosens. Bioelectron. 2020, 169, 112651. [Google Scholar] [CrossRef] [PubMed]
- Saengsawang, N.; Ruang-Areerate, T.; Kesakomol, P.; Thita, T.; Mungthin, M.; Dungchai, W. Development of a Fluorescent Distance-Based Paper Device Using Loop-Mediated Isothermal Amplification to Detect in Urine. Analyst 2021, 145, 8077–8086. [Google Scholar] [CrossRef] [PubMed]
- Choopara, I.; Suea-Ngam, A.; Teethaisong, Y.; Howes, P.D.; Schmelcher, M.; Leelahavanichkul, A.; Thunyaharn, S.; Wongsawaeng, D.; deMello, A.J.; Dean, D.; et al. Fluorometric Paper-Based, Loop-Mediated Isothermal Amplification Devices for Quantitative Point-of-Care Detection of Methicillin-Resistant (MRSA). ACS Sens. 2021, 6, 742–751. [Google Scholar] [CrossRef]
- Tavakoli, H.; Hirth, E.; Luo, M.; Sharma Timilsina, S.; Dou, M.; Dominguez, D.C.; Li, X. A Microfluidic Fully Paper-Based Analytical Device Integrated with Loop-Mediated Isothermal Amplification and Nano-Biosensors for Rapid, Sensitive, and Specific Quantitative Detection of Infectious Diseases. Lab Chip 2022, 22, 4693–4704. [Google Scholar] [CrossRef]
- Zhou, Q.; Pan, J.; Mo, L.; Luo, Z.; Qin, Z.; Dai, Z.; Yi, C. Fluorescent On-Site Detection of Multiple Pathogens Using Smartphone-Based Portable Device with Paper-Based Isothermal Amplification Chip. Microchim. Acta 2022, 189, 333. [Google Scholar] [CrossRef]
- Sen, A.; Morris, C.; Priye, A.; Broom, M. A Paper Based Microfluidic Platform Combining LAMP-CRISPR/Cas12a for Fluorometric Detection of Nucleic Acids. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, J.; Shen, Z.; Liu, Y.; Song, Y.; Liang, Y.; Li, Z.; Nie, L.; Fang, Y.; Zhao, Y. A Newly Developed Paper Embedded Microchip Based on LAMP for Rapid Multiple Detections of Foodborne Pathogens. BMC Microbiol. 2021, 21, 197. [Google Scholar] [CrossRef]
- Qian, J.; Zhang, Q.; Lu, M. Integration of on-Chip Lysis and Paper-Based Sensor for Rapid Detection of Viral and Exosomal RNAs. Biosens. Bioelectron. 2023, 226, 115114. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, N.M.; Linnes, J.C.; Fan, A.; Ellenson, C.K.; Pollock, N.R.; Klapperich, C.M. Paper-Based RNA Extraction, in Situ Isothermal Amplification, and Lateral Flow Detection for Low-Cost, Rapid Diagnosis of Influenza A (H1N1) from Clinical Specimens. Anal. Chem. 2015, 87, 7872–7879. [Google Scholar] [CrossRef] [PubMed]
- Garneret, P.; Coz, E.; Martin, E.; Manuguerra, J.-C.; Brient-Litzler, E.; Enouf, V.; González Obando, D.F.; Olivo-Marin, J.-C.; Monti, F.; van der Werf, S.; et al. Performing Point-of-Care Molecular Testing for SARS-CoV-2 with RNA Extraction and Isothermal Amplification. PLoS ONE 2021, 16, e0243712. [Google Scholar] [CrossRef] [PubMed]
