Abstract
Sepsis is defined as a systemic inflammatory dysfunction strictly associated with infectious diseases, which represents an important health issue whose incidence is continuously increasing worldwide. Nowadays, sepsis is considered as one of the main causes of death that mainly affects critically ill patients in clinical settings, with a higher prevalence in low-income countries. Currently, sepsis management still represents an important challenge, since the use of traditional techniques for the diagnosis does not provide a rapid response, which is crucial for an effective infection management. Biosensing systems represent a valid alternative due to their characteristics such as low cost, portability, low response time, ease of use and suitability for point of care/need applications. This review provides an overview of the infectious agents associated with the development of sepsis and the host biomarkers suitable for diagnosis and prognosis. Special focus is given to the new emerging biosensing technologies using electrochemical and optical transduction techniques for sepsis diagnosis and management.
1. Introduction
According to the European Society of Intensive Care Medicine and the Society of Critical Care Medicine (2016 SCCM/ESICM task force), sepsis is defined as “a life-threatening organ dysfunction caused by a dysregulated host response to infection”. This definition highlights the importance of the non-homeostatic host response to infection, the significantly higher lethality than straightforward infection and the need for urgent diagnosis []. Sepsis is a “time-dependent” pathology whose clinical outcome depends on the timing of the diagnosis and the effectiveness of clinical management from the very first hour []. In fact, if not recognized early and managed promptly, this condition can lead to septic shock, multiple organ failure and death [,]. Normally, the sepsis state is the result of the complex host defense response against the invasion from external pathogens such as bacteria and fungi (Figure 1a). After the pathogen invasion, the host defense system is activated to prevent the spreading and multiplication of foreign organisms inside the body which is followed by an inflammatory response, regulated by pro- and anti-inflammatory cytokines [].
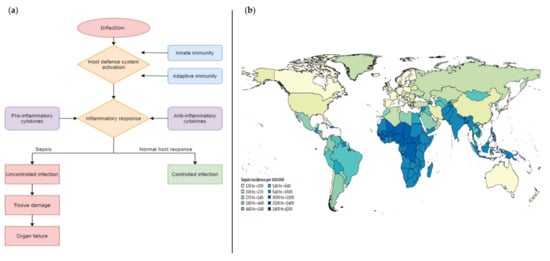
Figure 1.
(a) comparison of the schematic flow between sepsis and normal host response to an infection; (b) geographical incidence of sepsis worldwide []. Copyright © 1969, Elsevier.
Cytokine signals trigger the cells of the immune system, such as macrophages or neutrophils that rush to the infection site to eliminate the pathogen. This process involves several mechanisms and molecules, including acute phase proteins with several roles and the ultimate objective of controlling the infection.
However, the immune system dysregulated response leads to a situation where severe coagulation provokes microvascular thrombosis and organ dysfunction, which can potentially lead to chronic critical illness or death [].
Currently, sepsis management is one of the main challenges in clinical settings, especially in the management of critically ill patients. Indeed, it is estimated that each year more than 30 million cases are registered worldwide, with a 9–13% incidence increase each year and a mortality rate of 33–35% []. This pathology mainly affects adults aged 65 years or more; people with chronic medical conditions (such as diabetes, lung disease, cancer, and kidney disease), people with weakened immune system and children up to one year of age []. In addition, the incidence of sepsis varies by geographical area (Figure 1b) and substantially across regions. The epidemiological trends show the highest burden in sub-Saharan Africa, Oceania, south Asia, east Asia, and southeast Asia, where cases often exceed 600 per 100,000 inhabitants [], while the prevalence in European countries is about 90 cases per 100,000 inhabitants, leading to 1.4 million estimated cases per year and a mortality rate that fluctuates, depending on the area, between 20 and 40%.
Although the medical guidelines to prevent the infections that can lead to sepsis are continuously updated, sepsis cases are drastically increasing, mainly in low-income countries []. In this context, the importance of a rapid and easy diagnosis by means of point of care devices plays a crucial role in this field. Unfortunately, the traditional laboratory techniques are not suitable to respond to this issue, since they require a multi-step analysis, expensive instrumentation, trained personnel, and equipped laboratories. To cope with this need, in recent years lots of efforts have been spent in the development of new biosensing technologies [,,].
Indeed, the characteristics of biosensor devices, such as the speed of response, the portability and the ease of use can effectively improve the diagnosis by helping the physician in the medical decision-making, which would reduce the time and cost of diagnosis. In addition, the routine uses of such point of care devices in the clinical environment close to close to the ward could increase the probability of a patient’s survival.
In this review, the new emerging biosensing technologies for sepsis diagnosis and management are presented. First, an overview of the pathogens associated to the development of sepsis as well as the potential host biomarkers suitable for the diagnosis and prognosis will be provided, followed by a brief description of the traditional laboratory techniques. Finally, special focus will be given to the recent advancement in the development of biosensing technologies, and some examples will be presented.
2. Pathogens, Biomarkers, and Conventional Diagnostic Techniques
Nowadays, due to the complex and dysregulated host response, where different mechanisms are involved, there is not an unequivocal biomarker for sepsis identification [,]. Indeed, the detection of a single biomarker has low clinical significance, and only the identification of several biomarkers and physiological parameters provides the physician with a framework of useful diagnostic elements for evaluating the patient’s condition.
From the clinicians’ routine point of view, in the case of suspicion of sepsis, a quick evaluation of wide physiological parameters is applied. This diagnostic methodology is called “qSOFA” (quick sequential organ failure assessment score), which is a score system based on respiratory, neurological, and hemodynamic variables. This method shows a better performance in specificity than other similar methos, such as “SIRS” (systemic inflammatory response syndrome) and “NEWS” (national early warning score), but worse results in sensitivity [].
At the same time, the microbiological analyses should be performed immediately to identify the pathogen and to rapidly orient the antibiotic administration []. Meanwhile, several biomarkers are analyzed and evaluated to acquire additional information about the infection (e.g., to distinguish between viral and bacterial infection) and determine the patient’s condition [,].
2.1. Pathogens
Not all pathogens can cause sepsis, as bacteria need specific features to overcome the defense barriers, survive, proliferate, and disseminate in the human body. Most of such pathogens are facultative aerobic or anaerobic microorganisms with effective defense systems against oxidative stress, such as the production of superoxide dismutase (SOD), catalase and glutathione peroxidases.
Different virulence factors have been identified, such as exotoxins for gram-positive and endotoxins (e.g., lipopolysaccharides and LPS) for gram-negative bacteria. Although gram-positive bacteria lack endotoxins, they invade host tissues more easily due to the presence of exposed peptidoglycans and a range of other toxic secreted products []. A list of sepsis-causing bacteria is reported in Table 1.

Table 1.
List of main pathogenic bacteria than can cause sepsis [].
2.2. Biomarkers
Hundreds of potential biomarkers have been proposed for the diagnosis and prognosis of septic patients. Generally, the main attributes of successful and effective biomarkers are high sensitivity, specificity, possibility of bed-side monitoring, and financial accessibility. However, these criteria are only met by a few parameters that can be potentially used in the clinical practice for a timely and reliable diagnosis. An important feature for an effective biomarker would be the capability to discriminate between inflammations of infectious or non-infectious origin, as this aspect significantly influences the success of therapies []. The main sepsis biomarkers and their characteristics are summarized below.
2.2.1. C-Reactive Protein (CRP)
CRP is a protein released in plasma by the liver whose concentration rises 24–38 h after inflammation develops. Physiologically, it binds to the lysophosphatidylcholine expressed on the surface of dead or dying cells (and some types of bacteria) to activate the complement system and stimulate the opsonization and phagocytosis
[]. The CRP concentration in healthy subjects is lower than 5 mg/L, and its level is diagnostically used to differentiate between viral and bacterial infections. CRP is not a specific biomarker for inflammation associated infections since its concentration is also elevated in many other pathological conditions
[].
2.2.2. Procalcitonin (PCT)
PCT is a calcitonin precursor protein that, in physiological conditions, is secreted by the C cells of the thyroid gland and stored in the Golgi apparatus. Consequently, plasma PCT is normally at trace levels but it increases during the sepsis state due to its production by macrophages and monocytic cells of different organs, especially of the liver. Procalcitonin acts as chemokine, modulating the induction of anti-inflammatory cytokines and inducing the production of nitric oxide synthase []. During bacterial infections in adults, PCT serum starts increasing 4 h after the onset of a systemic infection and peaks between 8 and 24 h, with an estimated half-life of approximately 22–29 h []. In viral infections, only a minimum increase in PCT concentration is observed [].
2.2.3. Lipopolysaccharide Binding Protein (LPB)
During the acute phase of inflammatory response, LBP is produced by the liver to help lipid A or bacterial lipopolysaccharide to bind a cluster of proteins on monocytes and macrophages [,]. Under physiological conditions, serum concentrations fluctuate between 5 and 15 μg/mL [], while during sepsis average values become 30–40 μg/mL within 24 h []. A meta-analysis performed by Chen K-F et al. showed a weak sensitivity and specificity for sepsis [], even if its prognostic importance has been demonstrated [].
