Advancing Healthcare: Synergizing Biosensors and Machine Learning for Early Cancer Diagnosis
Abstract
1. Introduction
2. Overview of Machine Learning Algorithms
2.1. Supervised Machine Learning
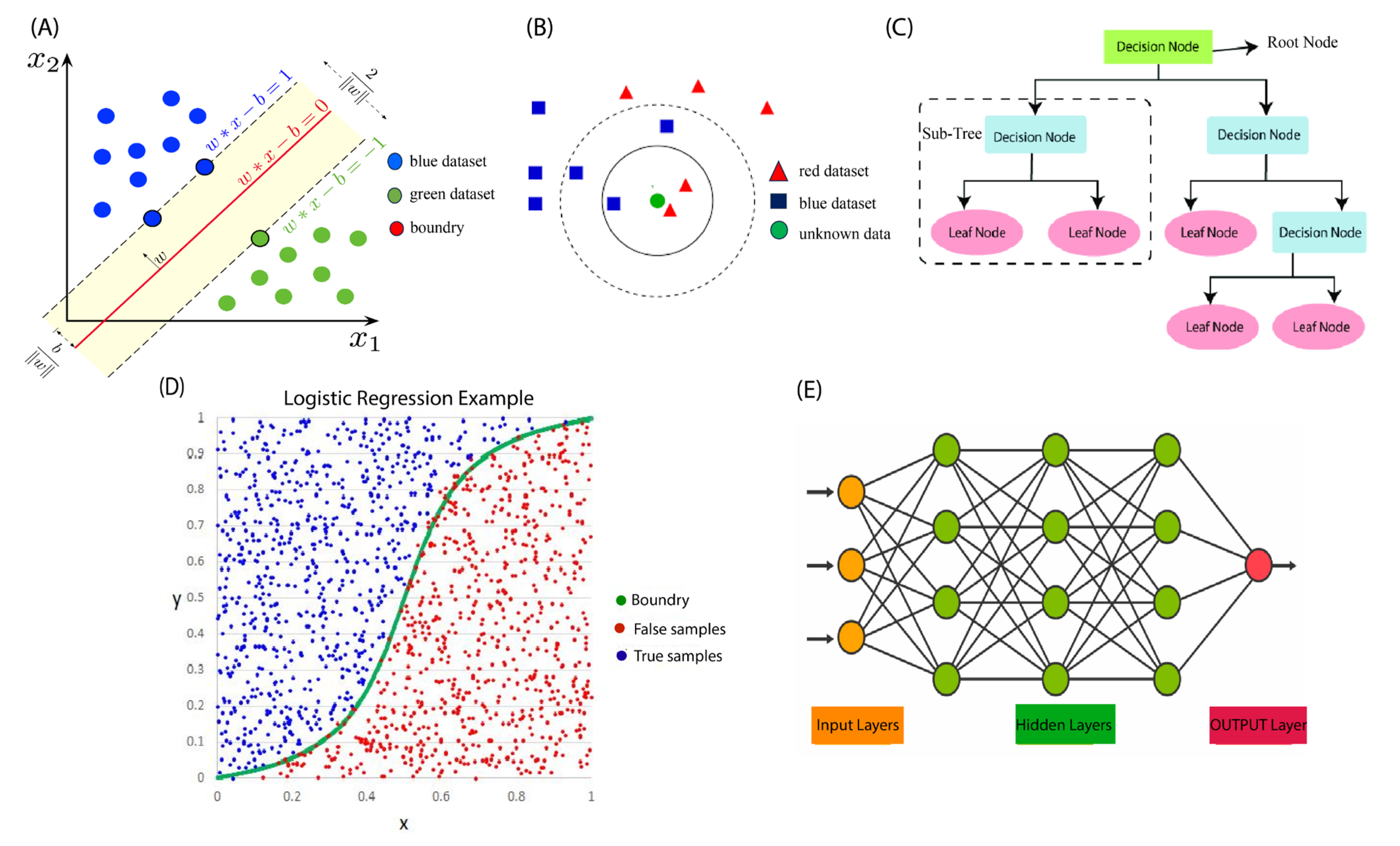
2.1.1. Support Vector Machines (SVMs)
2.1.2. K-Nearest Neighbor (KNN)
2.1.3. Decision Tree (DT)
2.1.4. Gaussian Naïve Bayes (GNB)
2.1.5. Logistic Regression (LR)
2.1.6. Random Forest (RF)
2.1.7. Artificial Neural Network (ANN)
2.2. Unsupervised Machine Learning
2.3. Machine Learning Figures of Merits
3. Lab-on-a-Chip in Cancer Detection
3.1. Optical Detection
3.1.1. Breast Cancer

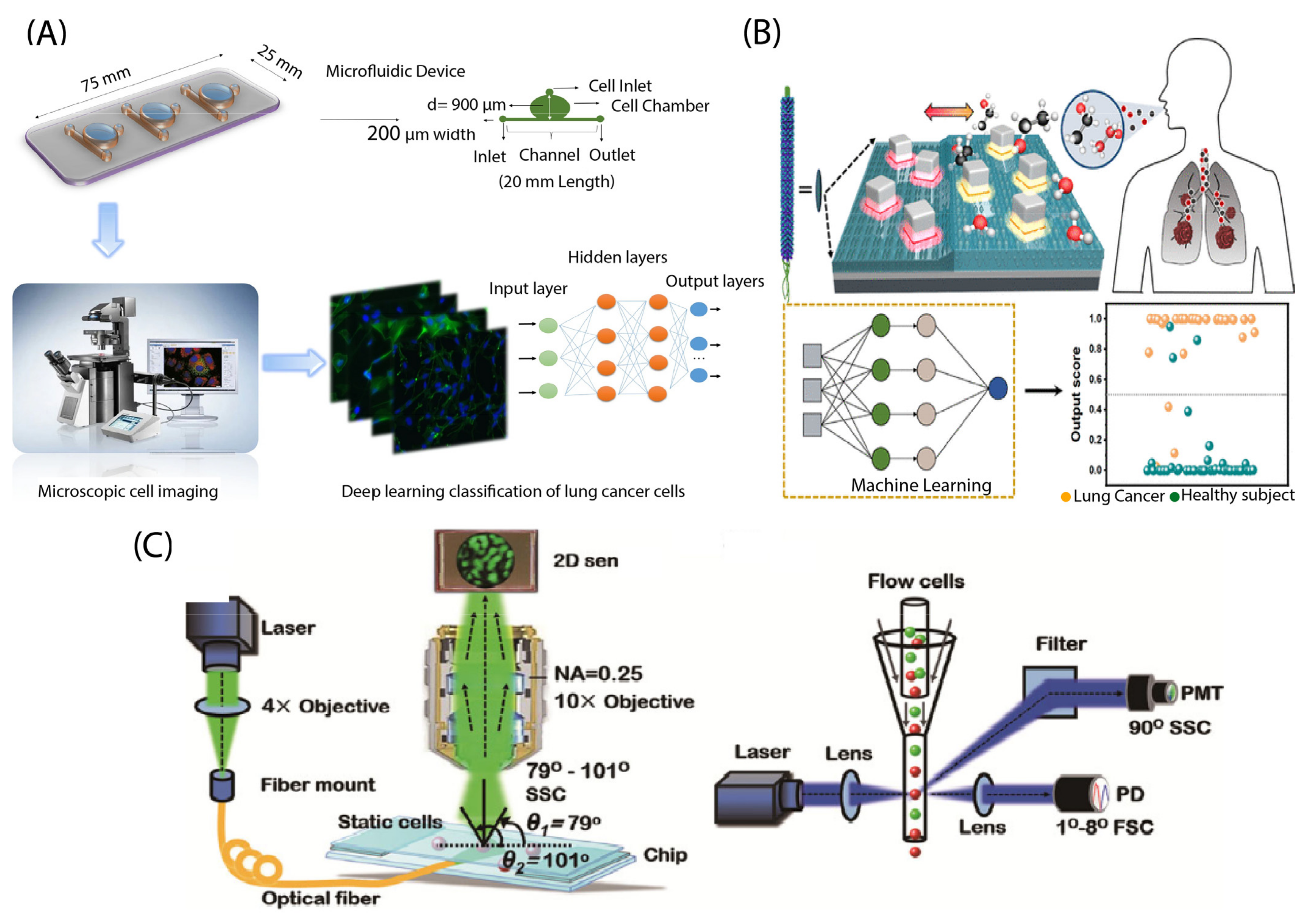
3.1.2. Lung Cancer
3.1.3. Gastrointestinal Cancer
3.1.4. Gynecological Cancer
3.1.5. Prostate Cancer
3.1.6. Brain Cancer
3.1.7. Hematological Cancer
3.2. Electrical Detection
3.2.1. Breast Cancer

3.2.2. Lung Cancer
3.2.3. Liver Cancer
3.2.4. Pancreatic Cancer
3.2.5. Hematological Cancer
3.2.6. Head and Neck Cancer
3.2.7. Gynecological Cancer
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 6 May 2023).
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Kumar, S.; Kaushik, B.K. Recent advancements in optical biosensors for cancer detection. Biosens. Bioelectron. 2022, 197, 113805. [Google Scholar] [CrossRef] [PubMed]
- Sinha, T. Tumors: Benign and malignant. Cancer Ther. Oncol. Int. J. 2018, 10, 52–54. [Google Scholar] [CrossRef]
- Weinberg, R.A. How cancer arises. Sci. Am. 1996, 275, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, R.; Soon, R.H.; Zhang, P.; Jiang, K.; Lim, C.T. Cancer diagnosis: From tumor to liquid biopsy and beyond. Lab Chip 2019, 19, 11–34. [Google Scholar] [CrossRef]
- Crulhas, B.P.; Basso, C.R.; Castro, G.R.; Pedrosa, V.A. Recent advances based on a sensor for cancer biomarker detection. ECS J. Solid State Sci. Technol. 2021, 10, 047004. [Google Scholar] [CrossRef]
- Alharthi, S.D.; Bijukumar, D.; Prasad, S.; Khan, A.M.; Mathew, M.T. Evolution in biosensors for cancers biomarkers detection: A review. J. Bio-Tribo-Corros. 2021, 7, 42. [Google Scholar] [CrossRef]
- Khanmohammadi, A.; Aghaie, A.; Vahedi, E.; Qazvini, A.; Ghanei, M.; Afkhami, A.; Hajian, A.; Bagheri, H. Electrochemical biosensors for the detection of lung cancer biomarkers: A review. Talanta 2020, 206, 120251. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Gao, X.; Chen, Y.; Liu, T. Nanotechnology in cancer diagnosis: Progress, challenges and opportunities. J. Hematol. Oncol. 2019, 12, 137. [Google Scholar] [CrossRef]
- Shahbazi, N.; Zare-Dorabei, R.; Naghib, S.M. Design of a ratiometric plasmonic biosensor for herceptin detection in HER2-positive breast cancer. ACS Biomater. Sci. Eng. 2022, 8, 871–879. [Google Scholar] [CrossRef]
- Mwesige, B.; Yeo, S.-G.; Yoo, B.C. Circulating tumor cells: Liquid biopsy for early detection of cancer. Soonchunhyang Med. Sci. 2019, 25, 1–9. [Google Scholar] [CrossRef][Green Version]
- de Vries, M.; Jager, P.; Suurmeijer, A.; Plukker, J.; van Ginkel, R.; Hoekstra, H. Sentinel lymph node biopsy for melanoma: Prognostic value and disadvantages in 300 patients. Ned. Tijdschr. Geneeskd. 2005, 149, 1845–1851. [Google Scholar]
- Yeasmin, S.; Ammanath, G.; Ali, Y.; Boehm, B.O.; Yildiz, U.H.; Palaniappan, A.; Liedberg, B. Colorimetric urinalysis for on-site detection of metabolic biomarkers. ACS Appl. Mater. Interfaces 2020, 12, 31270–31281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, J.; Cheng, Y.; Chen, J.; Zhao, H.; Ren, X. Urine analysis has a very broad prospect in the future. Front. Anal. Sci. 2022, 1, 13. [Google Scholar] [CrossRef]
- Wang, L. Screening and biosensor-based approaches for lung cancer detection. Sensors 2017, 17, 2420. [Google Scholar] [CrossRef]
