Optimized Identification of Advanced Chronic Kidney Disease and Absence of Kidney Disease by Combining Different Electronic Health Data Resources and by Applying Machine Learning Strategies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Classification of CKD and NKD by ICD-10 Billing Codes
2.3. Laboratory and Demographic Data
- -
- Numerical variables: age, eGFR at admission, eGFR at discharge, eGFR over index hospital stay. Measurements of albumin in urine were available in less than 5% of the cohort and therefore excluded from further analysis.
- -
- Categorical variable: sex.
2.4. Classification of CKD and NKD by Blood Creatinine and eGFR
2.5. Classification of CKD and NKD by Manual Review
2.6. Dataset for the Machine Learning Methods
- Numerical variables: age; first eGFR of the index hospital stay; last eGFR of the index hospital stay; time difference between the first and last blood measurement of the index hospital stay as an indicator for the length of hospital stay; mean eGFR over index hospital stay; mean eGFR over all available laboratory values.
- Due to the varying distribution of eGFR measurements, additionally derived numerical variables were defined for usage in ML algorithms: the ratio between the number of hospital visits with eGFR measurements and the number of total visits; the ratio between the number of total eGFR measurements and hospital visits with eGFR measurements; the ratio between the number of eGFR measurements lower than 60 mL/min/1.73 m2 and hospital visits with eGFR measurements.
- Categorical variables: sex; occurrence of AKI and AKI recovery over laboratory history; occurrence of AKI and AKI recovery over index stay.
2.7. Classification of CKD and NKD Using Machine Learning Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, J.; Wang, F.; Saran, R.; He, Z.; Zhao, M.H.; Li, Y.; Zhang, L.; Bragg-Gresham, J. Mortality risk of chronic kidney disease: A comparison between the adult populations in urban China and the United States. PLoS ONE 2018, 13, e0193734. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Bowe, B.; Mokdad, A.H.; Xian, H.; Yan, Y.; Li, T.; Maddukuri, G.; Tsai, C.Y.; Floyd, T.; Al-Aly, Z. Analysis of the Global Burden of Disease study highlights the global, regional, and national trends of chronic kidney disease epidemiology from 1990 to 2016. Kidney Int. 2018, 94, 567–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kidney Disease: Improving Global Outcomes (KDIGO) CKD Work Group. KDIGO 2012 Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease. Kidney Int. Suppl. 2013, 3, 1–150. [Google Scholar]
- Anderson, J.; Glynn, L.G. Definition of chronic kidney disease and measurement of kidney function in original research papers: A review of the literature. Nephrol. Dial. Transplant. 2011, 26, 2793–2798. [Google Scholar] [CrossRef] [PubMed]
- Jalal, K.; Anand, E.J.; Venuto, R.; Eberle, J.; Arora, P. Can billing codes accurately identify rapidly progressing stage 3 and stage 4 chronic kidney disease patients: A diagnostic test study. BMC Nephrol. 2019, 20, 260. [Google Scholar] [CrossRef] [Green Version]
- Vlasschaert, M.E.; Bejaimal, S.A.; Hackam, D.G.; Quinn, R.; Cuerden, M.S.; Oliver, M.J.; Iansavichus, A.; Sultan, N.; Mills, A.; Garg, A.X. Validity of administrative database coding for kidney disease: A systematic review. Am. J. Kidney Dis. 2011, 57, 29–43. [Google Scholar] [CrossRef]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.L.; Castro, A.F., 3rd; Feldman, H.I.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Greene, T.; et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Jurkovitz, C.; Shatkay, H. Co-occurrence of medical conditions: Exposing patterns through probabilistic topic modeling of snomed codes. J. Biomed. Inform. 2018, 82, 31–40. [Google Scholar] [CrossRef]
- Singh, B.; Singh, A.; Ahmed, A.; Wilson, G.A.; Pickering, B.W.; Herasevich, V.; Gajic, O.; Li, G. Derivation and validation of automated electronic search strategies to extract Charlson comorbidities from electronic medical records. Mayo Clin. Proc. 2012, 87, 817–824. [Google Scholar] [CrossRef] [Green Version]
- Upadhyaya, S.G.; Murphree, D.H., Jr.; Ngufor, C.G.; Knight, A.M.; Cronk, D.J.; Cima, R.R.; Curry, T.B.; Pathak, J.; Carter, R.E.; Kor, D.J. Automated Diabetes Case Identification Using Electronic Health Record Data at a Tertiary Care Facility. Mayo Clin. Proc. Innov. Qual. Outcomes 2017, 1, 100–110. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Lou, Y.S.; Tsai, D.J.; Lee, C.C.; Hsu, C.J.; Wu, D.C.; Wang, M.C.; Fang, W.H. Projection Word Embedding Model With Hybrid Sampling Training for Classifying ICD-10-CM Codes: Longitudinal Observational Study. JMIR Med. Inform. 2019, 7, e14499. [Google Scholar] [CrossRef] [PubMed]