- Dinh, V.P.; Lee, N.Y. Fabrication of a Fully Integrated Paper Microdevice for Point-of-Care Testing of Infectious Disease Using Safranin O Dye Coupled with Loop-Mediated Isothermal Amplification. Biosens. Bioelectron. 2022, 204, 114080. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Mao, K.; Ran, F.; Xu, P.; Zhao, Y.; Zhang, X.; Zhou, H.; Yang, Z.; Zhang, H.; Jiang, G. Paper Device Combining CRISPR/Cas12a and Reverse-Transcription Loop-Mediated Isothermal Amplification for SARS-CoV-2 Detection in Wastewater. Environ. Sci. Technol. 2022, 56, 13245–13253. [Google Scholar] [CrossRef]
- Cardona-Ospina, J.A.; Villalba-Miranda, M.F.; Palechor-Ocampo, L.A.; Mancilla, L.I.; Sepúlveda-Arias, J.C. A Systematic Review of FTA Cards® as a Tool for Viral RNA Preservation in Fieldwork: Are They Safe and Effective? Prev. Vet. Med. 2019, 172, 104772. [Google Scholar] [CrossRef]
- Da Cunha Santos, G. FTA Cards for Preservation of Nucleic Acids for Molecular Assays: A Review on the Use of Cytologic/Tissue Samples. Arch. Pathol. Lab. Med. 2018, 142, 308–312. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.Q. Rolling Replication of Short DNA Circles. Proc. Natl. Acad. Sci. USA 1995, 92, 4641–4645. [Google Scholar] [CrossRef]
- Garafutdinov, R.R.; Sakhabutdinova, A.R.; Gilvanov, A.R.; Chemeris, A.V. Rolling Circle Amplification as a Universal Method for the Analysis of a Wide Range of Biological Targets. Russ. J. Bioorganic Chem. 2021, 47, 1172–1189. [Google Scholar] [CrossRef]
- Mohsen, M.G.; Kool, E.T. The Discovery of Rolling Circle Amplification and Rolling Circle Transcription. Acc. Chem. Res. 2016, 49, 2540–2550. [Google Scholar] [CrossRef]
- Monsur Ali, M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Chris Le, X.; Zhao, W. Rolling Circle Amplification: A Versatile Tool for Chemical Biology, Materials Science and Medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, H.; Cao, H.; Jiang, Y.; Mao, K.; Yang, Z. Rolling Circle Amplification as an Efficient Analytical Tool for Rapid Detection of Contaminants in Aqueous Environments. Biosensors 2021, 11, 352. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhang, C.; Liu, F.; Chen, Q.; Yang, Y.; Wang, Y.; Chen, G. Establishment of Double Probes Rolling Circle Amplification Combined with Lateral Flow Dipstick for Rapid Detection of Chattonella marina. Harmful Algae 2020, 97, 101857. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, C.; Yang, Y.; Yang, Y.; Wang, Y.; Chen, G. Detection of Prorocentrum minimum by Hyperbranched Rolling Circle Amplification Coupled with Lateral Flow Dipstick. Environ. Sci. Pollut. Res. 2020, 27, 44995–45007. [Google Scholar] [CrossRef]