- Altintas, Z.; Tothill, I. Biomarkers and biosensors for the early diagnosis of lung cancer. Sens. Actuators B Chem. 2013, 188, 988–998. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, B.; Hu, Q.; Hong, Y.; Wallace, A.; Reynolds, K.; Ramsey, C.; Maeder, A.; Reed, R.; Tang, Y.; et al. Detection of biomarkers in body fluids using bioprobes based on aggregation-induced emission fluorogens. Mater. Chem. Front. 2020, 4, 2548–2570. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, A.; Ahmed, A.; Sundramoorthy, A.K.; Furukawa, H.; Arya, S.; Khosla, A. Recent advances in electrochemical biosensors: Applications, challenges, and future scope. Biosensors 2021, 11, 336. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, J.; Liu, T.; Luo, Y.; Loh, X.J.; Chen, X. Machine learning-reinforced noninvasive biosensors for healthcare. Adv. Healthc. Mater. 2021, 10, 2100734. [Google Scholar] [CrossRef]
- Bax, C.; Taverna, G.; Eusebio, L.; Sironi, S.; Grizzi, F.; Guazzoni, G.; Capelli, L. Innovative diagnostic methods for early prostate cancer detection through urine analysis: A review. Cancers 2018, 10, 123. [Google Scholar] [CrossRef]
- Azab, M.Y.; Hameed, M.F.O.; Obayya, S.S. Overview of Optical Biosensors for Early Cancer Detection: Fundamentals, Applications and Future Perspectives. Biology 2023, 12, 232. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Gandhi, S. A concise review on potential cancer biomarkers and advanced manufacturing of smart platform-based biosensors for early-stage cancer diagnost ics. Biosens. Bioelectron. X 2022, 11, 100178. [Google Scholar]
- Tothill, I.E. Biosensors for cancer markers diagnosis. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2009; Volume 20, pp. 55–62. [Google Scholar]
- Queralto, N.; Berliner, A.N.; Goldsmith, B.; Martino, R.; Rhodes, P.; Lim, S.H. Detecting cancer by breath volatile organic compound analysis: A review of array-based sensors. J. Breath Res. 2014, 8, 027112. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Casas, B.; Galdámez-Martínez, A.; Gutiérrez-Flores, J.; Ibañez, A.B.; Panda, P.K.; Santana, G.; de la Vega, H.A.; Suar, M.; Rodelo, C.G.; Kaushik, A.; et al. Bio-acceptable 0D and 1D ZnO nanostructures for cancer diagnostics and treatment. Mater. Today 2021, 50, 533–569. [Google Scholar] [CrossRef]
- Gao, Q.; Lee, W.-Y. Urinary metabolites for urological cancer detection: A review on the application of volatile organic compounds for cancers. Am. J. Clin. Exp. Urol. 2019, 7, 232. [Google Scholar]
- Amethiya, Y.; Pipariya, P.; Patel, S.; Shah, M. Comparative analysis of breast cancer detection using machine learning and biosensors. Intell. Med. 2022, 2, 69–81. [Google Scholar] [CrossRef]
- Cui, F.; Yue, Y.; Zhang, Y.; Zhang, Z.; Zhou, H.S. Advancing biosensors with machine learning. ACS Sens. 2020, 5, 3346–3364. [Google Scholar] [CrossRef]
- Kokabi, M.; Donnelly, M.; Xu, G. Benchmarking Small-Dataset Structure-Activity-Relationship Models for Prediction of Wnt Signaling Inhibition. IEEE Access 2020, 8, 228831–228840. [Google Scholar] [CrossRef]
- Kokabi, M.; Sui, J.; Gandotra, N.; Khamseh, A.P.; Scharfe, C.; Javanmard, M. Nucleic Acid Quantification by Multi-Frequency Impedance Cytometry and Machine Learning. Biosensors 2023, 13, 316. [Google Scholar] [CrossRef]
- Kokabi, H.; Najafi, M.; Jazayeri, S.A.; Jahanian, O. Performance optimization of RCCI engines running on landfill gas, propane and hydrogen through the deep neural network and genetic algorithm. Sustain. Energy Technol. Assess. 2023, 56, 103045. [Google Scholar] [CrossRef]
- Fujiyoshi, H.; Hirakawa, T.; Yamashita, T. Deep learning-based image recognition for autonomous driving. IATSS Res. 2019, 43, 244–252. [Google Scholar] [CrossRef]