- Batool, R.; Khattak, A.M.; Kim, T.-S.; Lee, S. Automatic extraction and mapping of discharge summary’s concepts into SNOMED CT. In Proceedings of the 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Osaka, Japan, 3–7 July 2013. [Google Scholar]
- Tang, B.; Cao, H.; Wu, Y.; Jiang, M.; Xu, H. Recognizing clinical entities in hospital discharge summaries using Structural Support Vector Machines with word representation features. BMC Med. Inform. Decis. Mak. 2013, 13 (Suppl. 1), S1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahu, S.K.; Anand, A.; Oruganty, K.; Gattu, M. Relation extraction from clinical texts using domain invariant convolutional neural network. In Proceedings of the 15th Workshop on Biomedical Natural Language Processing, BioNLP@ACL 2016, Berlin, Germany, 12 August 2016; pp. 206–215. [Google Scholar]
- Xiao, J.; Ding, R.; Xu, X.; Guan, H.; Feng, X.; Sun, T.; Zhu, S.; Ye, Z. Comparison and development of machine learning tools in the prediction of chronic kidney disease progression. J. Transl. Med. 2019, 17, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polat, H.; Danaei Mehr, H.; Cetin, A. Diagnosis of Chronic Kidney Disease Based on Support Vector Machine by Feature Selection Methods. J. Med. Syst. 2017, 41, 55. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, Z.; Zhu, R.; Xiang, Y.; Harrington, P.B. Diagnosis of patients with chronic kidney disease by using two fuzzy classifiers. Chemom. Intell. Lab. Syst. 2016, 153, 140–145. [Google Scholar] [CrossRef]
- Alexander Arman, S. Diagnosis Rule Extraction from Patient Data for Chronic Kidney Disease Using Machine Learning. Int. J. Biomed. Clin. Eng. IJBCE 2016, 5, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Elhoseny, M.; Shankar, K.; Uthayakumar, J. Intelligent Diagnostic Prediction and Classification System for Chronic Kidney Disease. Sci. Rep. 2019, 9, 9583. [Google Scholar] [CrossRef]
- Almansour, N.A.; Syed, H.F.; Khayat, N.R.; Altheeb, R.K.; Juri, R.E.; Alhiyafi, J.; Alrashed, S.; Olatunji, S.O. Neural network and support vector machine for the prediction of chronic kidney disease: A comparative study. Comput. Biol. Med. 2019, 109, 101–111. [Google Scholar] [CrossRef]
- Winter, A.; Staubert, S.; Ammon, D.; Aiche, S.; Beyan, O.; Bischoff, V.; Daumke, P.; Decker, S.; Funkat, G.; Gewehr, J.E.; et al. Smart Medical Information Technology for Healthcare (SMITH). Methods Inf. Med. 2018, 57, e92–e105. [Google Scholar] [CrossRef] [Green Version]
- Hahn, U.; Matthies, F.; Lohr, C.; Loffler, M. 3000PA-Towards a National Reference Corpus of German Clinical Language. Stud. Health Technol. Inform. 2018, 247, 26–30. [Google Scholar]
- Lohr, C.; Luther, S.; Matthies, F.; Modersohn, L.; Ammon, D.; Saleh, K.; Henkel, A.G.; Kiehntopf, M.; Hahn, U. CDA-Compliant Section Annotation of German-Language Discharge Summaries: Guideline Development, Annotation Campaign, Section Classification. AMIA Annu. Symp. Proc. 2018, 2018, 770–779. [Google Scholar] [PubMed]
- Quan, H.; Sundararajan, V.; Halfon, P.; Fong, A.; Burnand, B.; Luthi, J.C.; Saunders, L.D.; Beck, C.A.; Feasby, T.E.; Ghali, W.A. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med. Care 2005, 43, 1130–1139. [Google Scholar] [CrossRef] [PubMed]
- James, M.T.; Levey, A.S.; Tonelli, M.; Tan, Z.; Barry, R.; Pannu, N.; Ravani, P.; Klarenbach, S.W.; Manns, B.J.; Hemmelgarn, B.R. Incidence and Prognosis of Acute Kidney Diseases and Disorders Using an Integrated Approach to Laboratory Measurements in a Universal Health Care System. JAMA Netw. Open 2019, 2, e191795. [Google Scholar] [CrossRef] [PubMed]
- Kidney Disease: Improving Global Outcomes AKI Work Group. KDIGO clinical practice guideline for acute kidney injury. Kidney Int. Suppl. 2012, 2, 1–138. [Google Scholar]
- Duff, S.; Murray, P.T. Defining Early Recovery of Acute Kidney Injury. Clin. J. Am. Soc. Nephrol. 2020, 15. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Lin, ear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Hagan, M.T.; Demuth, H.B.; Beale, M. Neural Network Design, 1st ed.; PWS Pub.: Boston, MA, USA, 1996. [Google Scholar]
- Boehmke, B.; Greenwell, B.M. Hands-on Machine Learning with R; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Saito, T.; Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef] [Green Version]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2019; Available online: http://www.rstudio.com/ (accessed on 12 September 2020).
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Chan, C.-H.; Chan, G.C.; Leeper, T.J.; Becker, J. Rio: A Swiss-Army Knife for Data File I/O; R package version 0.5.16; 2018. Available online: https://cran.r-project.org/web/packages/rio/index.html (accessed on 12 September 2020).