- Jain, S.; Dandy, D.S.; Geiss, B.J.; Henry, C.S. Padlock Probe-Based Rolling Circle Amplification Lateral Flow Assay for Point-of-Need Nucleic Acid Detection. Analyst 2021, 146, 4340–4347. [Google Scholar] [CrossRef]
- Lee, H.N.; Lee, J.; Kang, Y.K.; Lee, J.H.; Yang, S.; Chung, H.J. A Lateral Flow Assay for Nucleic Acid Detection Based on Rolling Circle Amplification Using Capture Ligand-Modified Oligonucleotides. BioChip J. 2022, 16, 441–450. [Google Scholar] [CrossRef]
- Li, W.; Peng, W.; Zhang, Y.; Liu, P.; Gong, X.; Liu, H.; Chang, J. A Lateral Flow Strip Biosensor Platform Based on Cascade Nucleic Acid Amplification Technology for Ultrasensitive Detection of OSCC-Associated Salivary MicroRNA. Anal. Chim. Acta 2022, 1221, 340112. [Google Scholar] [CrossRef]
- Dong, N.; Wang, W.; Qin, Y.; Wang, Y.; Shan, H. Sensitive Lateral Flow Assay for Bisulfite-Free DNA Methylation Detection Based on the Restriction Endonuclease GlaI and Rolling Circle Amplification. Anal. Chim. Acta 2022, 1227, 340307. [Google Scholar] [CrossRef]
- He, F.; Lv, X.; Li, X.; Yao, M.; Li, K.; Deng, Y. Fluorescent Microspheres Lateral Flow Assay Integrated with Smartphone-Based Reader for Multiple microRNAs Detection. Microchem. J. 2022, 179, 107551. [Google Scholar] [CrossRef]
- Zhu, Z.; Guo, Y.; Wang, C.; Yang, Z.; Li, R.; Zeng, Z.; Li, H.; Zhang, D.; Yang, L. An Ultra-Sensitive One-Pot RNA-Templated DNA Ligation Rolling Circle Amplification-Assisted CRISPR/Cas12a Detector Assay for Rapid Detection of SARS-CoV-2. Biosens. Bioelectron. 2023, 228, 115179. [Google Scholar] [CrossRef]
- Kim, T.-Y.; Kim, S.; Jung, J.H.; Woo, M.-A. Paper-Based Radial Flow Assay Integrated to Portable Isothermal Amplification Chip Platform for Colorimetric Detection of Target DNA. BioChip J. 2023, 17, 263–273. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, J.; Yang, H.; Yu, Z.; Xu, Y.; Liu, S.; Dai, Z.; Zou, X. A Chromogenic Reaction-Free Distance-Based Paper Device for Facile Detection of microRNA via Viscosity Amplification and Surface Hydrophobicity Modulation. Sens. Actuators B Chem. 2022, 359, 131570. [Google Scholar] [CrossRef]