- Varkonyi, A.; Mosavi, A. Learning in Robotics. Int. J. Comput. Appl. 2017, 157, 8–11. [Google Scholar] [CrossRef]
- El Naqa, I.; Murphy, M.J. What Is Machine Learning? Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Alloghani, M.; Al-Jumeily, D.; Mustafina, J.; Hussain, A.; Aljaaf, A.J. A systematic review on supervised and unsupervised machine learning algorithms for data science. In Supervised and Unsupervised Learning for Data Science; Springer: Berlin/Heidelberg, Germany, 2020; pp. 3–21. [Google Scholar]
- Nasteski, V. An overview of the supervised machine learning methods. Horizons B 2017, 4, 51–62. [Google Scholar] [CrossRef]
- Berry, M.W.; Mohamed, A.; Yap, B.W. Supervised and Unsupervised Learning for Data Science; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Poellmann, M.J.; Bu, J.; Liu, S.; Wang, A.Z.; Seyedin, S.N.; Chandrasekharan, C.; Hong, H.; Kim, Y.; Caster, J.M.; Hong, S. Nanotechnology and machine learning enable circulating tumor cells as a reliable biomarker for radiotherapy responses of gastrointestinal cancer patients. Biosens. Bioelectron. 2023, 226, 115117. [Google Scholar] [CrossRef] [PubMed]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef] [PubMed]
- Raji, H.; Tayyab, M.; Sui, J.; Mahmoodi, S.R.; Javanmard, M. Biosensors and machine learning for enhanced detection, stratification, and classification of cells: A review. Biomed. Microdevices 2022, 24, 26. [Google Scholar] [CrossRef]
- Sun, S.; Huang, R. An adaptive k-nearest neighbor algorithm. In Proceedings of the 2010 Seventh International Conference on Fuzzy Systems and Knowledge Discovery, Yantai, China, 10–12 August 2010; IEEE: Piscataway, NJ, USA, 2010; Volume 1, pp. 91–94. [Google Scholar]
- Laaksonen, J.; Oja, E. Classification with learning k-nearest neighbors. In Proceedings of the International Conference on Neural Networks (ICNN’96), Washington, DC, USA, 3–6 June 1996; IEEE: Piscataway, NJ, USA, 1996; Volume 3, pp. 1480–1483. [Google Scholar]
- Ozaki, K.; Shimbo, M.; Komachi, M.; Matsumoto, Y. Using the mutual k-nearest neighbor graphs for semi-supervised classification on natural language data. In Proceedings of the Fifteenth Conference on Computational Natural Language Learning, Portland, OR, USA, 23–24 June 2011; pp. 154–162. [Google Scholar]
- Khateeb, N.; Usman, M. Efficient heart disease prediction system using K-nearest neighbor classification technique. In Proceedings of the International Conference on Big Data and Internet of Thing, London, UK, 20–22 December 2017; pp. 21–26. [Google Scholar]
- Myles, A.J.; Feudale, R.N.; Liu, Y.; Woody, N.A.; Brown, S.D. An introduction to decision tree modeling. J. Chemom. 2004, 18, 275–285. [Google Scholar] [CrossRef]
- Jahromi, A.H.; Taheri, M. A non-parametric mixture of Gaussian naive Bayes classifiers based on local independent features. In Proceedings of the 2017 Artificial Intelligence and Signal Processing Conference (AISP), Shiraz, Iran, 25–27 October 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 209–212. [Google Scholar]
- Bisong, E.; Bisong, E. Logistic regression. In Building Machine Learning and Deep Learning Models on Google Cloud Platform: A Comprehensive Guide for Beginners; Springer: Berlin/Heidelberg, Germany, 2019; pp. 243–250. [Google Scholar]
- Goswami, M.; Sebastian, N.J. Performance Analysis of Logistic Regression, KNN, SVM, Naïve Bayes Classifier for Healthcare Application during COVID-19. In Innovative Data Communication Technologies and Application: Proceedings of ICIDCA 2021; Springer: Singapore, 2022; pp. 645–658. [Google Scholar]
- Kulkarni, A.D.; Lowe, B. Random Forest Algorithm for Land Cover Classification. 2016. Available online: https://scholarworks.uttyler.edu/compsci_fac/1/ (accessed on 6 May 2023).