- Wickham, H. The Split-Apply-Combine Strategy for Data Analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; Team, R.C. Nlme: Linear and Nonlinear Mixed Effects Models; R package version 3.1-142; 2019. Available online: https://CRAN.R-project.org/package=nlme (accessed on 12 September 2020).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Sour. Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sing, T.; Sander, O.; Beerenwinkel, N.; Lengauer, T. ROCR: Visualizing classifier performance in R. Bioinformatics 2005, 21, 3940–3941. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Caret: Classification and Regression Training; R package version 6.0-86; 2020. Available online: https://cran.r-project.org/web/packages/caret/index.html (accessed on 12 September 2020).
- Diamantidis, C.J.; Hale, S.L.; Wang, V.; Smith, V.A.; Scholle, S.H.; Maciejewski, M.L. Lab-based and diagnosis-based chronic kidney disease recognition and staging concordance. BMC Nephrol. 2019, 20, 357. [Google Scholar] [CrossRef]
- Stevens, L.A.; Li, S.; Wang, C.; Huang, C.; Becker, B.N.; Bomback, A.S.; Brown, W.W.; Burrows, N.R.; Jurkovitz, C.T.; McFarlane, S.I.; et al. Prevalence of CKD and comorbid illness in elderly patients in the United States: Results from the Kidney Early Evaluation Program (KEEP). Am. J. Kidney Dis. 2010, 55, S23–S33. [Google Scholar] [CrossRef] [Green Version]
- Konstantinidis, I.; Nadkarni, G.N.; Yacoub, R.; Saha, A.; Simoes, P.; Parikh, C.R.; Coca, S.G. Representation of Patients With Kidney Disease in Trials of Cardiovascular Interventions: An Updated Systematic Review. JAMA Intern. Med. 2016, 176, 121–124. [Google Scholar] [CrossRef] [Green Version]
- Ronksley, P.E.; Tonelli, M.; Quan, H.; Manns, B.J.; James, M.T.; Clement, F.M.; Samuel, S.; Quinn, R.R.; Ravani, P.; Brar, S.S.; et al. Validating a case definition for chronic kidney disease using administrative data. Nephrol. Dial. Transplant. 2012, 27, 1826–1831. [Google Scholar] [CrossRef] [Green Version]