- Soares, R.R.G.; Varela, J.C.; Neogi, U.; Ciftci, S.; Ashokkumar, M.; Pinto, I.F.; Nilsson, M.; Madaboosi, N.; Russom, A. Sub-Attomole Detection of HIV-1 Using Padlock Probes and Rolling Circle Amplification Combined with Microfluidic Affinity Chromatography. Biosens. Bioelectron. 2020, 166, 112442. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, R.; Liang, F.; Deng, C.; Seidi, F.; Xiao, H. Fluorescent Paper-Based Analytical Devices for Ultra-Sensitive Dual-Type RNA Detections and Accurate Gastric Cancer Screening. Biosens. Bioelectron. 2022, 197, 113781. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chao, Y.; Guo, Y.; Zhang, F.; Mao, C.; Guan, C.; Chen, G.; Feng, C. Paper-Based Netlike Rolling Circle Amplification (NRCA) for Ultrasensitive and Visual Detection of SARS-CoV-2. Sens. Actuators B Chem. 2022, 358, 131460. [Google Scholar] [CrossRef]
- Bialy, R.M.; Ali, M.M.; Li, Y.; Brennan, J.D. Protein-Mediated Suppression of Rolling Circle Amplification for Biosensing with an Aptamer-Containing DNA Primer. Chem. Eur. J. 2020, 26, 5085–5092. [Google Scholar] [CrossRef]
- Sun, Y.; Chang, Y.; Zhang, Q.; Liu, M. An Origami Paper-Based Device Printed with DNAzyme-Containing DNA Superstructures for Escherichia coli Detection. Micromachines 2019, 10, 531. [Google Scholar] [CrossRef]
- Shah, K.G.; Kumar, S.; Yager, P. Near-Digital Amplification in Paper Improves Sensitivity and Speed in Biplexed Reactions. Sci. Rep. 2022, 12, 14618. [Google Scholar] [CrossRef]
- Qian, J.; Zhang, Q.; Lai, J.C.; Wang, Y.; Lu, M. Rapid MicroRNA Detection Using Paper-Based Isothermal Amplification. In Proceedings of the 2021 IEEE Sensors, Sydney, NSW, Australia, 31 October–3 November 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Miao, X.C.Y.M. Visual Detection of Shigella in Milk by Competitive Annealing-Mediated Isothermal Amplification with Paper-Based DNA Extraction Method. Int. J. Food Sci. Technol. 2022, 57, 6055–6063. [Google Scholar] [CrossRef]
- Sun, Q.; Cao, M.; Zhang, X.; Wang, M.; Ma, Y.; Wang, J. A Simple and Low-Cost Paper-Based Colorimetric Method for Detecting and Distinguishing the GII.4 and GII.17 Genotypes of Norovirus. Talanta 2021, 225, 121978. [Google Scholar] [CrossRef]
- Cao, M.; Sun, Q.; Zhang, X.; Ma, Y.; Wang, J. Detection and Differentiation of Respiratory Syncytial Virus Subgroups A and B with Colorimetric Toehold Switch Sensors in a Paper-Based Cell-Free System. Biosens. Bioelectron. 2021, 182, 113173. [Google Scholar] [CrossRef] [PubMed]
- Karlikow, M.; da Silva, S.J.R.; Guo, Y.; Cicek, S.; Krokovsky, L.; Homme, P.; Xiong, Y.; Xu, T.; Calderón-Peláez, M.-A.; Camacho-Ortega, S.; et al. Field Validation of the Performance of Paper-Based Tests for the Detection of the Zika and Chikungunya Viruses in Serum Samples. Nat. Biomed. Eng. 2022, 6, 246–256. [Google Scholar] [CrossRef]
