Unveiling Wound Healing Properties of Biostimulated Walnut Kernel Extracts via Epithelial Mesenchymal Transition: Switching a Nutritional Matrix into a Therapeutic Remedy
Abstract
1. Introduction
2. Materials and Methods
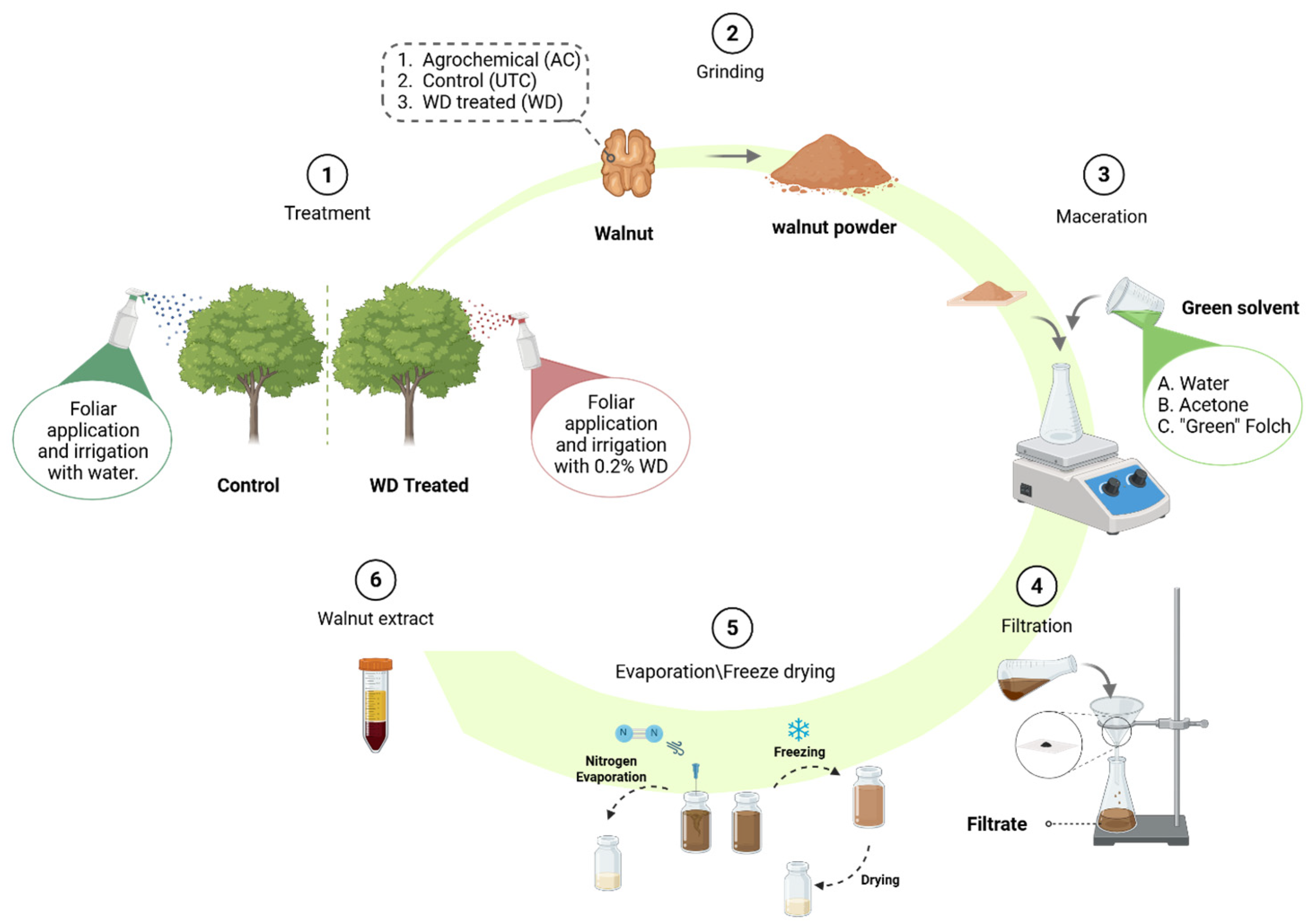
2.1. Experimental Scheme and Sample Recovery
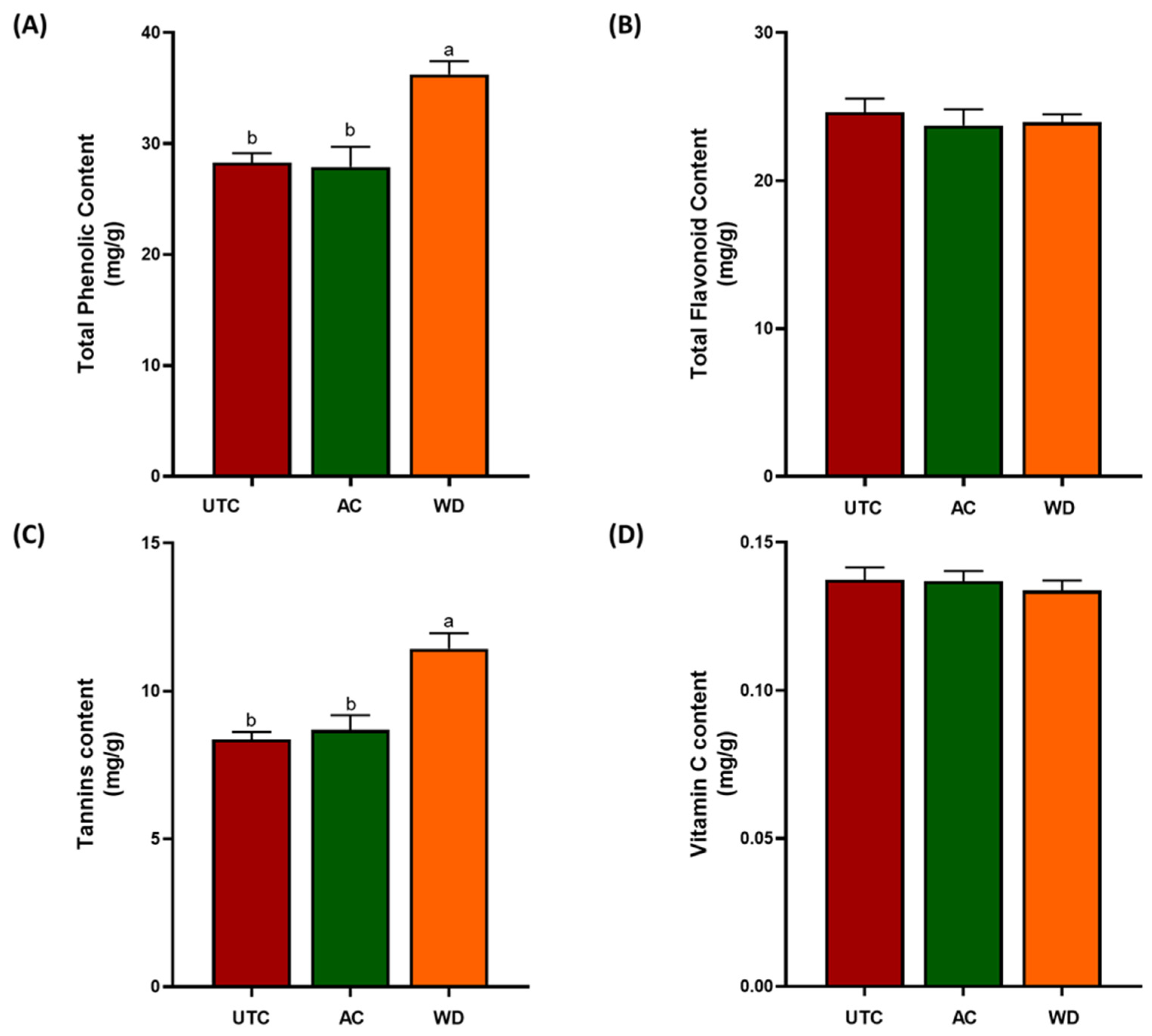