- Kern, E.F.; Maney, M.; Miller, D.R.; Tseng, C.L.; Tiwari, A.; Rajan, M.; Aron, D.; Pogach, L. Failure of ICD-9-CM codes to identify patients with comorbid chronic kidney disease in diabetes. Health Serv. Res. 2006, 41, 564–580. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Salgado, J.; Bernabeu-Wittel, M.; Aguilera-Gonzalez, C.; Goicoechea-Salazar, J.A.; Larrocha, D.; Nieto-Martin, M.D.; Moreno-Gavino, L.; Ollero-Baturone, M. Concordance between the Clinical Definition of Polypathological Patient versus Automated Detection by Means of Combined Identification through ICD-9-CM Codes. J. Clin. Med. 2019, 8, 613. [Google Scholar] [CrossRef] [Green Version]
- Chase, H.S.; Radhakrishnan, J.; Shirazian, S.; Rao, M.K.; Vawdrey, D.K. Under-documentation of chronic kidney disease in the electronic health record in outpatients. J. Am. Med. Inform. Assoc. 2010, 17, 588–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Boussard, T.; Monda, K.L.; Crespo, B.C.; Riskin, D. Real world evidence in cardiovascular medicine: Ensuring data validity in electronic health record-based studies. J. Am. Med. Inform. Assoc. 2019, 26, 1189–1194. [Google Scholar] [CrossRef] [PubMed]
- Nadkarni, G.N.; Gottesman, O.; Linneman, J.G.; Chase, H.; Berg, R.L.; Farouk, S.; Nadukuru, R.; Lotay, V.; Ellis, S.; Hripcsak, G.; et al. Development and validation of an electronic phenotyping algorithm for chronic kidney disease. AMIA Annu. Symp. Proc. 2014, 2014, 907–916. [Google Scholar] [PubMed]