- Wu, Q.; Suo, C.; Brown, T.; Wang, T.; Teichmann, S.A.; Bassett, A.R. INSIGHT: A Population-Scale COVID-19 Testing Strategy Combining Point-of-Care Diagnosis with Centralized High-put Sequencing. Sci. Adv. 2021, 7, eabe5054. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Shen, J.; Liu, Y.; Huang, A.; Zhang, H.; Chen, F.; Zhou, D.; Zhou, Y.; Hao, G. Rapid and Effective Detection of Macrobrachium rosenbergii Nodavirus Using a Combination of Nucleic Acid Sequence-Based Amplification Test and Immunochromatographic Strip. J. Invertebr. Pathol. 2023, 198, 107921. [Google Scholar] [CrossRef] [PubMed]
- Zasada, A.A.; Mosiej, E.; Prygiel, M.; Polak, M.; Wdowiak, K.; Formińska, K.; Ziółkowski, R.; Żukowski, K.; Marchlewicz, K.; Nowiński, A.; et al. Detection of SARS-CoV-2 Using Reverse Transcription Helicase Dependent Amplification and Reverse Transcription Loop-Mediated Amplification Combined with Lateral Flow Assay. Biomedicines 2022, 10, 2329. [Google Scholar] [CrossRef] [PubMed]
- Rosenbohm, J.M.; Klapperich, C.M.; Cabodi, M. Tunable Duplex Semiquantitative Detection of Nucleic Acids with a Visual Lateral Flow Immunoassay Readout. Anal. Chem. 2022, 94, 3956–3962. [Google Scholar] [CrossRef]
- Shanmugakani, R.K.; Wu, M. An Isothermal Amplification-Coupled Dipstick for the Rapid Detection of COVID-19. J. Med. Microbiol. 2022, 71, 001519. [Google Scholar] [CrossRef]
- Zhao, J.; Fang, S.; Liu, Y.; Zeng, L.; He, Z. A Lateral Flow Biosensor Based on Gold Nanoparticles Detects Four Hemorrhagic Fever Viruses. Anal. Meth. 2020, 12, 5613–5620. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Zheng, J.; Yu, Y.; Huang, X.; Wei, J.; Mukama, O.; Wang, S.; Qin, Q. A Lateral Flow Biosensor for Rapid Detection of Singapore Grouper Iridovirus (SGIV). Aquaculture 2021, 541, 736756. [Google Scholar] [CrossRef]
- Liu, X.; Bu, S.; Wei, H.; Wang, Z.; Yu, S.; Li, Z.; Hao, Z.; He, X.; Wan, J. Visual Assay of Escherichia coli O157:H7 Based on an Isothermal Strand Displacement and Hybrid Chain Reaction Amplification Strategy. Anal. Meth. 2021, 13, 3379–3385. [Google Scholar] [CrossRef]
- Feng, S.; Chen, H.; Hu, Z.; Wu, T.; Liu, Z. Ultrasensitive Detection of miRNA via CRISPR/Cas12a Coupled with Strand Displacement Amplification Reaction. ACS Appl. Mater. Interfaces 2023, 15, 28933–28940. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Fang, J.; Fu, W. Argonaute-Based Isothermal Amplification Assay for Ultrasensitive and Specific RNA Detection. Anal. Chem. 2023, 95, 8291–8298. [Google Scholar] [CrossRef] [PubMed]
- Kropocheva, E.V.; Lisitskaya, L.A.; Agapov, A.A.; Musabirov, A.A.; Kulbachinskiy, A.V.; Esyunina, D.M. Prokaryotic Argonaute Proteins as a Tool for Biotechnology. Mol. Biol. 2022, 56, 854. [Google Scholar] [CrossRef]