- Agatonovic-Kustrin, S.; Beresford, R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Available online: https://en.wikipedia.org/wiki/Support_vector_machine (accessed on 6 May 2023).
- KNN. Available online: https://en.wikipedia.org/wiki/K-nearest_neighbors_algorithm (accessed on 6 May 2023).
- DT Algorithm. Available online: https://www.javatpoint.com/machine-learning-decision-tree-classification-algorithm (accessed on 6 May 2023).
- LR. Available online: https://machine-learning.paperspace.com/wiki/logistic-regression (accessed on 6 May 2023).
- ANN. Available online: https://www.tibco.com/reference-center/what-is-a-neural-network (accessed on 6 May 2023).
- Hoo, Z.H.; Candlish, J.; Teare, D. What is an ROC curve? Emerg. Med. J. 2017, 34, 357–359. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J. Optical biosensors: An exhaustive and comprehensive review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef]
- García-Hernández, L.A.; Martínez-Martínez, E.; Pazos-Solís, D.; Aguado-Preciado, J.; Dutt, A.; Chávez-Ramírez, A.U.; Korgel, B.; Sharma, A.; Oza, G. Optical Detection of Cancer Cells Using Lab-on-a-Chip. Biosensors 2023, 13, 439. [Google Scholar] [CrossRef] [PubMed]
- Salahandish, R.; Ghaffarinejad, A.; Naghib, S.M.; Majidzadeh-A, K.; Zargartalebi, H.; Sanati-Nezhad, A. Nano-biosensor for highly sensitive detection of HER2 positive breast cancer. Biosens. Bioelectron. 2018, 117, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Kaur, H.; Gautam, N.; Mantha, A.K. Biosensors for breast cancer diagnosis: A review of bioreceptors, biotransducers and signal amplification strategies. Biosens. Bioelectron. 2017, 88, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Piliarik, M.; Homola, J. Surface plasmon resonance (SPR) sensors: Approaching their limits? Opt. Express 2009, 17, 16505–16517. [Google Scholar] [CrossRef] [PubMed]
- Gade, A.; Sharma, A.; Srivastava, N.; Flora, S. Surface plasmon resonance: A promising approach for label-free early cancer diagnosis. Clin. Chim. Acta 2022, 527, 79–88. [Google Scholar] [CrossRef]
- Kumar, A.; Verma, P.; Jindal, P. Surface plasmon resonance sensor based on MXene coated PCF for detecting the cancer cells with machine learning approach. Microelectron. Eng. 2023, 267, 111897. [Google Scholar] [CrossRef]
- Verma, P.; Kumar, A.; Jindal, P. Machine Learning Approach for SPR based Photonic Crystal Fiber Sensor for Breast Cancer Cells Detection. In Proceedings of the 2022 IEEE 7th Forum on Research and Technologies for Society and Industry Innovation (RTSI), Paris, France, 24–26 August 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 7–12. [Google Scholar]
- Li, H.; Jiang, B.; Li, J. Recent advances in dopamine-based materials constructed via one-pot co-assembly strategy. Adv. Colloid Interface Sci. 2021, 295, 102489. [Google Scholar] [CrossRef]
- Jin, Y.; Du, N.; Huang, Y.; Shen, W.; Tan, Y.; Chen, Y.Z.; Dou, W.T.; He, X.P.; Yang, Z.; Xu, N.; et al. Fluorescence analysis of circulating exosomes for breast cancer diagnosis using a sensor array and deep learning. ACS Sens. 2022, 7, 1524–1532. [Google Scholar] [CrossRef]
- Pala, M.A.; Çimen, M.E.; Akgül, A.; Yıldız, M.Z.; Boz, A.F. Fractal dimension-based viability analysis of cancer cell lines in lens-free holographic microscopy via machine learning. Eur. Phys. J. Spec. Top. 2022, 231, 1023–1034. [Google Scholar] [CrossRef]
- Yang, G.; Xiao, Z.; Tang, C.; Deng, Y.; Huang, H.; He, Z. Recent advances in biosensor for detection of lung cancer biomarkers. Biosens. Bioelectron. 2019, 141, 111416. [Google Scholar] [CrossRef]
- Hashemzadeh, H.; Shojaeilangari, S.; Allahverdi, A.; Rothbauer, M.; Ertl, P.; Naderi-Manesh, H. A combined microfluidic deep learning approach for lung cancer cell high throughput screening toward automatic cancer screening applications. Sci. Rep. 2021, 11, 9804. [Google Scholar] [CrossRef] [PubMed]
- Siu, D.M.; Lee, K.C.M.; Lo, M.C.K.; Stassen, S.V.; Wang, M.; Zhang, I.Z.Q.; So, H.K.H.; Chan, G.C.F.; Cheah, K.S.E.; Wong, K.K.Y.; et al. Deep-learning-assisted biophysical imaging cytometry at massive throughput delineates cell population heterogeneity. Lab Chip 2020, 20, 3696–3708. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.; Chung, J.H.; Bak, G.H.; Kim, Y.H.; Kim, M.; Kim, Y.J.; Kwon, R.J.; Choi, E.J.; Kim, K.H.; Kim, Y.S.; et al. Multiarray Biosensor for Diagnosing Lung Cancer Based on Gap Plasmonic Color Films. ACS Sens. 2022, 8, 167–175. [Google Scholar] [CrossRef]
- Wei, H.; Xie, L.; Liu, Q.; Shao, C.; Wang, X.; Su, X. Automatic Classification of Label-Free Cells from Small Cell Lung Cancer and Poorly Differentiated Lung Adenocarcinoma with 2D Light Scattering Static Cytometry and Machine Learning. Cytom. Part A 2018, 95, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Khalid, S.; Khalil, T.; Nasreen, S. A survey of feature selection and feature extraction techniques in machine learning. In Proceedings of the 2014 Science and Information Conference, London, UK, 27–29 August 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 372–378. [Google Scholar]