2.2.4. D-Dimer (DD)
Circulating D-dimer, a degradation product of cross-linked fibrin, is widely used as a fibrin-related marker for diagnostic and prognostic purposes, but its prognostic value in sepsis, either alone or in combination with other biomarkers, needs further validation. Since DD formation depends on coagulation and fibrinolysis, it may yield negative results in conditions associated with pronounced fibrinolytic inhibition such as sepsis. A recent study showed that the correction of DD for thrombin and plasmin generation may represent a new prognostic marker in septic patients
[].
2.2.5. Interleukins (ILs) and Other Cytokines
ILs are a group of cytokines, i.e., proteins acting as signal molecules both between cells of the immune system and between these cells and different organs and tissues. In particular, IL-6, IL-8, and IL-10 are used to diagnose sepsis, to assess the level of inflammatory response and to help the prognosis. IL-6 is a proinflammatory cytokine produced by cells such as monocytes, fibroblasts, endothelial cells, keratinocytes, T-lymphocytes, and tumor cells. It is released into the bloodstream 4–6 h after the increase in LPS, whereas IL-6 concentration decreases after 24–48 h of the presence of viable bacteria.
IL-8 is the main chemokine produced by macrophages and endothelial cells. It is considered a good predictive marker of sepsis in pediatric patients, but not for adults [].
IL-10 is an anti-inflammatory cytokine produced by macrophages, monocytes, neutrophils, T and B lymphocytes, and mesangial cells. High levels of both IL-6 and IL-10 are related to mortality of septic patients [].
Other cytokines are involved in sepsis and septic shock, such as TRAIL and IP-10, whose levels are significantly increased in septic patients []. TRAIL is a potent inducer of apoptosis, whose levels are associated with in-hospital mortality, organ dysfunction, and septic shock []. All these cytokines allow for a quantitative assessment of the severity of sepsis.
2.2.6. Surface Markers of Circulating Leukocytes
Several studies point out the importance of surface markers of circulating leukocytes, such as Cluster of Differentiation 64 (CD64), for the diagnosis of sepsis in neonatal and adult patients [,,].
CD64 is a type of integral membrane protein that binds monomeric IgG-type antibodies with high affinity [,]. Currently, there is a standard test called Trillium Diagnostic’s Leuko64 for the determination of the expression of CD64 on neutrophils, which represents a positive step in the sepsis mosaic [].
2.2.7. Fibronectin (FN)
FN is a high-molecular weight glycoprotein that plays an important role in cell adhesion and migration, anti-infection, hemostasis, injury repair and maintenance of microvascular integrity.
In typical physiological conditions, the FN plasma concentration is about 200–600 μg/mL (0.4–1.2 μM), but this value decreases in patients with severe infection and is closely related to the severity of sepsis [].
2.2.8. Lactate Dehydrogenase (LDH)
LDH is an enzyme catalyzing the conversion of pyruvate to lactate by reducing NAD+ to NADH []. Increased LDH levels in serum indicate tissue injury, hypoxia, necrosis, malignancies, hemolysis, but it is also associated with mortality in septic patients [,]. Erez et al. reported LDH as an independent parameter for predicting the mortality of any hospitalized patient []. According to Zein et al., LDH levels that do not stabilize within the first 48 h of inflammation are a significant indicator of mortality in patients with severe sepsis [].
2.2.9. MicroRNAs (miRNAs)
MiRNAs are small (20–24 nucleotides) RNA molecules that regulate gene expression. miRNA genes are estimated to represent only about 1% of the human genome but are thought to regulate up to 60% of all protein-coding genes. MicroRNAs belong to complex networks regulating gene expression in physiological and pathophysiological processes. The disruption of highly regulated mechanisms such as development aging, cell death may be associated to the aberrant miRNA expression, interestingly this abnormal expression can also be identified in diseases associated with infection and sepsis. The expression of IL-6, TNFα and other sepsis biomarkers is in fact regulated by miRNAs. Consequently, circulating miRNAs could be used as diagnostic biomarkers of sepsis, providing rapid information about infections compared to the traditional microbiological methods. Nevertheless, further studies should be performed to improve the understanding of miRNA concentrations in septic patients [].
2.3. Traditional Laboratory Techniques
A rapid and effective diagnosis of sepsis is fundamental in clinical settings, since each hour of delay in the identification and administration of antimicrobial therapy, drastically increases the mortality of the patient []. Currently, the traditional techniques used are classified in culture-based approaches, molecular techniques, and serological analysis. In the following paragraphs, the main techniques used to diagnose the infection and its associated pathogens and biomarkers will be discussed (Figure 2).
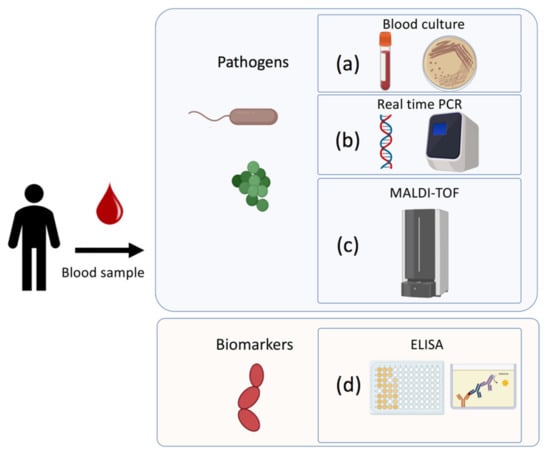
Figure 2.
Traditional techniques to detect pathogens and biomarkers associated to sepsis: (a) Blood culture-based approach for microorganism identification; (b) DNA/RNA amplification-based technique (RT-PCR); (c) Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry; (d) Immunoenzymatically serological assay for biomarkers detection.
2.3.1. Blood Cultures (BCs)
The blood culture-based approaches are the historical and traditional ones, encompassing several laboratory identification methods (e.g., gram staining, biochemical tests, etc.), that provide information on the bacterial species and are usually flanked by antibiotic susceptibility testing []. BCs are usually performed by automated instruments/tests (BacT/ALERT 3D bioMérieux, Marcy l’Etoile, France, or BACTEC Becton Dickinson B. V., Breda, Netherlands) that continuously monitor bacterial growth []. However, the long time needed for the definitive identification of the organism responsible for the bacteremia and its antibiotic susceptibility testing (usually more than 1 day) delays the administration of proper antibiotic and supportive treatments []. In addition to this issue, some studies reported that BCs tests are not suitable for the neonatal patient, because they require a high sample volume (at least 5 mL), and these techniques can fail in the identification of slow-growing pathogens as is case of previous antimicrobial treatment [,]. For this reason, the usefulness of blood cultures sampling at admission in emergency departments has been recently questioned [], and new molecular techniques are becoming the new routine tests.
2.3.2. Molecular Methods
Recently developed molecular diagnostic techniques are able to rapidly provide information on the infecting pathogen with high sensitivity and level of confidence []. The molecular methods generally employed can be classified in nucleic acid amplification-based (based on polymerase chain reaction (PCR)) and in protein-based techniques such as Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF).
Nucleic Acid Amplification-Based Techniques
Amplification methods allow for the enhancement of the detection signal in complex samples and the identification of target sequences associated to a bacterial species, or antibiotic resistance genes. Real-time quantitative PCR (RT-qPCR) amplifies and simultaneously detects the presence of nucleic acids in a sample. The technique is easy to perform for trained staff and is characterized by high specificity and sensitivity. For this reason, it is considered a suitable alternative to culture-based methods in a clinical microbiology laboratory. RT-qPCR relies on the use of specific oligonucleotide primers to amplify a DNA substrate, a polymerase, an intercalating fluorescence probe and precise thermal step cycles. In general, it only takes two hours to overcome the amplicon threshold needed to obtain a recordable signal. Two major drawbacks of these techniques are the risk of contamination, making a certain dose of expertise necessary, and the presence of false positives, due to amplification of non-target genes for the aspecific annealing of primers []. Importantly, this technique can be performed directly on whole blood samples (usually 500 µL of volume) allowing for a rapid identification of the pathogen. The direct diagnosis from whole blood samples circumvents the drawbacks of blood culturing approaches, in particular for slow-growing bacteria or non-culturable microorganisms, and in cases when the patient has already received antimicrobials [].
From a commercial point of view, multiplex PCR assays on whole blood are already available. SeptiFAST (Roche Diagnostics, Mannheim, Germany) was the first commercial multiplex assay for the detection of pathogens directly from blood and consequently it is mostly studied. It is important to mention that molecular diagnostic tests are expensive if compared to culture-based and phenotypic methods but are generally less laborious and faster. In a cost-effectiveness study, SeptiFAST was assessed to provide a significant economic saving [].
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF)
MALDI-TOF is now a routine method in several laboratories as it allows for easier and faster diagnosis of human pathogens than conventional phenotypic and molecular identification methods, with reliability and cost-effectiveness []. This technique allows for the identification of species according to their unique proteomic profile, which is obtained from post-culture samples [,]. In addition, it has been demonstrated that it is able to rapidly detect antibiotic resistant strains as shown by Kempf et al., with the spectrum of susceptible strain showing a peak matching with the drug’s spectrum, while resistant strains show different peaks corresponding to degradation products of the drug [].
Although MALDI-TOF is a promising technique for the identification of bacteria and for a rapid evaluation of antimicrobial resistance, a major drawback is that the reference database must be regularly extended to allow for matching of uncommon strains or species. MALDI-TOF MS is characterized with a high sensitivity and specificity, and the cost of the reagents is low, but many laboratories cannot afford the initial investment for the instrument. Moreover, the technique is suitable for high-throughput analyses reducing costs [,].