- Wei, W.Q.; Leibson, C.L.; Ransom, J.E.; Kho, A.N.; Caraballo, P.J.; Chai, H.S.; Yawn, B.P.; Pacheco, J.A.; Chute, C.G. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. J. Am. Med. Inform. Assoc. 2012, 19, 219–224. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.Q.; Leibson, C.L.; Ransom, J.E.; Kho, A.N.; Chute, C.G. The absence of longitudinal data limits the accuracy of high-throughput clinical phenotyping for identifying type 2 diabetes mellitus subjects. Int. J. Med. Inform. 2013, 82, 239–247. [Google Scholar] [CrossRef] [Green Version]
- Delanaye, P.; Glassock, R.J.; De Broe, M.E. Epidemiology of chronic kidney disease: Think (at least) twice! Clin. Kidney J. 2017, 10, 370–374. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.Q.; Teixeira, P.L.; Mo, H.; Cronin, R.M.; Warner, J.L.; Denny, J.C. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J. Am. Med. Inform. Assoc. 2016, 23, e20–e27. [Google Scholar] [CrossRef]
- Salekin, A.; Stankovic, J. Detection of Chronic Kidney Disease and Selecting Important Predictive Attributes. In Proceedings of the 2016 IEEE International Conference on Healthcare Informatics (ICHI), Chicago, IL, USA, 4–7 October 2016; pp. 262–270. [Google Scholar]
- Rashidian, S.; Hajagos, J.; Moffitt, R.A.; Wang, F.; Noel, K.M.; Gupta, R.R.; Tharakan, M.A.; Saltz, J.H.; Saltz, M.M. Deep Learning on Electronic Health Records to Improve Disease Coding Accuracy. AMIA Summits Transl. Sci. Proc. 2019, 2019, 620–629. [Google Scholar]
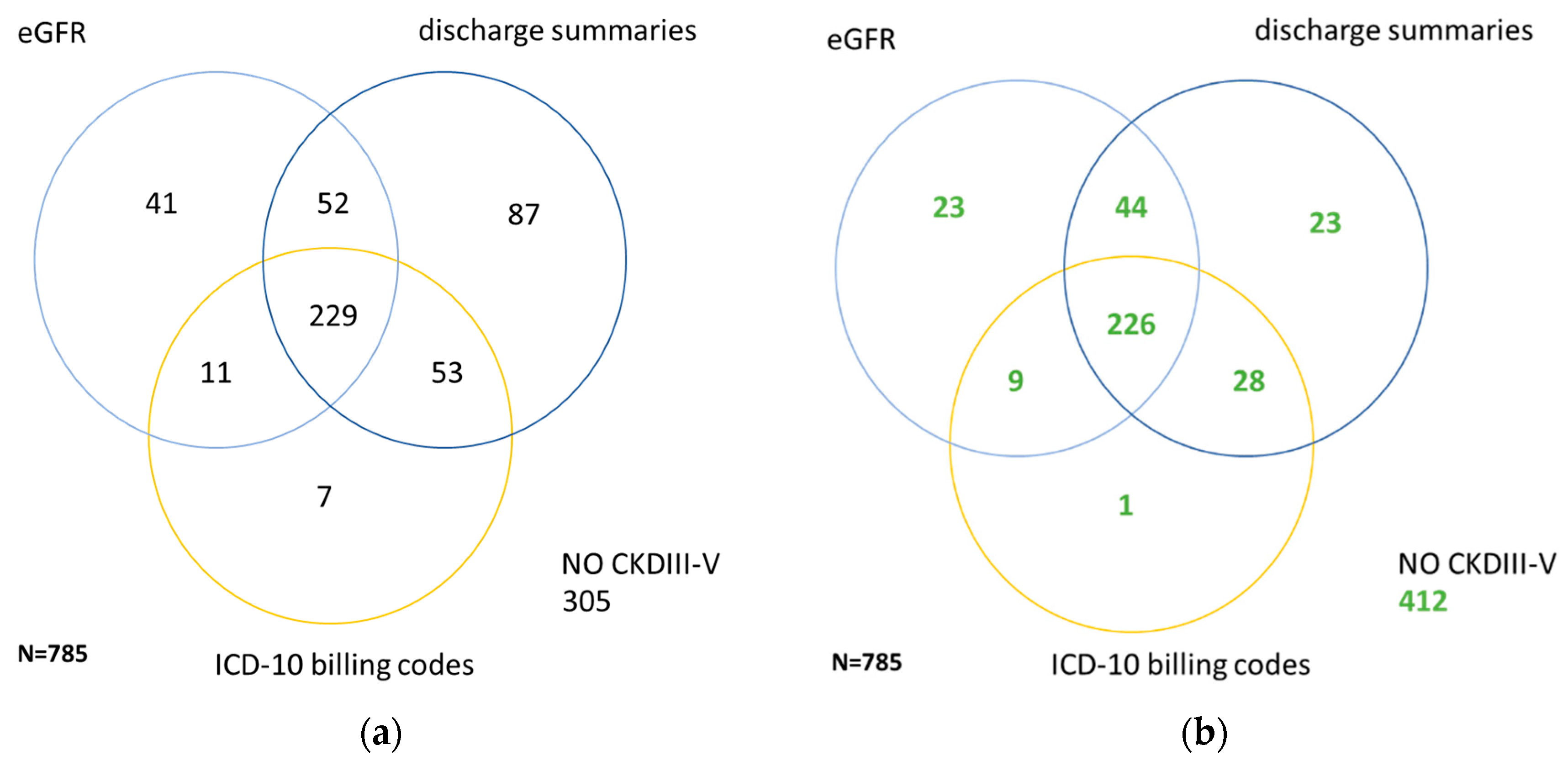
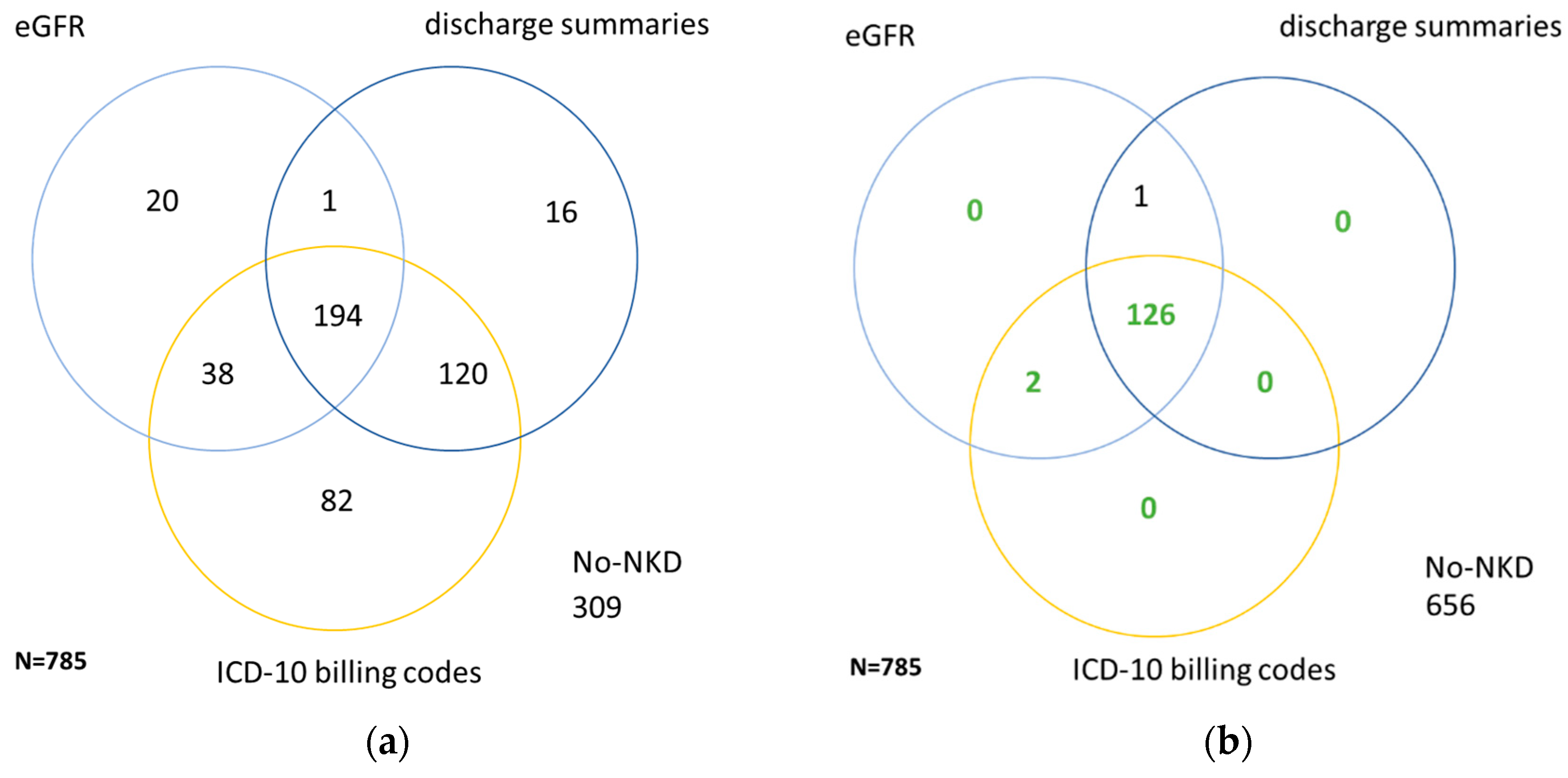

| Characteristics | Cohort (n = 785) | CKD ≥ III (n = 373) | NKD (n = 129) |
|---|---|---|---|
| Age, years, mean [SD] | 74.6 [12.2] | 77.9 [10] | 68.4 [13.7] |
| Sex, male | 476 (60.6%) | 215 (57.6%) | 79 (61.2%) |
| eGFR at admission, median, [quartiles], mL/min/1.73 m2 | (n = 780) 1 49.6 [28.6–77.3] | (n = 372) 1 28.9 [18.1–41.8] | 88.6 [78.5–99.6] |
| (n = 748) | |||
| Charlson morbidity category ≥1 | 711 (95.3%) | 366 (98.1%) | 113 (87.6%) |
| ≥3 | 387 (49.3%) | 224 (60.1%) | 36 (27.9%) |