- Lisitskaya, L.; Aravin, A.A.; Kulbachinskiy, A. DNA Interference and Beyond: Structure and Functions of Prokaryotic Argonaute Proteins. Nat. Commun. 2018, 9, 5165. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.-R.; Wang, F.; Liu, C.-Y.; Li, J.; Shan, C.; Wang, K.; Wang, Y.; Li, P.-F.; Li, X.-M. Sensitive Naked-Eye Detection of Telomerase Activity Based on Exponential Amplification Reaction and Lateral Flow Assay. Anal. Bioanal. Chem. 2022, 414, 6139–6147. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Zhao, W.; Deng, Y.; Sun, Y.; Qiu, C.; Wu, B.; Bao, J.; Chen, Z.; Yu, L. Universal Point-of-Care Detection of Proteins Based on Proximity Hybridization-Mediated Isothermal Exponential Amplification. Analyst 2022, 147, 1709–1715. [Google Scholar] [CrossRef]
- Zhou, T.; Huang, R.; Huang, M.; Shen, J.; Shan, Y.; Xing, D. CRISPR/Cas13a Powered Portable Electrochemiluminescence Chip for Ultrasensitive and Specific MiRNA Detection. Adv. Sci. 2020, 7, 1903661. [Google Scholar] [CrossRef]
- Hillary, V.E.; Ceasar, S.A. A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Mol. Biotechnol. 2022, 65, 311–325. [Google Scholar] [CrossRef]
- Kavuri, N.R.; Ramasamy, M.; Qi, Y.; Mandadi, K. Applications of CRISPR/Cas13-Based RNA Editing in Plants. Cells 2022, 11, 2665. [Google Scholar] [CrossRef]
- Walker, G.T.; Linn, C.P.; Nadeau, J.G. DNA Detection by Strand Displacement Amplification and Fluorescence Polarization with Signal Enhancement Using a DNA Binding Protein. Nucleic Acids Res. 1996, 24, 348–353. [Google Scholar] [CrossRef]
- Mao, R.; Qi, L.; Li, J.; Sun, M.; Wang, Z.; Du, Y. Competitive Annealing Mediated Isothermal Amplification of Nucleic Acids. Analyst 2018, 143, 639–642. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, B.B.; Veigas, B.; Baptista, P.V. Isothermal Amplification of Nucleic Acids: The Race for the Next “Gold Standard”. Front. Sens. 2021, 2, 752600. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, L.; Ma, R.; Cong, L.; Wu, Z.; Wei, Y.; Xue, S.; Zheng, W.; Tang, S. Rapid Detection of Salmonella with Recombinase Aided Amplification. J. Microbiol. Meth. 2017, 139, 202–204. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.; Xu, Y.; Kong, H. Helicase-Dependent Isothermal DNA Amplification. EMBO Rep. 2004, 5, 795–800. [Google Scholar] [CrossRef]
- Reid, M.S.; Le, X.C.; Zhang, H. Exponential Isothermal Amplification of Nucleic Acids and Assays for Proteins, Cells, Small Molecules, and Enzyme Activities: An EXPAR Example. Angew. Chem. Int. Ed. 2018, 57, 11856–11866. [Google Scholar] [CrossRef] [PubMed]