2.3.3. Serological Methods
The diagnosis and management of sepsis relies on heterogeneous information including biomarker levels, which are usually assessed through immunoassay. These include a plethora of qualitative or quantitative analytical techniques for the detection and measurement of many clinically relevant analytes.
The immunochemical techniques rely on the ability of antibodies to specifically bind antigens such as proteins, carbohydrates, and other molecules []. The commonly used immunochemical assay is the enzyme-linked immunosorbent assay (ELISA) in its ‘’sandwich’’ strategy []. In this assay an aliquot of sample containing the analyte is added into a polystyrene microtiter plate where a known amount of antigen-specific antibody is bound. After washing, an enzyme-labeled antibody is added, forming a “sandwich complex” triple layer of Antibody-Antigen-Antibody-enzyme. After washing away the unbound antibody, the enzyme substrate is added and an amount of colored product forms that is proportional to the amount of analyte in the sample [].
Despite its clear advantages, ELISA has some limitations, such as the laborious procedure and the insufficient level of sensitivity towards certain biomolecules []. Lastly, to detect a given antibody or antigen, a known reciprocal antigen or antibody must be generated [].
3. Biosensor as an Alternative Device for Sepsis
A biosensor is an analytical device that converts chemical/biochemical information into a useful analytical signal [,,]. It is always composed of two basic elements: a bioreceptor, a selective and specific biological recognition element such as enzyme(s), DNA, antibodies among others; and a transducer, which converts the receptor-analyte interaction into an analytical (i.e., optical or electrical) signal whose intensity is directly or inversely proportional to the analyte concentration [].
Biosensors can be classified based on their applications, and more in general the biosensor should fulfil the following characteristics such as: low cost, portability, low response time, ease of use and suitability for point of care/need applications []. Even though many different types of biosensors have been described, in this review we have examined the most recent emerging electrochemical and optical biosensors developed for the early detection of sepsis.
3.1. Electrochemical Biosensors
Electrochemical biosensors (Figure 3 and Figure 4) combine the sensitivity and the low response time of electroanalytical methods with the selectivity and specificity of the biological recognition element.
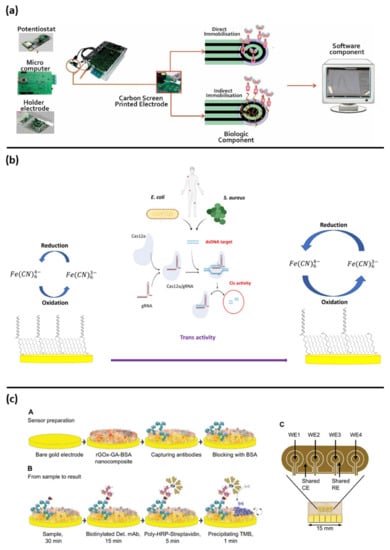
Figure 3.
Examples of electrochemical biosensors approaches for sepsis diagnosis: (a) representation of all components of point of care biosensor for CRP detection, reprinted from [] Creative common CC BY 4.0; (b) assay schematic of CRISPR/cas12a based biosensor for E. coli and S. aureus detection, reprinted with permission of [], copyright 2021 Elsevier B.V.; (c) fabrication steps and electrode schematic for PCT and CRP detection, reprinted with permission [], copyright 2021 Wiley-VCH GmbH.
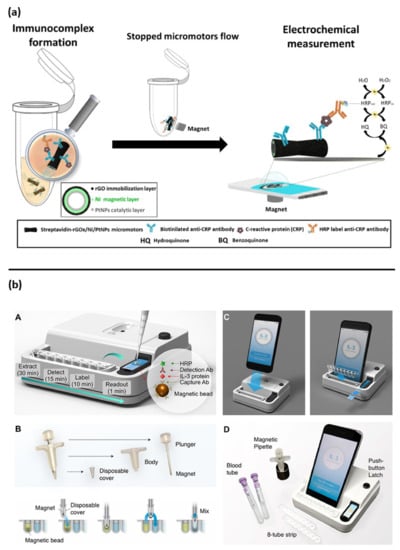
Figure 4.
Other examples of electrochemical biosensor approaches for sepsis diagnosis: (a) magnetic micromotors-based assay immunoassay for CRP detection, reprinted with permission [] Copyright 2020 Elsevier B.V; (b) assay schematic of point of care integrated sensor for cytokines detection, reprinted with permission of [] Copyright © 2018, American Chemical Society.
Electrochemical sensing usually requires a working electrode (WE), a reference electrode (RE), and a counter/auxiliary electrode (CE). Reactions are detected only near to the WE surface, which has a key role in determining the detection ability thanks to its dimensions, nano/materials and bio element and modification [,]. Electrochemical sensors are generally classified as amperometric, potentiometric, impedimetric and conductometric sensors, according to the electroanalytical technique they use [,,]. Furthermore, these biosensors are suitable for the miniaturization and integration in microfluidic and low-cost point of care devices [,,]. Herein, we report the emerging electrochemical biosensors developed for sepsis diagnosis (Table 2).

Table 2.
List of electrochemical biosensors.
3.1.1. Procalcitonin (PCT), C-Reactive Protein (CRP) Detection
Different emerging biosensors have been described in literature to detect PCT and CRP. These systems used a label-free approach for a single analyte to a multi-analyte detection platform, taking the advantage of the latest improvements of the engineering and the nano-(bio)technologies fields.
In 2017, Lim et al. developed a new label-free biosensor for the rapid detection of PCT based on recently discovered PCT-Binding Protein 3 (BP3 peptide) and electrochemical impedance spectroscopy (EIS) as transduction technique []. The peptide was immobilized onto a gold electrode and detection was performed in a buffer solution.
Another label-free approach was proposed by Guillem et al., for the CRP detection. The authors showed an interesting low-cost point of care device associated to open-source electronic readout elements (Figure 3a). The device was based on a carbon screen printed electrode functionalized with antibodies. This was able to detect the analyte in a small sample volume (50 µL), both in buffer and in spiked plasma [].
The use of nanotechnologies was described by Ge and collaborators. They developed a label-free electrochemical immunosensor based on the synergic effect of two different nanostructures the AuPtCu nanodendrites coupled with graphene-wrapped Co nanoparticles encapsulated in 3D N-doped carbon nanobrushes used to modify the electrode surface. These nanostructures have improved both the antibody loading capacity of the electrode and the mass/electron transport catalyzing the H2O2 reduction, reaching an ultra-low LOD around 0.011 pg/mL in diluted serum samples [].
Others similar works based on nanomaterials strategies are listed in Table 2 [,].
Interesting new approaches for PCT and CRP detection are based on magnetic-assisted workflow improving the sensitivity and selectivity of the biosensor. Águeda Molinero-Fernández et al. (2020) have developed a biosensor with a sandwich immunoassay configuration for the detection of PCT using magnetic beads []. Streptavidin coated magnetic beads (MBs) were functionalized with anti PCT antibodies, then disposable screen-printed carbon electrodes (SPE-C, on-drop detection) and electro-kinetically driven microfluidic chips with integrated Au electrodes (EMC-Au) were used. The amperometric measurements demonstrated a lower LOD with the EMC-Au electrode. In a lateral study, the same group also described a magnetic assisted immunoassay for the detection of CRP in neonatal septic patients, in a very small plasma sample volume (<10 µL), using an innovative system based on micromotors (Figure 4a) []. In that study, bubble-propelled micromotors converted chemical energy into autonomous propulsion, moving within the sample and binding the analyte. Micromotors consisted of an inner catalytic layer of platinum nanoparticles, which allowed the reaction responsible for propulsion, and an outer layer of reduced graphene oxide (rGO) functionalized with anti-CRP antibodies to bind the analyte. The CRP detection was based on an amperometric sandwich assay by using functionalized micromotors, a secondary antibody conjugated with an enzyme (Horseradish peroxidase: HRP) and a screen-printed carbon electrode. Micromotors offer an important advantage compared to traditional microbeads, since moving the bioreceptor within the sample increases the chances of interaction with the analyte and hence the sensitivity, which is usually limited by an inefficient transport of the analyte towards the bioreceptor.
With the aim to improve the fundamental diagnostic information for medical doctors’ decisions, multianalyte devices able to simultaneously detect PCT and CRP biomarkers have been developed. Ambalika Sanjeev Tanak and et al. have shown the possibility to detect both of PCT and CRP at the same time []. Such dual marker biosensing strategy consisted of two gold interdigitated electrodes on a flexible polyimide substrate coated with a thin film of ZnO, functionalized with specific antibodies. PCT and CRP were measured in human serum and whole blood. A very impressive and useful output was provided from Zupančič et al. The authors described an electrochemical immune biosensor able to detect PCT and CRP in buffer solution as well as in serum and whole blood samples collected from clinical patients. The biosensor was able to discriminate between the infected and non-infected groups. These results agreed with a standard ELISA kit and shown the possibility to use this sensor as a POC device. The sensor was based on a gold surface functionalized electrode with a 3D nanocomposite containing crosslinked bovine serum albumin (BSA) doped with conductive reduced graphene oxide nano flakes (rGOx) nanomaterials functionalized with antibodies; this used a sandwich assay approach for the detection of the biomarker of interest (Figure 3c) [].