| Median | 2 | 3 | 2 |
| Myocardial infarction | 128 (16.3%) | 75 (20.1%) | 11 (8.5%) |
| Chronic heart failure | 419 (54.4) | 247 (66.2%) | 33 (25.6%) |
| Peripheral vascular disease | 131 (16.7%) | 75 (20.1%) | 17 (13.2%) |
| Cerebrovascular disease | 51 (6.5%) | 28 (7.5%) | 7 (5.4%) |
| Dementia | 31 (3.9%) | 18 (4.8%) | 4 (3.1%) |
| Chronic pulmonary disease | 183 (23.3%) | 73 (16.9%) | 23 (17.8%) |
| Rheumatic diseases | 13 (1.7%) | 4 (1.1%) | 3 (2.3%) |
| Peptic ulcer disease | 21 (2.7%) | 11 (2.9%) | 1 (0.8%) |
| Hemiplegia or paraplegia | 29 (3.7%) | 8 (2.1%) | 6 (4.7%) |
| Liver disease | 137 (17.5%) | 44 (11.8%) | 35 (25.1%) |
| Diabetes mellitus | 332 (42.3%) | 152 (40.7%) | 51 (39.5%) |
| Any malignancy | 137 (17.5%) | 32 (8.6%) | 38 (29.5%) |
| Hypertension | 567 (72.3%) | 270 (72.4%) | 93 (72.1%) |
| Major cause for admission | |||
| Infectious diseases | 58 (7.4%) | 28 (7.5%) | 6 (4.7%) |
| Oncology disorders | 119 (15.2%) | 30 (8.0%) | 34 (26.4%) |
| Cardiovascular | 315 (40.1%) | 192 (51.5%) | 40 (31.0%) |
| Diseases | |||
| Pulmonary diseases | 82 (10.4%) | 25 (6.7%) | 12 (9.3%) |
| Gastrointestinal | 118 (15.0%) | 35 (9.4%) | 27 (20.9%) |
| and liver diseases | |||
| Kidney diseases | 47 (6.0%) | 36 (9.7%) | 2 (1.6%) |
| other | 46 (5.9%) | 27 (7.2%) | 8 (6.2%) |
| Characteristics | Reference Standard (n = 373) | eGFR (n = 333) | Discharge Summaries (n = 421) | ICD-10 Billing Codes (n = 300) |
|---|---|---|---|---|
| Age, years, mean [SD] | 77.9 [10] | 78.0 [9.7] | 76.4 [10.9] | 77.2 [10.3] |
| Sex, male | 215 (57.6%) | 189 (56.8%) | 258 (61.3%) | 182 (60.7%) |
| eGFR at admission, median, [quartiles], mL/min/1.73 m2 | (n = 372) 1 28.9 [18.1–41.8] | 26.8 [17.5–39.4] | (n = 420) 1 32.9 [19.6–50] | 25.7 [15.2–39.6] |
| Charlson morbidity category ≥1 | 366 (98.1%) | 326 (97.9%) | 413 (98.1%) | 297 (99%) |