- Brink, A.A.; Vervoort, M.B.; Middeldorp, J.M.; Meijer, C.J.; van den Brule, A.J. Nucleic Acid Sequence-Based Amplification, a New Method for Analysis of Spliced and Unspliced Epstein-Barr Virus Latent Transcripts, and Its Comparison with Reverse Transcriptase PCR. J. Clin. Microbiol. 1998, 36, 3164–3169. [Google Scholar] [CrossRef]
- Mumtaz, Z.; Rashid, Z.; Ali, A.; Arif, A.; Ameen, F.; AlTami, M.S.; Yousaf, M.Z. Prospects of Microfluidic Technology in Nucleic Acid Detection Approaches. Biosensors 2023, 13, 584. [Google Scholar] [CrossRef]
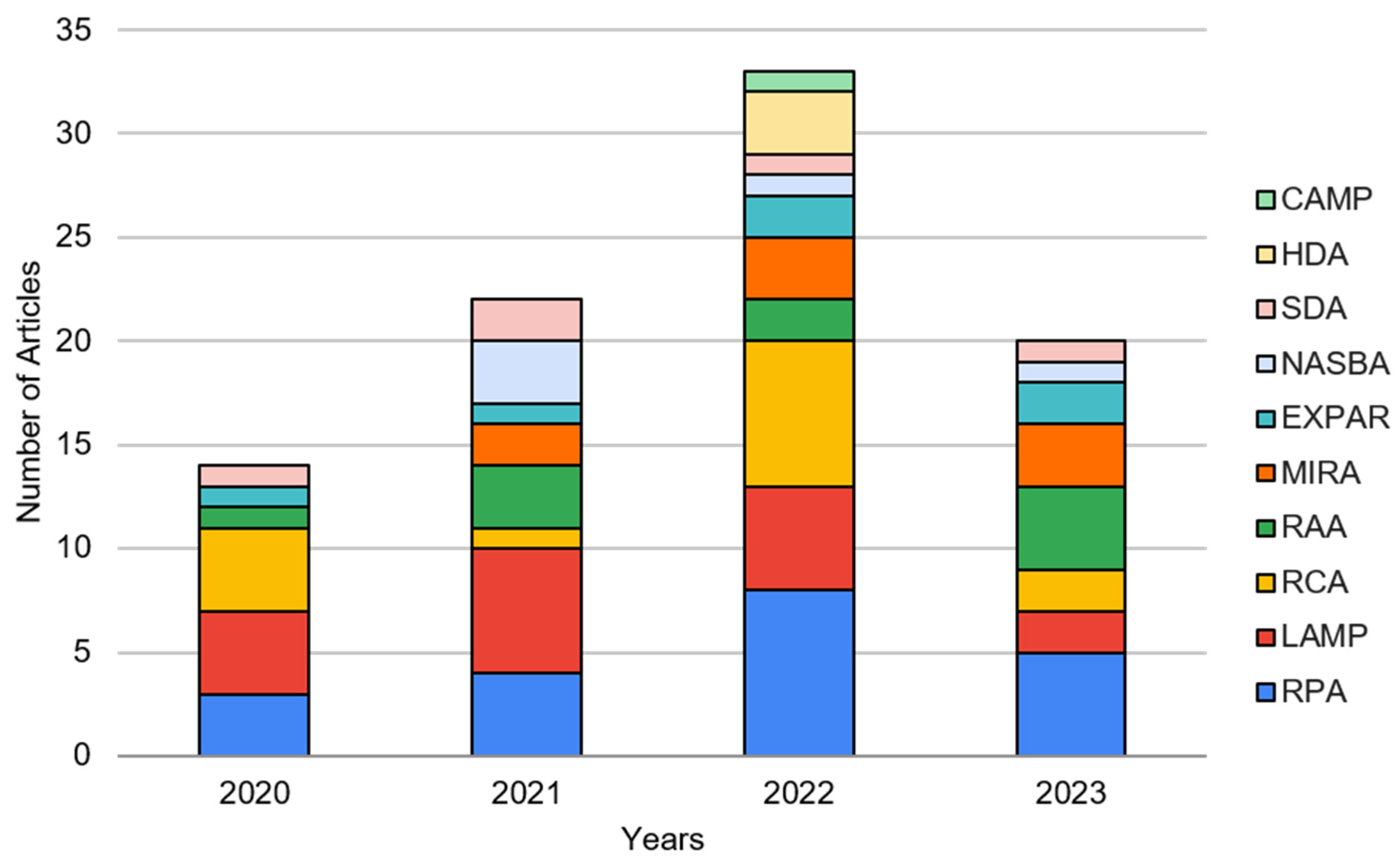

| Extraction | Amplification + Detection | Detection | Fully Integrated | Total | |
| RPA | 4 | 3 | 14 | 1 | 22 |
| RAA | 0 | 0 | 10 | 0 | 10 |
| MIRA | 1 | 0 | 8 | 0 | 9 |
| LAMP | 0 | 6 | 6 | 5 | 17 |
| RCA | 0 | 2 | 12 | 0 | 14 |
| NASBA | 0 | 0 | 5 | 0 | 5 |
| HDA | 0 | 0 | 3 | 0 | 3 |
| SDA | 0 | 1 | 4 | 0 | 5 |
| EXPAR | 0 | 1 | 4 | 1 | 6 |
| CAMP | 1 | 0 | 0 | 0 | 1 |
| Total | 6 | 13 | 66 | 7 | 92 |
| Colorimetric | Fluorometric | Flow Distance | SERS | Other | Total | |
| RPA | 14 (14) | 2 | 0 | 1 | 1 | 18 (14) |
| RAA | 9 (9) | 0 | 1 | 0 | 0 | 10 (9) |
| MIRA | 8 (8) | 0 | 0 | 0 | 0 | 8 (8) |
| LAMP | 7 (5) | 11 | 2 | 0 | 0 | 20 (5) |
| RCA | 8 (7) | 5 (1) | 1 | 0 | 0 | 14 (8) |
| NASBA | 5 (2) | 0 | 0 | 0 | 0 | 5 (2) |
| HDA | 3 (3) | 0 | 0 | 0 | 0 | 3 (3) |
| SDA | 3 (3) | 1 | 1 | 0 | 0 | 5 (3) |
| EXPAR | 3 (3) | 2 | 0 | 0 | 1 | 6 (3) |
| CAMP | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 60 (54) | 21 (1) | 5 | 1 | 2 | 89 (55) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reynolds, J.; Loeffler, R.S.; Leigh, P.J.; Lopez, H.A.; Yoon, J.-Y. Recent Uses of Paper Microfluidics in Isothermal Nucleic Acid Amplification Tests. Biosensors 2023, 13, 885. https://doi.org/10.3390/bios13090885
Reynolds J, Loeffler RS, Leigh PJ, Lopez HA, Yoon J-Y. Recent Uses of Paper Microfluidics in Isothermal Nucleic Acid Amplification Tests. Biosensors. 2023; 13(9):885. https://doi.org/10.3390/bios13090885
Chicago/Turabian StyleReynolds, Jocelyn, Reid S. Loeffler, Preston J. Leigh, Hannah A. Lopez, and Jeong-Yeol Yoon. 2023. "Recent Uses of Paper Microfluidics in Isothermal Nucleic Acid Amplification Tests" Biosensors 13, no. 9: 885. https://doi.org/10.3390/bios13090885
APA StyleReynolds, J., Loeffler, R. S., Leigh, P. J., Lopez, H. A., & Yoon, J.-Y. (2023). Recent Uses of Paper Microfluidics in Isothermal Nucleic Acid Amplification Tests. Biosensors, 13(9), 885. https://doi.org/10.3390/bios13090885