3.1.2. Cytokines
Since cytokines are involved in several mechanisms during the host response to infection, they are used as convenient biomarkers for sepsis detection (Section 2.2.5). Among this class of biomarkers, most of the developed biosensors are referred to detect the interleukins (Table 2).
Russell et al. [] developed a microelectrode for the real time electrochemical detection of IL-6 using a needle shaped silicon substrate bearing eight gold disc electrodes functionalized with an antibody for IL-6. Measurements were carried out by using EIS and DPV techniques. The study demonstrated the possibility to detect IL-6 in clinically relevant samples without the need of complex electrode modifications or labelling steps.
Chen et al. proposed a label-free capacitive immuno- nano-biosensor based on a gold interdigitated electrode modified with longitudinal zeolite and iron oxide-complexed nanocomposite functionalized with antibodies to diagnose IL-3. The biosensor was able to detect IL-3 with a LOD of 3 pg/mL in a spiked human serum.
Specifically, for IL-3 detection, Min at al. have described an interesting magneto electrochemical sensor, integrated with a simple smartphone readout for POC applications (Figure 4b). This assay was able to rapidly detect the IL-3 in (<1 h) with a LOD of <10 pg/mL in human plasma samples [].
The diagnostic information would definitively improve if more biomarkers could be detected simultaneously. In this sense, similarly to what was described for other biomarkers, methods to detect multiple different interleukins simultaneously have been explored. Ambalika S. Tanak et al. [] demonstrated a novel multiarray point of care device that directly monitored a panel of five cytokine biomarkers (e.g., IL-6, IL-8, IL-10, TRAIL and IP-10). The device enclosed an array of gold electrodes coated with a nanofilm of semiconductive ZnO, functionalized with specific antibodies. The binding interaction when using plasma samples was registered by using EIS. This device was able to determine different information about patient’s condition. In fact, the combination of pro- and anti-inflammatory markers (e.g., IL-6, IL-8, and IL-10) can reveal the host immune response during the early stages of sepsis, while the detection of TRAIL and IP-10 may provide information about the origin of the infection, since these two biomarkers are differentially expressed in viral and bacterial infections []. In 2022, the same research group have tested and validated the previously biosensor developed named Direct Electrochemical Technique Targeting (DETecT) sepsis device with a 124 sepsis patients’ samples. Therefore, they have detected not only the interleukins group but were added the PCT and CRP biomarkers. The data was compared and resulted to agree with the LUMINEX standard method, opening the possibility to use this device to obtain a lot of information for sepsis diagnoses [].
3.1.3. Pathogens
As reported in Section 2.2, despite the urgent need for a quick pathogen identification, the traditional laboratory diagnostic techniques are rather slow. In response to this issue, a study conducted by Gao et al. (2016) [] reported a multiplex electrochemical biosensor for the rapid identification of pathogens in blood samples. This biosensor detected the species-specific sequences of the 16S ribosomal RNA of both gram-positive and -negative bacteria such as S. aureus, E. coli, P. aeruginosa and P. mirabilis. Gold electrodes were deposited on a plastic substrate, where each chip was composed of 16 electrodes. A sandwich strategy was used to detect the analyte on every gold electrode, and different thiolated oligonucleotide probes (universal and specific ones) were immobilized on the electrode surfaces. After the interaction between capture probe and 16S rRNA, an HRP targeted DNA was added as a secondary detection probe to catalyze the redox reaction of H2O2, whose current signal was amperometrically registered by a multichannel potentiostat.
Another interesting approach was proposed by Sharma et al. [] who reported the use of a molecularly imprinted polymer (MIP) as the recognition element to detect a Klebsiella pneumoniae. The MIP was synthetized by using polypyrrole (PPy) a conductive polymer and the bacterium served as a template. Difference pulse voltammetry DPV was used as the transduction technique and the sensor was able to detect the bacteria in buffer solution with a LOD of 1.35 CFU/mL.
Based on the new and recently discovered clustered regularly interspaced short palindromic repeats (CRISPR) and associated protein systems (Cas) named CRISPR/Cas system, Bonini et al. developed a label-free electrochemical biosensor for the bacterial DNA detection. The authors shown the possibility to detect E. coli and S. aureus bacterial DNA from clinical isolates. This biosensor was based on a DNA functionalized gold electrode and took advantage of the programmability of the Cas12a/gRNA enzyme using its primary and collateral activities for the specific bacterial detection and the signal amplification, respectively (Figure 3b) [,].
3.2. Optical Biosensors
An optical biosensor is a compact analytical device containing a biorecognition sensing element integrated with an optical transducer []. An optical transduction can be achieved by measuring the light power absorbed or emitted by a component of the sensing layer at a specific wavelength. Optical methods need a component in the sensing layer that absorbs or emits light: whenever this condition is not fulfilled, optical signaling labels have to be used. An alternative transduction is represented by the optical monitoring of a physical property (e.g., the refractive index) of the sensing layer that varies upon the interaction with the analyte. These transduction techniques do not need an optical label and are denoted as label-free methods [,,,].
Recent optical biosensors developed for sepsis diagnosis together with their characteristics, grouped by analyte, are shown in (Table 3).

Table 3.
List of optical biosensors and their main characteristics.
3.2.1. Procalcitonin (PCT), C-Reactive Protein (CRP) Detection and Interleukins
In 2017, Wang et al. [] developed a label-free biosensor using a fiber optic and surface plasmon resonance (SPR) for the specific detection of CRP. Such biosensor included a multi-mode fiber as the optical waveguide coated with a gold film in which plasmons could be generated. The Au layer of the sensor was functionalized by using dopamine as a crosslinking agent to immobilize the anti-CRP monoclonal antibody used as the selective ligand (Figure 5a). When the analyte bound the antibody, the change in refractive index of the medium through which the plasmonic wave was propagating shifted the reflectance angle. The magnitude of the shift depended on the amount of captured analyte, and the shift could be measured in almost real time []. This sensor showed good selectivity and consistency during specificity and performance tests.
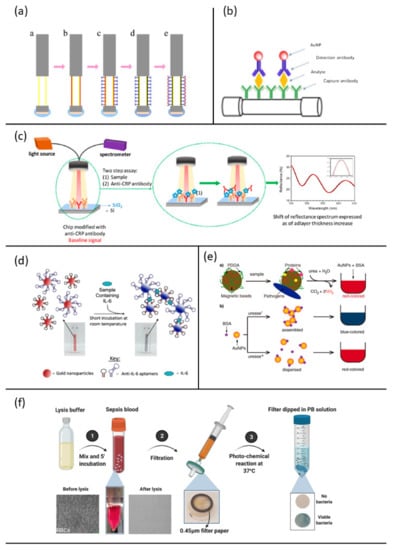
Figure 5.
Examples of optical biosensor approaches for sepsis diagnosis: (a) steps for fabrication of the fiber optic SPR biosensor fort the CRP detection, reprinted with permission of [] Creative common CC BY 4.0; (b) schematic sandwich assay for PCT detection, reprinted with permission of [] Copyright 2019 Elsevier B.V.; (c) schematic sensing principle for CRP detection, reprinted from [] Creative common CC BY 4.0; (d) schematic of the aptamer-gold nanoparticle-based assay for the detection of mouse IL-6, reprinted from [] Creative common CC BY 4.0; (e) schematic representation of the method for detect urease positive bacteria involved in sepsis, reprinted with permission of [], Copyright © 2019, American Chemical Society; (f) schematic showing the working principle of selective isolation and detection of bacteria from whole blood involved in sepsis, reprinted [], Creative common CC BY 4.0.
Functionalized optic fibers were also chosen by Chiang et al. (2019) []. Unlike the method previously reported, this sensor was based on localized surface plasmon resonance (LSPR), which allows an easier and less expensive fabrication. In addition, LSPR is less prone to errors in experimental data due to a smaller decay length which makes this less sensitive to bulk effects and external variables [,]. The fiber optic nanogold-linked immunosorbent assay was developed employing an immobilized capture probe on the fiber core surface and a detection probe conjugated to gold nanoparticles in a solution (Figure 5b). The introduction of a sample containing both analyte and detection probe in the microfluidics of the biosensor chip led to the formation of a sandwich-like complex between capture probe-analyte-detection probe on the fiber core surface, which induced the absorption of the fiber optic evanescent wave. This technique provided a fast response, required low-cost instrumentation and showed a lower LOD for PCT when compared to commercial assays for the same analyte.
Tsounidi et al. [] developed a compact bench-top bioanalytical system for CRP determination in human blood samples, using White Light Reflectance Spectroscopy (WLRS) (Figure 5c). This label-free two-site sandwich immunoassay was able to detect up to 1 ng/mL of the target analyte in 12 min and its dynamic range covered normal values of CRP in plasma and acute inflammation cases. The protein was first bound on a chip immobilized capture antibody and then reacted with the detection antibody. Goat polyclonal antibody (GC019) was used for both capture and detection. WLRS detected the increase in thickness on the silicon chip where the capture antibody was anchored. This fast technique provided accurate bioanalytical results, real time signal monitoring and low cost of consumables and instrumentation. Furthermore, results obtained from analyses of the same samples using standard diagnostic laboratory methods were comparable to those achieved by this system.