| ≥3 | 224 (60.1%) | 198 (59.5%) | 257 (61.1%) | 220 (73.3%) |
| Median | 3 | 3 | 3 | 3 |
| Chracteristics | Reference Standard (n = 129) | eGFR (n = 253) | Discharge Summaries (n = 334) | ICD-10 Billing Codes (n = 437) |
|---|---|---|---|---|
| Age, years, mean [SD] | 68.4 [13.7] | 69.3 [13.3] | 72.9 [13.3] | 73.3 [13.0] |
| Sex, male | 79 (61.2%) | 161 (63.6%) | 196 (58.7%) | 265 (60.6%) |
| eGFR at admission, median, [quartiles], mL/min/1.73m2 | 88.6 [78.6–99.3] | 84.5 [75.7–96.2] | 76.0 *,1 [53.8–89.5] | 69.9 *,2 [50.0–87.7] |
| Charlson morbidity score ≥1 | 113 (87.6%) | 232 (91.7%) | 308 (92.2%) | 403 (92.2%) |
| ≥3 | 36 (27.9%) | 91 (36.0%) | 116 (34.7%) | 145 (33.2%) |
| Median | 2 | 2 | 2 | 2 |
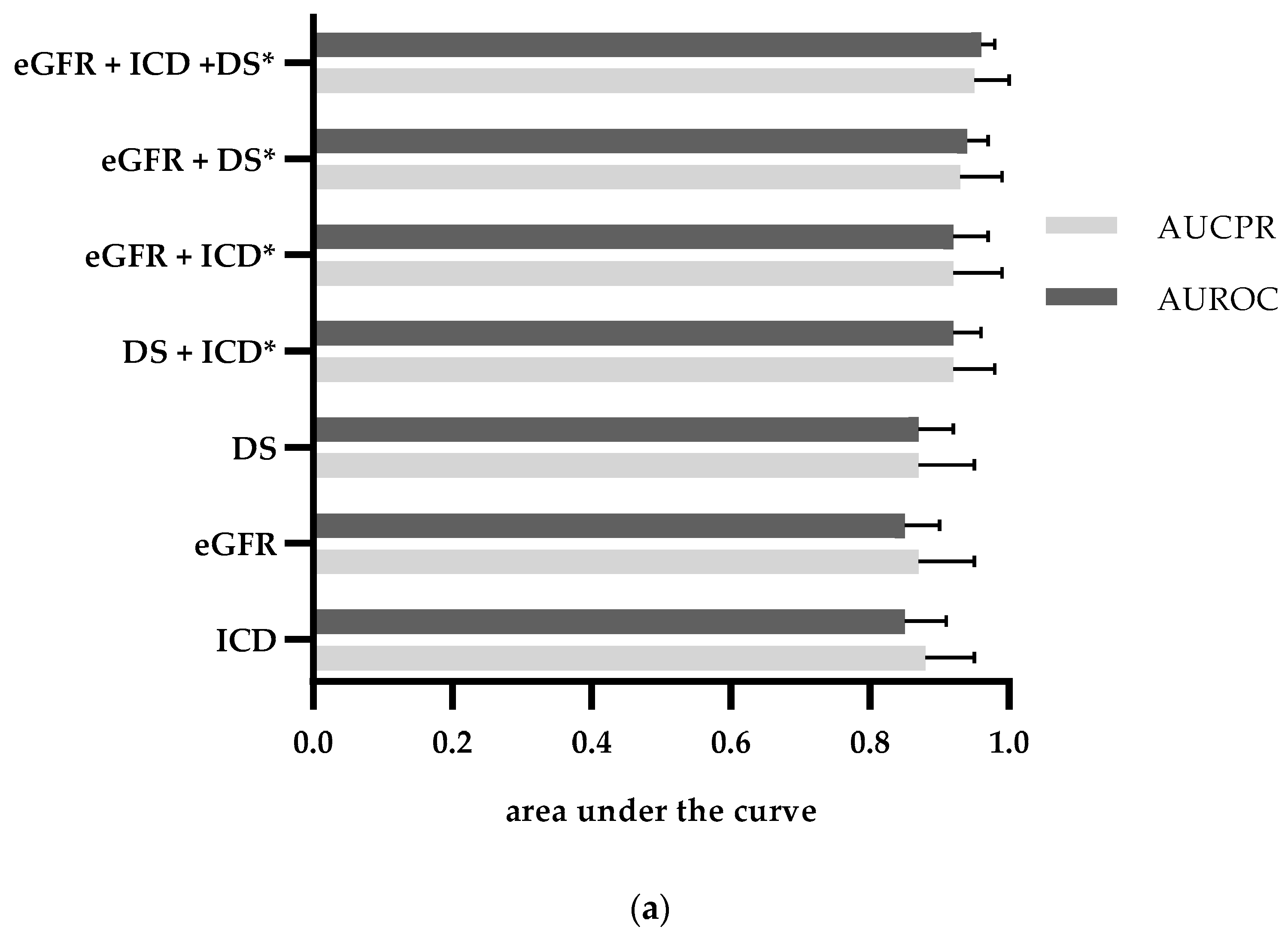
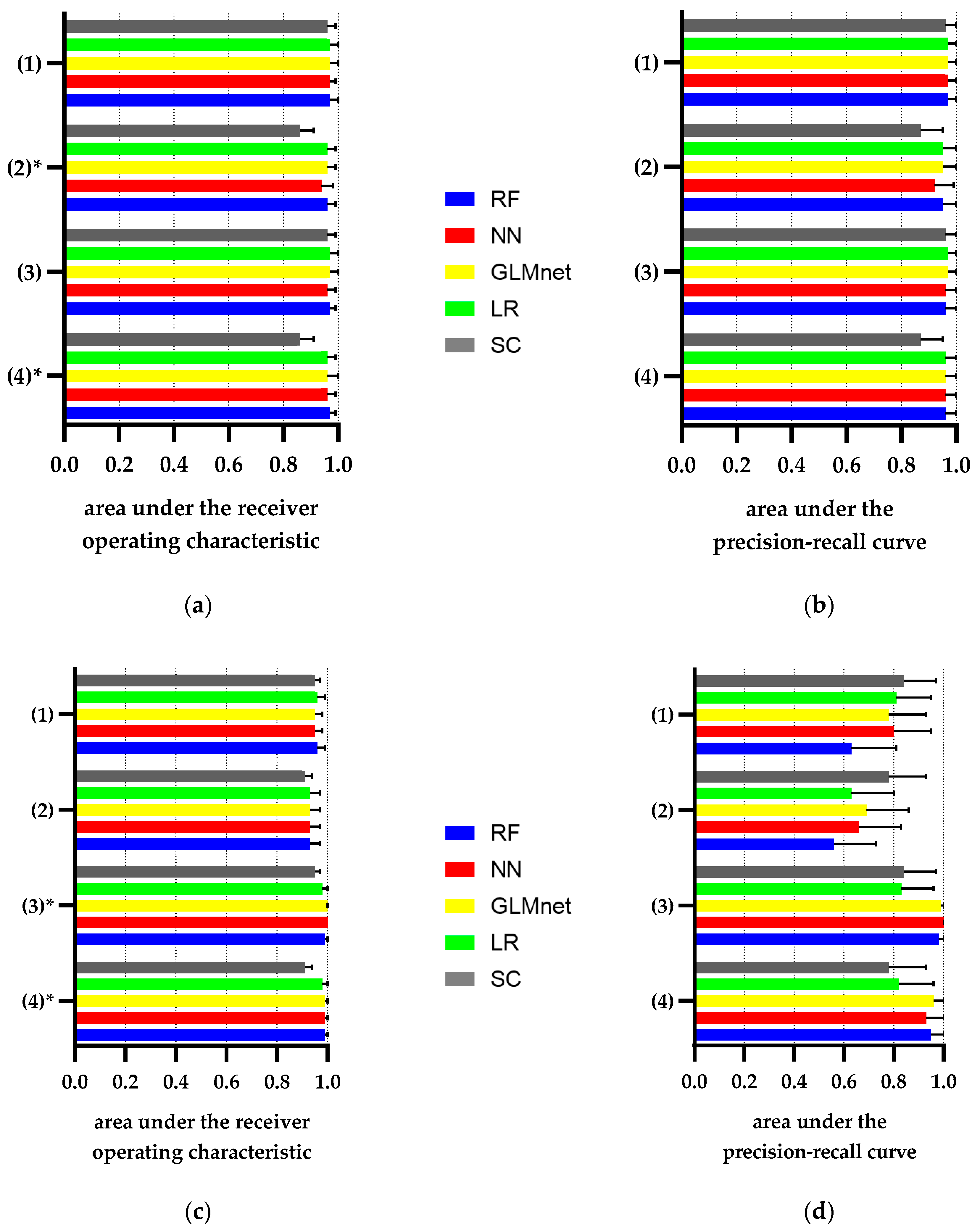
| Category | Sensitivity | Specificity | PPV | NPV | AUROC (CI) | AUCPR (CI) |
|---|---|---|---|---|---|---|
| ICD-10 billing codes | 0.71 | 0.91 | 0.88 | 0.78 | 0.81 (0.78–0.84) | 0.86 (0.83–0.90) |
| Discharge summary | 0.86 | 0.76 | 0.76 | 0.86 | 0.81 (0.78–0.84) | 0.84 (0.81–0.88) |
| eGFR <60 mL/min/1.73 m2 during Index hospital stay | 0.81 | 0.92 | 0.91 | 0.84 | 0.87 (0.84–0.90) | 0.90 (0.87–0.93) |
| eGFR_at_admission <60 mL/min/1.73 m2 | 0.96 | 0.75 | 0.77 | 0.95 | 0.85 (0.83–0.87) | 0.88 (0.84–0.91) |
| eGFR_at_discharge <60 mL/min/1.73 m2 | 0.91 | 0.82 | 0.82 | 0.91 | 0.86 (0.84–0.89) | 0.89 (0.85–0.92) |
| Category | Sensitivity | Specificity | PPV | NPV | AUROC (CI) | AUPR (CI) |
|---|---|---|---|---|---|---|
| ICD-10 billing codes | 0.99 | 0.53 | 0.29 | 1 | 0.76 | 0.64 |
| (0.74–0.78) | (0.56–0.73) | |||||
| Discharge summary | 0.98 | 0.68 | 0.38 | 1 | 0.83 | 0.68 |
| (0.81–0.86) | (0.60–0.76) | |||||
| eGFR ≥ 60 mL/min/1.73m2 during Index hospital stay | 1.00 | 0.82 | 0.52 | 1 | 0.91 (0.89–0.92) | 0.75 (0.68–0.83) |