- Ahmad, A.; Sala, F.; Paiè, P.; Candeo, A.; D’Annunzio, S.; Zippo, A.; Frindel, C.; Osellame, R.; Bragheri, F.; Bassi, A.; et al. On the robustness of machine learning algorithms toward microfluidic distortions for cell classification via on-chip fluorescence microscopy. Lab Chip 2022, 22, 3453–3463. [Google Scholar] [CrossRef]
- Lin, C.; Liang, S.; Li, Y.; Peng, Y.; Huang, Z.; Li, Z.; Yang, Y.; Luo, X. Localized plasmonic sensor for direct identifying lung and colon cancer from the blood. Biosens. Bioelectron. 2022, 211, 114372. [Google Scholar] [CrossRef]
- Park, J.; Hwang, M.; Choi, B.; Jeong, H.; Jung, J.H.; Kim, H.K.; Hong, S.; Park, J.H.; Choi, Y. Exosome classification by pattern analysis of surface-enhanced Raman spectroscopy data for lung cancer diagnosis. Anal. Chem. 2017, 89, 6695–6701. [Google Scholar] [CrossRef]
- Ko, J.; Bhagwat, N.; Yee, S.S.; Ortiz, N.; Sahmoud, A.; Black, T.; Aiello, N.M.; McKenzie, L.; O’Hara, M.; Redlinger, C.; et al. Combining machine learning and nanofluidic technology to diagnose pancreatic cancer using exosomes. ACS Nano 2017, 11, 11182–11193. [Google Scholar] [CrossRef]
- Li, P.; Chen, J.; Chen, Y.; Song, S.; Huang, X.; Yang, Y.; Li, Y.; Tong, Y.; Xie, Y.; Li, J.; et al. Construction of Exosome SORL1 Detection Platform Based on 3D Porous Microfluidic Chip and its Application in Early Diagnosis of Colorectal Cancer. Small 2023, 19, 2207381. [Google Scholar] [CrossRef]
- Cheng, N.; Fu, J.; Chen, D.; Chen, S.; Wang, H. An antibody-free liver cancer screening approach based on nanoplasmonics biosensing chips via spectrum-based deep learning. NanoImpact 2021, 21, 100296. [Google Scholar] [CrossRef]
- D’Orazio, M.; Murdocca, M.; Mencattini, A.; Casti, P.; Filippi, J.; Antonelli, G.; Di, G.D.; Comes, M.C.; Di, N.C.; Sangiuolo, F.; et al. Machine learning phenomics (MLP) combining deep learning with time-lapse-microscopy for monitoring colorectal adenocarcinoma cells gene expression and drug-response. Sci. Rep. 2022, 12, 8545. [Google Scholar] [CrossRef] [PubMed]
- Saren, G.; Zhu, L.; Han, Y. Quantitative Detection of Gastrointestinal Tumor Markers Using a Machine Learning Algorithm and Multicolor Quantum Dot Biosensor. Comput. Intell. Neurosci. 2022, 2022, 9022821. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.-M.; Chuang, Y.W.; Yu, M.C.; Chen, C.H.; Yang, C.K.; Huang, S.T.; Lin, C.L.; Shu, K.H.; Kao, C.H. Risk of cancer in patients with polycystic kidney disease: A propensity-score matched analysis of a nationwide, population-based cohort study. Lancet Oncol. 2016, 17, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Cachat, F.; Renella, R. Risk of cancer in patients with polycystic kidney disease. Lancet Oncol. 2016, 17, e474. [Google Scholar] [CrossRef]
- Mencattini, A.; Rizzuto, V.; Antonelli, G.; Di Giuseppe, D.; D’Orazio, M.; Filippi, J.; Comes, M.C.; Casti, P.; Vives Corrons, J.L.; Garcia-Bravo, M.; et al. Machine Learning Microfluidic based platform: Integration of Lab-on-Chip devices and data analysis algorithms for Red Blood Cell plasticity evaluation in Pyruvate Kinase Disease monitoring. Sens. Actuators A Phys. 2023, 351, 114187. [Google Scholar] [CrossRef]
- Asare-Werehene, M.; Hunter, R.A.; Gerber, E.; Reunov, A.; Brine, I.; Chang, C.Y.; Chang, C.C.; Shieh, D.B.; Burger, D.; Anis, H.; et al. The application of an extracellular vesicle-based biosensor in early diagnosis and prediction of chemoresponsiveness in ovarian cancer. Cancers 2023, 15, 2566. [Google Scholar] [CrossRef]
- Liu, C.; Wang, Z.; Jia, J.; Liu, Q.; Su, X. High-content video flow cytometry with digital cell filtering for label-free cell classification by machine learning. Cytom. Part A 2022, 103, 325–334. [Google Scholar] [CrossRef]
- Kim, M.; Chen, C.; Wang, P.; Mulvey, J.J.; Yang, Y.; Wun, C.; Antman-Passig, M.; Luo, H.B.; Cho, S.; Long-Roche, K.; et al. Detection of ovarian cancer via the spectral fingerprinting of quantum-defect-modified carbon nanotubes in serum by machine learning. Nat. Biomed. Eng. 2022, 6, 267–275. [Google Scholar] [CrossRef]
- Pirone, D.; Xin, L.; Bianco, V.; Miccio, L.; Xiao, W.; Che, L.; Li, X.; Memmolo, P.; Pan, F.; Ferraro, P. Identification of drug-resistant cancer cells in flow cytometry combining 3D holographic tomography with machine learning. Sens. Actuators B Chem. 2023, 375, 132963. [Google Scholar] [CrossRef]
- Kim, H.; Park, S.; Jeong, I.G.; Song, S.H.; Jeong, Y.; Kim, C.S.; Lee, K.H. Noninvasive precision screening of prostate cancer by urinary multimarker sensor and artificial intelligence analysis. ACS Nano 2020, 15, 4054–4065. [Google Scholar] [CrossRef]