Giorgi-Coll et al. [] recently reported an aptamer-based optical assay for the proof-of concept determination of IL-6 []. The optical assay (Figure 5d) was based on the aggregation of gold nanoparticles coated with two complementary “sandwich style” aptamers, each with a different IL-6 target moiety.
Recognition and binding to the complementary aptamer pair from IL-6 caused the aggregation of the functionalized nanoparticles, thus shifting the maximum absorption from red to pink, which could be monitored visually.
As we have mentioned in the previous sections, the diagnostic information would improve if more biomarkers could be detected simultaneously, and optical biosensors have been recently developed in this direction.
Nuria Fabri-Faja et al. developed a phase-sensitive interferometric biosensor with a label-free microarray configuration [] for the simultaneous and rapid evaluation of different biomarkers such as: proteins (e.g., CRP and IL-6) and miRNAs. The sensor chip was based on lens-free interferometric microscopy and equipped with several metallic nanostructures to allow an efficient immobilization of probes such as antibodies for proteins or oligonucleotides for miRNAs. Proteins could be directly detected with this assay, whereas an additional amplification step was required for miRNAs. Despite its potential, further improvements are required for a clinical application of the device due to the limited dynamic range and LOD.
Lower LOD values were achieved by the Surface Enhanced Raman Scattering (SERS) based detection system developed by Kundu et al. []. The use of a AgNPs-laden black phosphorous-based SERS platform, allowed to reach a LOD of 1 pM and 100 fM for IL-3 and PCT, respectively. SERS is a technique based on the enhancement of the Raman scattering due to the presence of metallic nanostructures. In this case, Ag nanoparticles (AgNPs) were grown on black phosphorous (BP) flakes. Their arrangement led to an enhancement factor of 1014. The major advantages of this system were the possibility to identify different biomarkers from each other, due to the elevated signal selectivity to molecular structure (fingerprint features of Raman spectroscopy), and the chance of real-time monitoring. Unfortunately, no tests in real matrices were performed. Furthermore, these results were achieved with a bench-top instrument that is more performing than a portable one that is more suitable for point-of-care purposes.
Another interesting SERS-based method proposed by Zhou et al. [] a sandwich structure AgMNPs/IMs/CPs for the detection of Inflammatory Markers (IMs) such as CRP, IL-6 and PCT, was described. Unlike the previous study, Raman Reporters (RaRs) signals were herein enhanced (not those from the analytes). In particular, 2-mercaptopyridine (2-MPY), 4-nitrophenythiophenol (4-NTP) and 2-naphtiothiol (2-NT) were used as RaRs for CRP, IL-6 and PCT, respectively, since their Raman signals do not overlap with those of other substances present in blood serum. Ag magnetic nanoparticles (AgMNPs) modified with an internal standard (4-mercaptophenylacetonitrile) and a specific aptamer bound the IM; then, core porous shells modified with the aptamer and the RaRs formed the sandwich structure and the bound analytes were separated magnetically from the solution. LODs on the order of fg/mL were achieved for the three targets and the results were consistent with hospital analyses, showing recoveries above 96%. This method allowed the simultaneous, precise, quantitative detection of IMs in serum, providing a rapid screening, accurate evaluation, early monitoring, and diagnosis of sepsis.
3.2.2. Pathogens
Santopolo et al. (2019) [] developed a new rapid method for identifying urease-producing bacteria based on the detection of urease. The same principle used by Giorgi-Coll et al. for IL-6 detection was exploited for developing this assay: assembled and dispersed functionalized Au nanoparticles exhibit different wavelengths of maximum absorption.
This method substitutes the slow bacteriological culture steps with a 10 min capture procedure. The negatively charged bacteria and proteins are captured on magnetic beads coated with the positively charged polymer poly (diallyldimethylammonium chloride) (PDDA) (Figure 5e). Subsequently, the presence of urease enzymes bound to the beads was detected by adding urea, Au nanoparticles (AuNPs) and bovine serum albumin (BSA), which regulates the colloids aggregation in a manner that depends on pH. In fact, urease-positive bacteria hydrolyze urea to ammonia, increasing the pH and destabilizing the nanoparticles aggregations (red-shifted).
In contrast, urease-negative bacteria do not increase the pH upon the addition of urea, and the BSA triggers the assembly of gold nanoparticles (blue-colored test). This rapid assay can detect pathogens in urine at ultra-low concentrations and requires minimal infrastructure and instrumentation, since colors are easily differentiated by eye. These features make it an ideal solution for the rapid screening of urease-positive bacteria in decentralized healthcare schemes.
A label-free method for pathogen detection in sepsis was proposed by Narayana Iyengar et al. []. They developed a colorimetric test for bacteria detection in whole blood, achieving a LOD of 103 CFU/mL in less than 5 h. The assay was divided into two main steps: first, a lysis buffer was added to whole blood and the red blood cells lysis occurs without damaging bacteria; then, the solution was filtered through a 0.45 μm cellulose filter. Viable bacteria, trapped into the filter, were then dispersed in a culture media containing ferric citrate and ferricyanide, incubated at 37 °C and exposed to visible light irradiation. Consequent bacterial proliferation implied the metabolic production of Prussian Blue (PB) molecules that turn the solution from colorless to blue (Figure 5f). The assay results sensitive to both Gram-negative and Gram-positive bacteria and even for mixtures.
4. Conclusions
The early detection of sepsis still remains an open challenge in the clinical settings particularly in low-income countries. Sepsis diagnosis comprises the evaluation of the patient’s physiological parameters, the pathogen detection and the biomarkers associated to the host response. Each one of these steps provides useful information necessary to choose the correct treatment and consequently impact the management. Regarding the detection of biomarkers, the acquisition of only one biomarker does not provide useful information to the medical doctor. Indeed, recent studies have reported the difficulties encountered in the selection of a unique biomarker able to univocally represent the sepsis condition. It was stated that in order to obtain more accurate information the diagnosis should be based on the assessment of the levels of different biomarkers such as PCT, CRP and interleukins (e.g., IL-6 and IL-3).
In this context, efforts are being applied to the discovery of new biomarkers. During the diagnostic process, the response time associated to traditional techniques represents a crucial bottleneck, which strongly depends on the time required for sample collection and laboratory analysis procedures. Recent significant advances in the field of biosensors may contribute to reduce the response time for both pathogen and biomarker detection. Different strategies have been developed from the label-free to label-based optical and electrochemical assays, taking advantage of the recent improvements in nanobiotechnology.
Point of care systems able to simultaneously detect different biomarkers represent interesting alternatives as well. From recent works regarding biosensors for sepsis diagnosis presented in this review, it is evident that there is a lack of an integrated point of care system able to detect both pathogens and biomarkers. This gap may be filled in the near future thanks to the rapid and continue advancements in the physics, chemistry, biology, and engineering fields.
Author Contributions
Conceptualization, A.B. and A.G.C.; writing—original draft preparation, A.B., A.G.C. and N.P.; writing—review and editing, N.P., F.M.V., D.B. (Denise Biagini) and A.L.; supervision, T.L., A.T., D.B. (Daria Bottai) and F.D.F. All authors have read and agreed to the published version of the manuscript.
Funding
A.B was supported by the project CRISPRSENSE funded by Tuscany region, Italy (“DECRT. DIRIG. n. 21607 of 29-11-2021”).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singer, M.; Deutschman, C.S.; Seymour, C.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- O’Brien, J.M.; Ali, N.A.; Aberegg, S.K.; Abraham, E. Sepsis. Am. J. Med. 2007, 120, 1012–1022. [Google Scholar] [CrossRef]
- World Health Organization. Sepsis. Available online: https://www.who.int/news-room/fact-sheets/detail/sepsis (accessed on 23 July 2022).
- Paoli, C.J.; Reynolds, M.A.; Sinha, M.; Gitlin, M.; Crouser, E. Epidemiology and Costs of Sepsis in the United States—An Analysis Based on Timing of Diagnosis and Severity Level. Crit. Care Med. 2018, 46, 1889. [Google Scholar] [CrossRef]
- Abbas, A.K.; Lichtman, A.H.; Pillai, S. Cellular and Molecular Immunology; Elsevier Health Sciences: Philadelphia, PA, USA, 2014; ISBN 9780323757508. [Google Scholar]
- Braunwald, E.; Fauci, A.S.; Hauser, S.L.; Longo, D.L.; Jameson, J.L. Harrison’s Principles of Internal Medicine; McGraw-Hill Companies, Inc.: New York, NY, USA, 2005. [Google Scholar]
- Minasyan, H. Sepsis and Septic Shock: Pathogenesis and Treatment Perspectives. J. Crit. Care 2017, 40, 229–242. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention; National Center for Emerging and Zoonotic Infectious Diseases (NCEZID); Division of Healthcare Quality Promotion (DHQP). What Is Sepsis? Available online: https://www.cdc.gov/sepsis/what-is-sepsis.html (accessed on 23 July 2022).