| eGFR_at_admission ≥ 60 mL/min/1.73 m2 | 1.00 | 0.71 | 0.41 | 1.00 | 0.86 (0.84–0.87) | 0.70 (0.62–0.78) |
| eGFR_at_discharge ≥ 60 mL/min/1.73 m2 | 1.00 | 0.64 | 0.35 | 1.00 | 0.82 (0.80–0.84) | 0.68 (0.59–0.76) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weber, C.; Röschke, L.; Modersohn, L.; Lohr, C.; Kolditz, T.; Hahn, U.; Ammon, D.; Betz, B.; Kiehntopf, M. Optimized Identification of Advanced Chronic Kidney Disease and Absence of Kidney Disease by Combining Different Electronic Health Data Resources and by Applying Machine Learning Strategies. J. Clin. Med. 2020, 9, 2955. https://doi.org/10.3390/jcm9092955
Weber C, Röschke L, Modersohn L, Lohr C, Kolditz T, Hahn U, Ammon D, Betz B, Kiehntopf M. Optimized Identification of Advanced Chronic Kidney Disease and Absence of Kidney Disease by Combining Different Electronic Health Data Resources and by Applying Machine Learning Strategies. Journal of Clinical Medicine. 2020; 9(9):2955. https://doi.org/10.3390/jcm9092955
Chicago/Turabian StyleWeber, Christoph, Lena Röschke, Luise Modersohn, Christina Lohr, Tobias Kolditz, Udo Hahn, Danny Ammon, Boris Betz, and Michael Kiehntopf. 2020. "Optimized Identification of Advanced Chronic Kidney Disease and Absence of Kidney Disease by Combining Different Electronic Health Data Resources and by Applying Machine Learning Strategies" Journal of Clinical Medicine 9, no. 9: 2955. https://doi.org/10.3390/jcm9092955
APA StyleWeber, C., Röschke, L., Modersohn, L., Lohr, C., Kolditz, T., Hahn, U., Ammon, D., Betz, B., & Kiehntopf, M. (2020). Optimized Identification of Advanced Chronic Kidney Disease and Absence of Kidney Disease by Combining Different Electronic Health Data Resources and by Applying Machine Learning Strategies. Journal of Clinical Medicine, 9(9), 2955. https://doi.org/10.3390/jcm9092955