- Rodrigues, V.C.; Juliana, C.S.; Andrey, C.S.; Daniel, C.B.; Matias, E.M.; Lucas, C.R.; Leonardo, F.S.S.; Odemir, M.B.; Andre, L.C.; Rui, M.R.; et al. Electrochemical and optical detection and machine learning applied to images of genosensors for diagnosis of prostate cancer with the biomarker PCA3. Talanta 2021, 222, 121444. [Google Scholar] [CrossRef]
- Linh, V.T.N.; Lee, M.-Y.; Mun, J.; Kim, Y.; Kim, H.; Han, I.W.; Park, S.-G.; Choi, S.; Kim, D.-H.; Rho, J.; et al. 3D plasmonic coral nanoarchitecture paper for label-free human urine sensing and deep learning-assisted cancer screening. Biosens. Bioelectron. 2023, 224, 115076. [Google Scholar] [CrossRef] [PubMed]
- McRae, M.P.; Simmons, G.; Wong, J.; McDevitt, J.T. Programmable bio-nanochip platform: A point-of-care biosensor system with the capacity to learn. Acc. Chem. Res. 2016, 49, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.R.; Hassan, N.; Khan, R.; Kim, Y.-T.; Iqbal, S.M. Classification of cancer cells using computational analysis of dynamic morphology. Comput. Methods Programs Biomed. 2018, 156, 105–112. [Google Scholar] [CrossRef]
- Weindel, K.; Moringlane, J.R.; Marmé, D.; Weich, H.A. Detection and quantification of vascular endothelial growth factor/vascular permeability factor in brain tumor tissue and cyst fluid: The key to angiogenesis? Neurosurgery 1994, 35, 439–449. [Google Scholar] [CrossRef]
- Behnan, J.; Isakson, P.; Joel, M.; Cilio, C.; Langmoen, I.A.; Vik-Mo, E.O.; Badn, W. Recruited brain tumor-derived mesenchymal stem cells contribute to brain tumor progression. Stem Cells 2014, 32, 1110–1123. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Islam, M.T.; Rahman, T.; Chowdhury, M.E.H.; Tahir, A.; Kiranyaz, S.; Mat, K.; Beng, G.K.; Soliman, M.S. Brain Tumor Segmentation and Classification from Sensor-Based Portable Microwave Brain Imaging System Using Lightweight Deep Learning Models. Biosensors 2023, 13, 302. [Google Scholar] [CrossRef]
- Avelino, K.Y.; Oliveira, L.S.; Santos, M.R.; Lucena-Silva, N.; Andrade, C.A.; Oliveira, M.D. Electrochemical DNA Biosensor for Chronic Myelocytic Leukemia Based on Hybrid Nanostructure. Bioelectrochemistry 2022, 147, 108176. Available online: https://www.sciencedirect.com/science/article/abs/pii/S156753942200127X?via%3Dihub (accessed on 6 May 2023). [CrossRef]
- Koowattanasuchat, S.; Ngernpimai, S.; Matulakul, P.; Thonghlueng, J.; Phanchai, W.; Chompoosor, A.; Panitanarak, U.; Wanna, Y.; Intharah, T.; Chootawiriyasakul, K.; et al. Rapid detection of cancer DNA in human blood using cysteamine-capped AuNPs and a machine learning-enabled smartphone. RSC Adv. 2023, 13, 1301–1311. [Google Scholar] [CrossRef]
- Soumerai, J.D.; Mato, A.R.; Dogan, A.; Seshan, V.E.; Joffe, E.; Flaherty, K.; Carter, J.; Hochberg, E.; Barnes, J.A.; Hamilton, A.M.; et al. Zanubrutinib, obinutuzumab, and venetoclax with minimal residual disease-driven discontinuation in previously untreated patients with chronic lymphocytic leukaemia or small lymphocytic lymphoma: A multicentre, single-arm, phase 2 trial. Lancet Haematol. 2021, 8, e879–e890. [Google Scholar] [CrossRef]
- Alten, J.; Klapper, W.; Leuschner, I.; Eckert, C.; Beier, R.; Vallo, E.; Krause, M.; Claviez, A.; Vieth, S.; Bleckmann, K.; et al. Secondary histiocytic sarcoma may cause apparent persistence or recurrence of minimal residual disease in childhood acute lymphoblastic leukemia. Pediatr. Blood Cancer 2015, 62, 1656–1660. [Google Scholar] [CrossRef] [PubMed]
- Uslu, F.; Icoz, K.; Tasdemir, K.; Yilmaz, B. Automated quantification of immunomagnetic beads and leukemia cells from optical microscope images. Biomed. Signal Process. Control. 2019, 49, 473–482. [Google Scholar] [CrossRef]
- Sarkar, S.; Kang, W.; Jiang, S.; Li, K.; Ray, S.; Luther, E.; Ivanov, A.R.; Fu, Y.; Konry, T. Machine learning-aided quantification of antibody-based cancer immunotherapy by natural killer cells in microfluidic droplets. Lab Chip 2020, 20, 2317–2327. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mahjoubfar, A.; Chen, C.L.; Niazi, K.R.; Pei, L.; Jalali, B. Deep cytometry: Deep learning with real-time inference in cell sorting and flow cytometry. Sci. Rep. 2019, 9, 11088. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, K.; Rather, G.M.; Lin, Z.; Sui, J.; Xie, P.; Le, T.; Bertino, J.R.; Javanmard, M. Toward point-of-care assessment of patient response: A portable tool for rapidly assessing cancer drug efficacy using multifrequency impedance cytometry and supervised machine learning. Microsyst. Nanoeng. 2019, 5, 34. [Google Scholar] [CrossRef]
- Sountharrajan, S.; Karthiga, M.; Suganya, E.; Rajan, C. Automatic classification on bio medical prognosisof invasive breast cancer. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 2541. [Google Scholar]
- Yang, H.-Y.; Wang, Y.-C.; Peng, H.-Y.; Huang, C.-H. Breath biopsy of breast cancer using sensor array signals and machine learning analysis. Sci. Rep. 2021, 11, 103. [Google Scholar] [CrossRef]