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef]
- Levy, M.M.; Evans, L.E.; Rhodes, A. The Surviving Sepsis Campaign Bundle: 2018 Update. Intensive Care Med. 2018, 44, 925–928. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tripathy, S.; Jyoti, A.; Singh, S.G. Recent Advances in Biosensors for Diagnosis and Detection of Sepsis: A Comprehensive Review. Biosens. Bioelectron. 2019, 124–125, 205–215. [Google Scholar] [CrossRef]
- Reddy, B.; Hassan, U.; Seymour, C.; Angus, D.C.; Isbell, T.S.; White, K.; Weir, W.; Yeh, L.; Vincent, A.; Bashir, R. Point-of-Care Sensors for the Management of Sepsis. Nat. Biomed. Eng. 2018, 2, 640–648. [Google Scholar] [CrossRef]
- Tsounidi, D.; Petrou, P.S.; Raptis, I. Current Progress on Biosensors and Point-of-Care Devices for Sepsis Diagnosis. IEEE Sens. J. 2021, 21, 12840–12855. [Google Scholar] [CrossRef]
- Prucha, M.; Bellingan, G.; Zazula, R. Sepsis Biomarkers. Clin. Chim. Acta 2015, 440, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Parlato, M.; Cavaillon, J.M. Host Response Biomarkers in the Diagnosis of Sepsis: A General Overview. Methods Molecular Biology 2014, 1237, 149–211. [Google Scholar] [CrossRef]
- Oduncu, A.F.; Kıyan, G.S.; Yalçınlı, S. Comparison of QSOFA, SIRS, and NEWS Scoring Systems for Diagnosis, Mortality, and Morbidity of Sepsis in Emergency Department. Am. J. Emergency Medicine 2021, 48, 54–59. [Google Scholar] [CrossRef]
- Kundu, S. Overview of Sepsis and Sepsis Biomarker Detection. Master’s Thesis, Iowa State University, Ames, IA, USA, 2019. [Google Scholar]
- List of Microorganisms. Available online: https://www.ecdc.europa.eu/en/healthcare-associated-infections-acute-care-hospitals/database/microorganisms-and-antimicrobial-resistance/list (accessed on 23 July 2022).
- Tan, M.; Lu, Y.; Jiang, H.; Zhang, L. The Diagnostic Accuracy of Procalcitonin and C-Reactive Protein for Sepsis: A Systematic Review and Meta-Analysis. J. Cell Biochem. 2019, 120, 5852–5859. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Umemura, Y.; Hayashida, K.; Hara, Y.; Aihara, M.; Yamakawa, K. Diagnostic Value of Procalcitonin and Presepsin for Sepsis in Critically Ill Adult Patients: A Systematic Review and Meta-Analysis. J. Intensive Care 2019, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.F.; Chaou, C.H.; Jiang, J.Y.; Yu, H.W.; Meng, Y.H.; Tang, W.C.; Wu, C.C. Diagnostic Accuracy of Lipopolysaccharide-Binding Protein as Biomarker for Sepsis in Adult Patients: A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0153188. [Google Scholar] [CrossRef]
- García de Guadiana Romualdo, L.; Albaladejo Otón, M.D.; Rebollo Acebes, S.; Esteban Torrella, P.; Hernando Holgado, A.; Jiménez Santos, E.; Jiménez Sánchez, R.; Ortón Freire, A. Diagnostic Accuracy of Lipopolysaccharide-Binding Protein for Sepsis in Patients with Suspected Infection in the Emergency Department. Ann. Clin. Biochem. 2017, 55, 143–148. [Google Scholar] [CrossRef]
- Semeraro, F.; Ammollo, C.T.; Caironi, P.; Masson, S.; Latini, R.; Panigada, M.; Pesenti, A.; Semeraro, N.; Gattinoni, L.; Colucci, M. D-Dimer Corrected for Thrombin and Plasmin Generation Is a Strong Predictor of Mortality in Patients with Sepsis. Blood Transfus. 2020, 18, 304. [Google Scholar] [CrossRef]
- Raymond, S.L.; Hawkins, R.B.; Stortz, J.A.; Murphy, T.J.; Ungaro, R.; Dirain, M.L.; Nacionales, D.C.; Hollen, M.K.; Rincon, J.C.; Larson, S.D.; et al. Sepsis Is Associated with Reduced Spontaneous Neutrophil Migration Velocity in Human Adults. PLoS ONE 2018, 13, e0205327. [Google Scholar] [CrossRef]
- Schenck, E.J.; Ma, K.C.; Price, D.R.; Nicholson, T.; Oromendia, C.; Gentzler, E.R.; Sanchez, E.; Baron, R.M.; Fredenburgh, L.E.; Huh, J.W.; et al. Circulating Cell Death Biomarker TRAIL Is Associated with Increased Organ Dysfunction in Sepsis. JCI Insight 2019, 4, e127143. [Google Scholar] [CrossRef]
- Patnaik, R.; Azim, A.; Agarwal, V. Neutrophil CD64 a Diagnostic and Prognostic Marker of Sepsis in Adult Critically Ill Patients: A Brief Review. Indian J. Crit. Care Med. 2020, 24, 1242. [Google Scholar] [CrossRef]
- Gad, G.I.; Shinkar, D.M.; Kamel El-Din, M.M.; Nagi, H.M. The Utility of Soluble CD14 Subtype in Early Diagnosis of Culture-Proven Early-Onset Neonatal Sepsis and Prediction of Outcome. Am. J. Perinatol. 2020, 37, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Abd Elkareem, R.M.; Ahmed, H.M.; Meabed, M.H.; Elias, S.S.; Elmaraghy, M.A. Diagnostic Value of CD64 in Early Detection of Neonatal Sepsis. Comp. Clin. Path 2020, 29, 639–643. [Google Scholar] [CrossRef]
- Hulett, M.D.; Hogarth, P.M. The Second and Third Extracellular Domains of FcγRI (CD64) Confer the Unique High Affinity Binding of IgG2a. Mol. Immunol. 1998, 35, 989–996. [Google Scholar] [CrossRef]
- Tian, H.Y.; Chen, J.Y.; Lin, J.; Liang, Q.R.; Lei, Y.; Li, X.; Wu, Y.; Yang, L.Y.; Lin, X.H.; Liu, A.L.; et al. Sepsis Progression Monitoring via Human Serum Fibronectin Detection Based on Sandwich-Type Electrochemical Immunosensor. Anal. Chim. Acta 2020, 1100, 225–231. [Google Scholar] [CrossRef]
- Nelson, D.L.; Cox, M.M. I Principi di Biochimica di Lehninger; Zanichelli: Bologna, Italy, 2002. [Google Scholar]
- Ryoo, S.M.; Lee, J.; Lee, Y.S.; Lee, J.H.; Lim, K.S.; Huh, J.W.; Hong, S.B.; Lim, C.M.; Koh, Y.; Kim, W.Y. Lactate Level versus Lactate Clearance for Predicting Mortality in Patients with Septic Shock Defined by Sepsis-3. Crit. Care Med. 2018, 46, E489–E495. [Google Scholar] [CrossRef]
- Ghimenti, S.; Lomonaco, T.; Bellagambi, F.G.; Biagini, D.; Salvo, P.; Trivella, M.G.; Scali, M.C.; Barletta, V.; Marzilli, M.; di Francesco, F.; et al. Salivary Lactate and 8-Isoprostaglandin F2α as Potential Non-Invasive Biomarkers for Monitoring Heart Failure: A Pilot Study. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Lu, J.; Wei, Z.; Jiang, H.; Cheng, L.; Chen, Q.; Chen, M.; Yan, J.; Sun, Z. Lactate Dehydrogenase Is Associated with 28-Day Mortality in Patients with Sepsis: A Retrospective Observational Study. J. Surg. Res. 2018, 228, 314–321. [Google Scholar] [CrossRef]
- Zein, J.G.; Lee, G.L.; Tawk, M.; Dabaja, M.; Kinasewitz, G.T. Prognostic Significance of Elevated Serum Lactate Dehydrogenase (LDH) in Patients with Severe Sepsis. Chest 2004, 126, 873S. [Google Scholar] [CrossRef]
- Benz, F.; Roy, S.; Trautwein, C.; Roderburg, C.; Luedde, T. Circulating MicroRNAs as Biomarkers for Sepsis. Int. J. Mol. Sci. 2016, 17, 78. [Google Scholar] [CrossRef]
- Peker, N.; Couto, N.; Sinha, B.; Rossen, J.W. Diagnosis of Bloodstream Infections from Positive Blood Cultures and Directly from Blood Samples: Recent Developments in Molecular Approaches. Clin. Microbiol. Infect. 2018, 24, 944–955. [Google Scholar] [CrossRef]
- Loonen, A.J.M.; Wolffs, P.F.G.; Bruggeman, C.A.; van den Brule, A.J.C. Developments for Improved Diagnosis of Bacterial Bloodstream Infections. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1687–1702. [Google Scholar] [CrossRef]
- Cheng, M.P.; Stenstrom, R.; Paquette, K.; Stabler, S.N.; Akhter, M.; Davidson, A.C.; Gavric, M.; Lawandi, A.; Jinah, R.; Saeed, Z.; et al. Blood Culture Results Before and After Antimicrobial Administration in Patients With Severe Manifestations of Sepsis: A Diagnostic Study. Ann. Intern. Med. 2019, 171, 547–554. [Google Scholar] [CrossRef]
- Sautter, R.L.; Bills, A.R.; Lang, D.L.; Ruschell, G.; Heiter, B.J.; Bourbeau, P.P. Effects of Delayed-Entry Conditions on the Recovery and Detection of Microorganisms from BacT/ALERT and BACTEC Blood Culture Bottles. J. Clin. Microbiol. 2006, 44, 1245–1249. [Google Scholar] [CrossRef] [PubMed]
- Fenollar, F.; Raoult, D. Molecular Diagnosis of Bloodstream Infections Caused by Non-Cultivable Bacteria. Int. J. Antimicrob. Agents 2007, 30, 7–15. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Raoult, D. Direct Identification of Bacteria in Positive Blood Culture Bottles by Matrix-Assisted Laser Desorption Ionisation Time-of-Flight Mass Spectrometry. PLoS ONE 2009, 4, e8041. [Google Scholar] [CrossRef] [PubMed]
- Opota, O.; Jaton, K.; Greub, G. Microbial Diagnosis of Bloodstream Infection: Towards Molecular Diagnosis Directly from Blood. Clin. Microbiol. Infection 2015, 21, 323–331. [Google Scholar] [CrossRef]
- Mwaigwisya, S.; Assiri, R.A.M.; O’Grady, J. Emerging Commercial Molecular Tests for the Diagnosis of Bloodstream Infection. Expert Rev. Mol. Diagn. 2015, 15, 681–692. [Google Scholar] [CrossRef]
- Seng, P.; Rolain, J.M.; Fournier, P.E.; la Scola, B.; Drancourt, M.; Raoult, D. MALDI-TOF-Mass Spectrometry Applications in Clinical Microbiology. Future Microbiol. 2010, 5, 1733–1754. [Google Scholar] [CrossRef]
- Panda, A.; Kurapati, S.; Samantaray, J.C.; Srinivasan, A.; Khalil, S. MALDI-TOF Mass Spectrometry Proteomic Based Identification of Clinical Bacterial Isolates. Indian J. Med. Res. 2014, 140, 770. [Google Scholar]
- Chen, H.; Liu, K.; Li, Z.; Wang, P. Point of Care Testing for Infectious Diseases. Clin. Chim. Acta 2019, 493, 138–147. [Google Scholar] [CrossRef]
- Kempf, M.; Bakour, S.; Flaudrops, C.; Berrazeg, M.; Brunel, J.M.; Drissi, M.; Mesli, E.; Touati, A.; Rolain, J.M. Rapid Detection of Carbapenem Resistance in Acinetobacter Baumannii Using Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. PLoS ONE 2012, 7, e31676. [Google Scholar] [CrossRef]
- Rychert, J. Benefits and Limitations of MALDI-TOF Mass Spectrometry for the Identification of Microorganisms. J. Infect. Epidemiol. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- Rifai, N. Tietz Textbook of Clinical Chemistry and Molecular Diagnostics; Elsevier Health Sciences: St. Louis, MI, USA, 2017. [Google Scholar]
- Mancini, N. Diagnostic Methods and Protocols; Springer: New York, NY, USA, 2015. [Google Scholar]
- Hosseini, S.; Vázquez-Villegas, P.; Rito-Palomares, M.; Martinez-Chapa, S.O. Advantages, Disadvantages and Modifications of Conventional ELISA. In Enzyme-Linked Immunosorbent Assay (ELISA); Springer Briefs in Applied Sciences and Technology; Springer: Singapore, 2018; pp. 67–115. [Google Scholar] [CrossRef]
- Gan, S.D.; Patel, K.R. Enzyme Immunoassay and Enzyme-Linked Immunosorbent Assay. J. Investig. Dermatol. 2013, 133, 1–3. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Labuda, J.; Bowater, R.P.; Fojta, M.; Gauglitz, G.; Glatz, Z.; Hapala, I.; Havliš, J.; Kilar, F.; Kilar, A.; Malinovská, L.; et al. Terminology of Bioanalytical Methods (IUPAC Recommendations 2018). Pure Applied Chemistry 2018, 90, 1121–1198. [Google Scholar] [CrossRef]
- Pandey, C.M.; Malhotra, B.D. Biosensors: Fundamentals and Applications; Walter de Gruyter GmbH & Co KG: Berlin, Germany, 2019. [Google Scholar]
- Salvo, P.; Vivaldi, F.M.; Bonini, A.; Biagini, D.; Bellagambi, F.G.; Miliani, F.M.; Di Francesco, F.; Lomonaco, T. Biosensors for Detecting Lymphocytes and Immunoglobulins. Biosensors 2020, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Land, K.J.; Boeras, D.I.; Chen, X.S.; Ramsay, A.R.; Peeling, R.W. REASSURED Diagnostics to Inform Disease Control Strategies, Strengthen Health Systems and Improve Patient Outcomes. Nat. Microbiol. 2018, 4, 46–54. [Google Scholar] [CrossRef]
- Quesada-González, D.; Merkoçi, A. Nanomaterial-Based Devices for Point-of-Care Diagnostic Applications. Chem. Soc. Rev. 2018, 47, 4697–4709. [Google Scholar] [CrossRef]
- Bonini, A.; Vivaldi, F.M.; Herrera, E.; Melai, B.; Kirchhain, A.; Sajama, N.V.P.; Mattonai, M.; Caprioli, R.; Lomonaco, T.; Di Francesco, F.; et al. A Graphenic Biosensor for Real-Time Monitoring of Urea during Dialysis. IEEE Sens. J. 2020, 20, 4571–4578. [Google Scholar] [CrossRef]
- Poma, N.; Vivaldi, F.; Bonini, A.; Salvo, P.; Kirchhain, A.; Ates, Z.; Melai, B.; Bottai, D.; Tavanti, A.; di Francesco, F. Microbial Biofilm Monitoring by Electrochemical Transduction Methods. TrAC Trends Anal. Chem. 2021, 134, 116134. [Google Scholar] [CrossRef]
- Kirchhain, A.; Bonini, A.; Vivaldi, F.; Poma, N.; di Francesco, F. Latest Developments in Non-Faradic Impedimetric Biosensors: Towards Clinical Applications. TrAC Trends in Anal. Chem. 2020, 133, 116073. [Google Scholar] [CrossRef]
- Dincer, C.; Bruch, R.; Costa-Rama, E.; Fernández-Abedul, M.T.; Merkoçi, A.; Manz, A.; Urban, G.A.; Güder, F. Disposable Sensors in Diagnostics, Food, and Environmental Monitoring. Adv. Mater. 2019, 31, 1806739. [Google Scholar] [CrossRef]
- Vivaldi, F.M.; Dallinger, A.; Bonini, A.; Poma, N.; Sembranti, L.; Biagini, D.; Salvo, P.; Greco, F.; di Francesco, F. Three-Dimensional (3D) Laser-Induced Graphene: Structure, Properties, and Application to Chemical Sensing. ACS Appl. Mater. Interfaces 2021, 13, 30245–30260. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.M.; Ryu, M.Y.; Kim, J.H.; Cho, C.H.; Park, T.J.; Park, J.P. An Electrochemical Biosensor for Detection of the Sepsis-Related Biomarker Procalcitonin. RSC Adv. 2017, 7, 36562–36565. [Google Scholar] [CrossRef]
- Molinero-Fernández, Á.; Moreno-Guzmán, M.; López, M.Á.; Escarpa, A. Magnetic Bead-Based Electrochemical Immunoassays On-Drop and On-Chip for Procalcitonin Determination: Disposable Tools for Clinical Sepsis Diagnosis. Biosensors 2020, 10, 66. [Google Scholar] [CrossRef]
- Molinero-Fernández, Á.; Arruza, L.; López, M.Á.; Escarpa, A. On-the-Fly Rapid Immunoassay for Neonatal Sepsis Diagnosis: C-Reactive Protein Accurate Determination Using Magnetic Graphene-Based Micromotors. Biosens. Bioelectron. 2020, 158, 112156. [Google Scholar] [CrossRef]
- Guillem, P.; Bustos, R.H.; Garzon, V.; Munoz, A.; Juez, G. A Low-Cost Electrochemical Biosensor Platform for C-Reactive Protein Detection. Sens. Biosens. Res. 2021, 31, 100402. [Google Scholar] [CrossRef]
- Ge, X.Y.; Zhang, J.X.; Feng, Y.G.; Wang, A.J.; Mei, L.P.; Feng, J.J. Label-Free Electrochemical Biosensor for Determination of Procalcitonin Based on Graphene-Wrapped Co Nanoparticles Encapsulated in Carbon Nanobrushes Coupled with AuPtCu Nanodendrites. Microchim. Acta 2022, 189, 110. [Google Scholar] [CrossRef]
- Wang, X.Y.; Feng, Y.G.; Wang, A.J.; Mei, L.P.; Luo, X.; Xue, Y.; Feng, J.J. Facile Construction of Ratiometric Electrochemical Immunosensor Using Hierarchical PtCoIr Nanowires and Porous SiO2@Ag Nanoparticles for Accurate Detection of Septicemia Biomarker. Bioelectrochemistry 2021, 140, 107802. [Google Scholar] [CrossRef]
- Miao, J.; Du, K.; Li, X.; Xu, X.; Dong, X.; Fang, J.; Cao, W.; Wei, Q. Ratiometric Electrochemical Immunosensor for the Detection of Procalcitonin Based on the Ratios of SiO2-Fc–COOH–Au and UiO-66-TB Complexes. Biosens. Bioelectron. 2021, 171, 112713. [Google Scholar] [CrossRef]