- Elsheakh, D.N.; Mohamed, R.A.; Fahmy, O.M.; Ezzat, K.; Eldamak, A.R. Complete Breast Cancer Detection and Monitoring System by Using Microwave Textile Based Antenna Sensors. Biosensors 2023, 13, 87. [Google Scholar] [CrossRef]
- Joshi, K.; Joshi, K.; Javani, A.; Park, J.; Velasco, V.; Xu, B.; Razorenova, O.; Esfandyarpour, R. A machine learning-assisted nanoparticle-printed biochip for real-time single cancer cell analysis. Adv. Biosyst. 2020, 4, 2000160. [Google Scholar] [CrossRef]
- Bondancia, T.J.; Soares, A.C.; Popolin-Neto, M.; Gomes, N.O.; Raymundo, P.P.A.; Barud, H.S.; Machado, S.A.S.; Ribeiro, S.J.L.; Melendez, M.E.; Carvalho, A.L.; et al. Low-cost bacterial nanocellulose-based interdigitated biosensor to detect the p53 cancer biomarker. Biomater. Adv. 2022, 134, 112676. [Google Scholar] [CrossRef]
- Liang, M.; Tang, Q.; Zhong, J.; Ai, Y. Machine learning empowered multi-stress level electromechanical phenotyping for high-dimensional single cell analysis. Biosens. Bioelectron. 2023, 225, 115086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, P.; Ren, H.; Jia, P.; Ji, J.; Cao, L.; Yang, P.; Li, Y.; Liu, J.; Li, Z.; et al. Synthetic biology-powered biosensors based on CRISPR/Cas mediated cascade signal amplification for precise RNA detection. Chem. Eng. J. 2022, 446, 136864. [Google Scholar] [CrossRef]
- Van de Goor, R.; van Hooren, M.; Dingemans, A.-M.; Kremer, B.; Kross, K. Training and validating a portable electronic nose for lung cancer screening. J. Thorac. Oncol. 2018, 13, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Nazir, N.U.; Abbas, S.R. Identification of phenol 2,2-methylene bis, 6 [1,1-D] as breath biomarker of hepatocellular carcinoma (HCC) patients and its electrochemical sensing: E-nose biosensor for HCC. Anal. Chim. Acta 2023, 1242, 340752. [Google Scholar] [CrossRef]
- Salahi, A.; Honrado, C.; Moore, J.; Adair, S.; Bauer, T.W.; Swami, N.S. Supervised learning on impedance cytometry data for label-free biophysical distinction of pancreatic cancer cells versus their associated fibroblasts under gemcitabine treatment. Biosens. Bioelectron. 2023, 231, 115262. [Google Scholar] [CrossRef]
- Honrado, C.; Salahi, A.; Adair, S.J.; Moore, J.H.; Bauer, T.W.; Swami, N.S. Automated biophysical classification of apoptotic pancreatic cancer cell subpopulations by using machine learning approaches with impedance cytometry. Lab Chip 2022, 22, 3708–3720. [Google Scholar] [CrossRef]
- Ferguson, C.A.; Hwang, J.C.; Zhang, Y.; Cheng, X. Single-Cell Classification Based on Population Nucleus Size Combining Microwave Impedance Spectroscopy and Machine Learning. Sensors 2023, 23, 1001. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ding, Q.; Lin, W.; Weng, Y.; Feng, S.; Chen, R.; Chen, C.; Qiu, S.; Lin, D. Profiling of Tumor Cell-Delivered Exosome by Surface Enhanced Raman Spectroscopy-Based Biosensor for Evaluation of Nasopharyngeal Cancer Radioresistance. Adv. Healthc. Mater. 2023, 12, 2202482. [Google Scholar] [CrossRef]
- Braz, D.C.; Neto, M.P.; Shimizu, F.M.; Sá, A.C.; Lima, R.S.; Gobbi, A.L.; Melendez, M.E.; Arantes, L.M.R.B.; Carvalho, A.L.; Paulovich, F.V.; et al. Using machine learning and an electronic tongue for discriminating saliva samples from oral cavity cancer patients and healthy individuals. Talanta 2022, 243, 123327. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Wu, Y.; Gao, J.; Han, Y.; Chu, Y.; Qiang, L.; Qiu, J.; Gao, Y.; Wang, Y.; et al. High-Throughput, Living Single-Cell, Multiple Secreted Biomarker Profiling Using Microfluidic Chip and Machine Learning for Tumor Cell Classification. Adv. Healthc. Mater. 2022, 11, 2102800. [Google Scholar] [CrossRef]
- Feng, Y.; Cheng, Z.; Chai, H.; He, W.; Huang, L.; Wang, W. Neural network-enhanced real-time impedance flow cytometry for single-cell intrinsic characterization. Lab Chip 2022, 22, 240–249. [Google Scholar] [CrossRef] [PubMed]





| Authors | Cancer Cell Type | Biosensor Type | ML Algorithm | Results (%) |
|---|---|---|---|---|
| Kumar et al. [64] | Breast Cancer | (Surface plasmon resonance) SPR sensor | ANN | MSE = 0.01525 percentage error of 2% |
| Verma et al. [65] | Breast Cancer | SPR sensor | ANN | MSE = 0.116 |
| Jin et al. [67] | Breast Cancer | Fluorescence sensor | ANN | ACC = 100 |
| Pala et al. [68] | Breast Cancer | CMOS imaging sensor | ANN | ACC = 99.65 |
| Hashemzadeh et al. [70] | Lung Cancer | Olympus fluorescence microscope | ANN | ACC = 98.37 |
| Sui et al. [71] | Lung Cancer | Fluorescence sensor | CNN | ACC = 91–95 |
| Nguyen et al. [72] | Lung Cancer | Gap plasmonic color sensors | Convolutional neural network (CNN) | ACC = 89 |
| Wei et al. [73] | Lung Cancer | Two-dimensional (2D) light-scattering | SVM | ACC = 99.87 |
| Ahmad et al. [75] | hTERT-immortalized human mammary epithelial cells (IMEC WT) Xenograft-derived primary tumor cells (XD) Lung metastasis-derived cells (MD) | Fluorescence microscopy Image-based sensor | CNN | ACC = 99.4 |