- Tanak, A.S.; Jagannath, B.; Tamrakar, Y.; Muthukumar, S.; Prasad, S. Non-Faradaic Electrochemical Impedimetric Profiling of Procalcitonin and C-Reactive Protein as a Dual Marker Biosensor for Early Sepsis Detection. Anal. Chim. Acta X 2019, 3, 100029. [Google Scholar] [CrossRef]
- Zupančič, U.; Jolly, P.; Estrela, P.; Moschou, D.; Ingber, D.E. Graphene Enabled Low-Noise Surface Chemistry for Multiplexed Sepsis Biomarker Detection in Whole Blood. Adv. Funct. Mater. 2021, 31, 2010638. [Google Scholar] [CrossRef]
- Russell, C.; Ward, A.C.; Vezza, V.; Hoskisson, P.; Alcorn, D.; Steenson, D.P.; Corrigan, D.K. Development of a Needle Shaped Microelectrode for Electrochemical Detection of the Sepsis Biomarker Interleukin-6 (IL-6) in Real Time. Biosens. Bioelectron. 2019, 126, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gopinath, S.C.B.; Anbu, P. Longitudinal Zeolite-Iron Oxide Nanocomposite Deposited Capacitance Biosensor for Interleukin-3 in Sepsis Detection. Nanoscale Res. Lett. 2021, 16, 68. [Google Scholar] [CrossRef]
- Min, J.; Nothing, M.; Coble, B.; Zheng, H.; Park, J.; Im, H.; Weber, G.F.; Castro, C.M.; Swirski, F.K.; Weissleder, R.; et al. Integrated Biosensor for Rapid and Point-of-Care Sepsis Diagnosis. ACS Nano 2018, 12, 3378–3384. [Google Scholar] [CrossRef] [PubMed]
- Tanak, A.S.; Muthukumar, S.; Krishnan, S.; Schully, K.L.; Clark, D.V.; Prasad, S. Multiplexed Cytokine Detection Using Electrochemical Point-of-Care Sensing Device towards Rapid Sepsis Endotyping. Biosens. Bioelectron. 2021, 171, 112726. [Google Scholar] [CrossRef] [PubMed]
- Tanak, A.S.; Sardesai, A.; Muthukumar, S.; Krishnan, S.; Striegel, D.A.; Schully, K.L.; Clark, D.V.; Prasad, S. Multiplexed Host Immune Response Biosensor for Rapid Sepsis Stratification and Endotyping at Point-of-Care. Biosens. Bioelectron. X 2022, 10, 100144. [Google Scholar] [CrossRef]
- Gao, J.; Jeffries, L.; Mach, K.E.; Craft, D.W.; Thomas, N.J.; Gau, V.; Liao, J.C.; Wong, P.K. A Multiplex Electrochemical Biosensor for Bloodstream Infection Diagnosis. SLAS Technol. 2017, 22, 466–474. [Google Scholar] [CrossRef]
- Sharma, R.; Lakshmi, G.B.V.S.; Kumar, A.; Solanki, P. Polypyrrole Based Molecularly Imprinted Polymer Platform for Klebsiella Pneumonia Detection. ECS Sens. Plus 2022, 1, 010603. [Google Scholar] [CrossRef]
- Bonini, A.; Poma, N.; Vivaldi, F.; Biagini, D.; Bottai, D.; Tavanti, A.; di Francesco, F. A Label-Free Impedance Biosensing Assay Based on CRISPR/Cas12a Collateral Activity for Bacterial DNA Detection. J. Pharm. Biomed. Anal. 2021, 204, 114268. [Google Scholar] [CrossRef]
- Oved, K.; Cohen, A.; Boico, O.; Navon, R.; Friedman, T.; Etshtein, L.; Kriger, O.; Bamberger, E.; Fonar, Y.; Yacobov, R.; et al. A Novel Host-Proteome Signature for Distinguishing between Acute Bacterial and Viral Infections. PLoS ONE 2015, 10, e0120012. [Google Scholar] [CrossRef] [PubMed]
- Bonini, A.; Poma, N.; Vivaldi, F.; Kirchhain, A.; Salvo, P.; Bottai, D.; Tavanti, A.; di Francesco, F. Advances in Biosensing: The CRISPR/Cas System as a New Powerful Tool for the Detection of Nucleic Acids. J. Pharm. Biomed. Anal. 2021, 192, 113645. [Google Scholar] [CrossRef]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical Biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [PubMed]
- Vivaldi, F.; Salvo, P.; Poma, N.; Bonini, A.; Biagini, D.; del Noce, L.; Melai, B.; Lisi, F.; di Francesco, F. Recent Advances in Optical, Electrochemical and Field Effect PH Sensors. Chemosensors 2021, 9, 33. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J. Optical Biosensors: An Exhaustive and Comprehensive Review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.T.; Lee, Y.C.; Lai, Y.H.; Lim, J.C.; Huang, N.T.; Lin, C.T.; Huang, J.J. Review of Integrated Optical Biosensors for Point-of-Care Applications. Biosensors 2020, 10, 209. [Google Scholar] [CrossRef]
- Guo, X. Surface Plasmon Resonance Based Biosensor Technique: A Review. J. Biophotonics 2012, 5, 483–501. [Google Scholar] [CrossRef]
- Wang, W.; Mai, Z.; Chen, Y.; Wang, J.; Li, L.; Su, Q.; Li, X.; Hong, X. A Label-Free Fiber Optic SPR Biosensor for Specific Detection of C-Reactive Protein. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Chiang, C.Y.; Huang, T.T.; Wang, C.H.; Huang, C.J.; Tsai, T.H.; Yu, S.N.; Chen, Y.T.; Hong, S.W.; Hsu, C.W.; Chang, T.C.; et al. Fiber Optic Nanogold-Linked Immunosorbent Assay for Rapid Detection of Procalcitonin at Femtomolar Concentration Level. Biosens. Bioelectron. 2020, 151, 111871. [Google Scholar] [CrossRef]
- Giorgi-Coll, S.; Marín, M.J.; Sule, O.; Hutchinson, P.J.; Carpenter, K.L.H. Aptamer-Modified Gold Nanoparticles for Rapid Aggregation-Based Detection of Inflammation: An Optical Assay for Interleukin-6. Microchim. Acta 2020, 187, 13. [Google Scholar] [CrossRef]
- Fabri-Faja, N.; Calvo-Lozano, O.; Dey, P.; Terborg, R.A.; Estevez, M.C.; Belushkin, A.; Yesilköy, F.; Duempelmann, L.; Altug, H.; Pruneri, V.; et al. Early Sepsis Diagnosis via Protein and MiRNA Biomarkers Using a Novel Point-of-Care Photonic Biosensor. Anal. Chim. Acta 2019, 1077, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Santopolo, G.; Doménech-Sánchez, A.; Russell, S.M.; de La Rica, R. Ultrafast and Ultrasensitive Naked-Eye Detection of Urease-Positive Bacteria with Plasmonic Nanosensors. ACS Sens. 2019, 4, 961–967. [Google Scholar] [CrossRef]
- Zhou, X.; Li, P.; Wu, X.; Lin, X.; Zhao, L.; Huang, H.; Wu, J.; Cai, H.; Xu, M.; Zhou, H.; et al. Multifunctional Biosensor Constructed by Ag-Coating Magnetic-Assisted Unique Urchin Core Porous Shell Structure for Dual SERS Enhancement, Enrichment, and Quantitative Detection of Multi-Components Inflammatory Markers. Biosens. Bioelectron. 2022, 210, 114257. [Google Scholar] [CrossRef] [PubMed]
- Narayana Iyengar, S.; Dietvorst, J.; Ferrer-Vilanova, A.; Guirado, G.; Muñoz-Berbel, X.; Russom, A. Toward Rapid Detection of Viable Bacteria in Whole Blood for Early Sepsis Diagnostics and Susceptibility Testing. ACS Sens. 2021, 6, 3357–3366. [Google Scholar] [CrossRef] [PubMed]
- Kundu, A.; Rani, R.; Ahmad, A.; Kumar, A.; Raturi, M.; Gupta, T.; Khan, R.; Hazra, K.S. Ultrasensitive and Label-Free Detection of Prognostic and Diagnostic Biomarkers of Sepsis on a AgNP-Laden Black Phosphorous-Based SERS Platform. Sens. Diagn. 2022, 1, 449–459. [Google Scholar] [CrossRef]
- Tsounidi, D.; Koukouvinos, G.; Christianidis, V.; Legaki, E.; Giogli, V.; Panagiotopoulou, K.; Taka, S.; Ekaterinidi, Z.; Kakabakos, S.; Raptis, I.; et al. Development of a Point-of-Care System Based on White Light Reflectance Spectroscopy: Application in Crp Determination. Biosensors 2021, 11, 268. [Google Scholar] [CrossRef]
- Willets, K.A.; van Duyne, R.P. Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu. Rev. Phys. Chem. 2007, 58, 267–297. [Google Scholar] [CrossRef]
- Cao, J.; Galbraith, E.K.; Sun, T.; Grattan, K.T.V. Comparison of Surface Plasmon Resonance and Localized Surface Plasmon Resonance-Based Optical Fibre Sensors. J. Phys. Conf. Ser. 2011, 307, 012050. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).