| Lin et al. [76] | Lung and Colon Cancer | Localized plasmonic sensor | SVM | ACC = 85.72 |
| Park et al. [77] | Lung Cancer | Surface-enhanced Raman spectroscopy (SERS) | Principal component analysis (PCA) | Sensitivity = 95.3 |
| Ko et al. [78] | Pancreatic Cancer | Image-based multichannel nanofluidic system | LDA | AUC = 0.81 |
| Li et al. [79] | Colorectal Cancer | Image-based 3D porous microfluidic chip | RF | ACC = 91.4 |
| Cheng et al. [80] | Liver Cancer | SERS sensor | ANN | ACC = 91 |
| D’Orazio et al. [81] | Colorectal Cancer | image-based time-lapse microscopy | ANN | ACC = 86.77 |
| Saren et al. [82] | Gastrointestinal Cancer | Quantum dot (QD)-labeled biofilms | Principal component analysis (PCA) | ACC = 94 |
| Mencattini et al. [85] | PKD, which may cause Liver, Colon, and Kidney Cancer | Image-based time-lapse microscopy | ANN | ACC = 88 |
| Liu et al. [87] | Cervical Cancer | Image-based high-content VFC (video flow cytometry) | CNN + SVM | ACC = 90.8 |
| Kim et al. [88] | Ovarian Cancer | Nanosensor array | SVM | ACC = 95 |
| Pirone et al. [89] | Endometrial cancer | Holographic flow cytometry (DHFC) | LDA | ACC = 96 |
| Rodrigues et al. [91] | Prostate Cancer | Genosensors | SVM and LDA | ACC = 99.9 |
| Linh et al. [92] | Prostate and Pancreatic Cancers | SERS sensor | ANN | ACC = 99.4 |
| McRae et al. [93] | Prostate and Ovarian Cancer | Bio-nanochip sensor | ANN | AUC = 0.94 |
| Hasan et al. [94] | Brain Cancer | Image-based time-lapse images | SVM + RF + NBC | ACC > 82 |
| Hossain et al. [97] | Brain Cancer | Sensor-based microwave brain imaging (SMBI) | CNN | ACC = ~90 |
| Koowattanasuchat et al. [99] | Leukemia Cancer | Colorimetric biosensors | RF + SVM | ACC = 90 |
| Uslu et al. [102] | Lymphoma and Leukemia Cancer | Microscope images | RF | ACC = 87.4 |
| Sarkar et al. [103] | Hematological Cancer | Droplet microfluidics-based cytotoxicity imaging approach | ANN | ACC = 94 |
| Li et al. [104] | Epithelial Cancer | Image-based microfluidic channel | CNN | ACC > 95 |
| Authors | Cancer Cell Type | Biosensor Type | ML Algorithm | Result (%) |
|---|---|---|---|---|
| Ahuja et al. [105] | T47D cancer cells (Type of Breast cancer) | Microfluidic device impedance cytometry | SVM | ACC = 95.9 |
| Sountharrajan et al. [106] | Breast Cancer | Surface acoustic wave (SAW) biosensor | SVM | ACC = 79.25 |
| Yang et al. [107] | Breast Cancer | Nanotube sensors | Random forest (RF) | ACC = 91 |
| Elsheakh et al. [108] | Breast Cancer | Microwave textile-based antenna sensors | CatBoost (Type of gradient boosting) | ACC = 100 |
| Joshi et al. [109] | Breast Cancer | Microfluidic channel sensor | Quadratic discriminant analysis (QDA) | ACC > 95.3 |
| Bondancia et al. [110] | Breast Cancer | Immunosensor | DT | ACC = 90 |
| Liang et al. [111] | Breast Cancer Combination of electrical and optical-based sensors | Impedance-based sensor | (Linear discriminant analysis) LDA + SVM | ACC = 91.2 |
| Zhang et al. [112] | Lung Cancer | SHARK (Synthetic Enzyme Shift RNA Signal Amplifier Related Cas13a Knockdown Reaction) | SVM | ACC = 82.81 |
| Van de Goor et al. [113] | Lung Cancer | E-nose biosensor | ANN | ACC = 93 |
| Nazir and Abbas [114] | Liver Cancer | E-nose biosensor | Unsupervised ML | ACC = 86 |
| Salahi et al. [115] | Pancreatic Cancer | Microfluidic device impedance cytometry | SVM | ACC = 93.7 |
| Honrado et al. [116] | Pancreatic Cancer | Microfluidic device impedance cytometry | KNN | ACC = 98.4 |
| Ferguson et al. [117] | Jurkat Cells (Type of Leukemia Cancer) | Microfluidic device | RF + SVM | ACC = 96 |
| Wu et al. [118] | Nasopharyngeal Cancer | Surface-enhanced Raman spectroscopy | ANN | ACC = 92.4 |
| Braz et al. [119] | Oral Cancer | E-tongue biosensor | RF + SVM | ACC = 80 |
| Wang et al. [120] | Ovarian, Kidney, Breast, Lymph Cancer | Microfluidic chip | K-means | ACC = 95 |
| Feng et al. [121] | Breast, Cervical, Lung, Leukemia Cancer | Impedance flow cytometry (IFC) | ANN | ACC = 91.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokabi, M.; Tahir, M.N.; Singh, D.; Javanmard, M. Advancing Healthcare: Synergizing Biosensors and Machine Learning for Early Cancer Diagnosis. Biosensors 2023, 13, 884. https://doi.org/10.3390/bios13090884
Kokabi M, Tahir MN, Singh D, Javanmard M. Advancing Healthcare: Synergizing Biosensors and Machine Learning for Early Cancer Diagnosis. Biosensors. 2023; 13(9):884. https://doi.org/10.3390/bios13090884
Chicago/Turabian StyleKokabi, Mahtab, Muhammad Nabeel Tahir, Darshan Singh, and Mehdi Javanmard. 2023. "Advancing Healthcare: Synergizing Biosensors and Machine Learning for Early Cancer Diagnosis" Biosensors 13, no. 9: 884. https://doi.org/10.3390/bios13090884
APA StyleKokabi, M., Tahir, M. N., Singh, D., & Javanmard, M. (2023). Advancing Healthcare: Synergizing Biosensors and Machine Learning for Early Cancer Diagnosis. Biosensors, 13(9), 884. https://doi.org/10.3390/bios13090884