2.2. Antioxidant Compounds
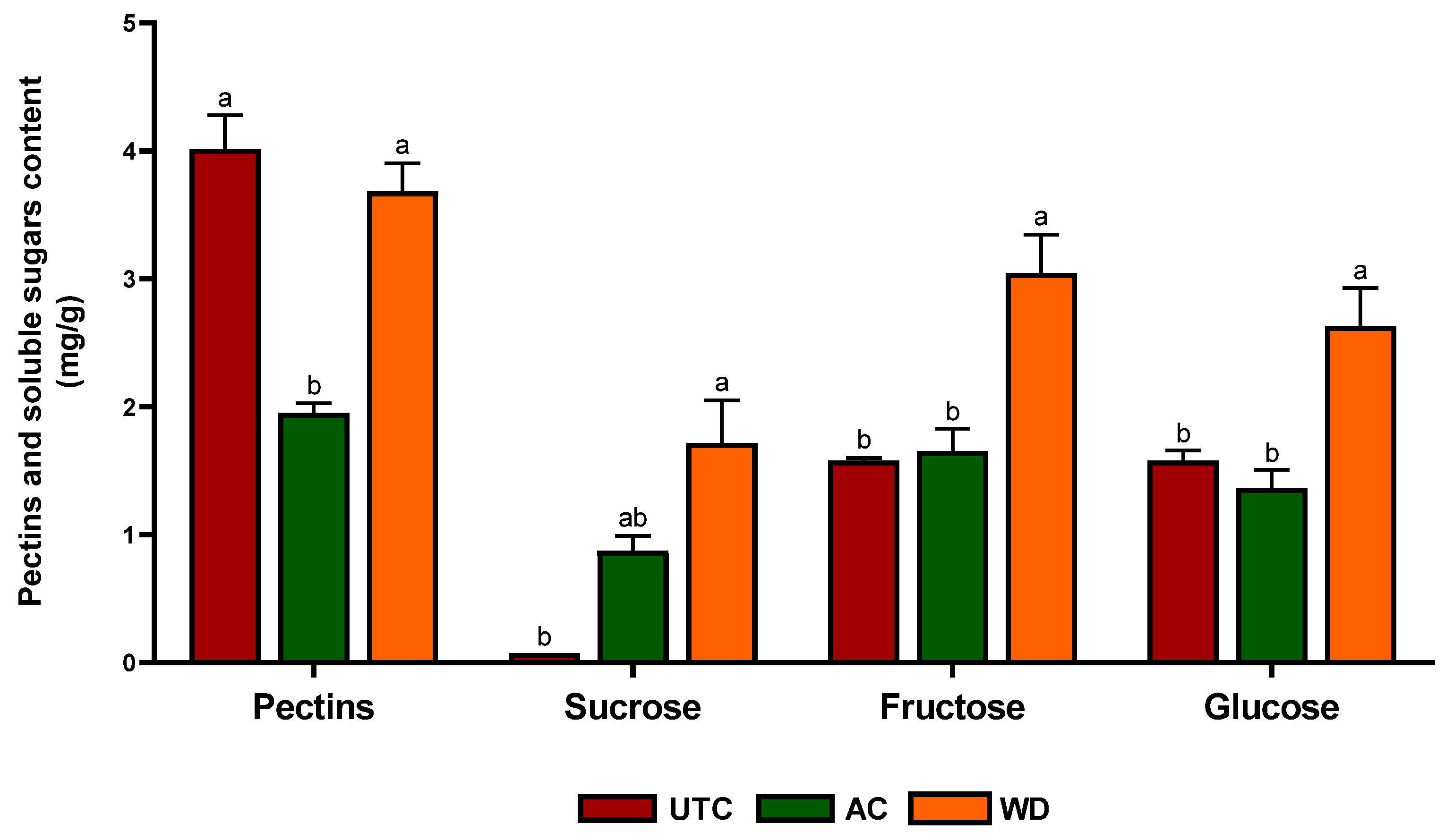
2.3. Soluble Sugars and Pectin Content
2.4. Total Soluble Protein and Free Amino Acids Content
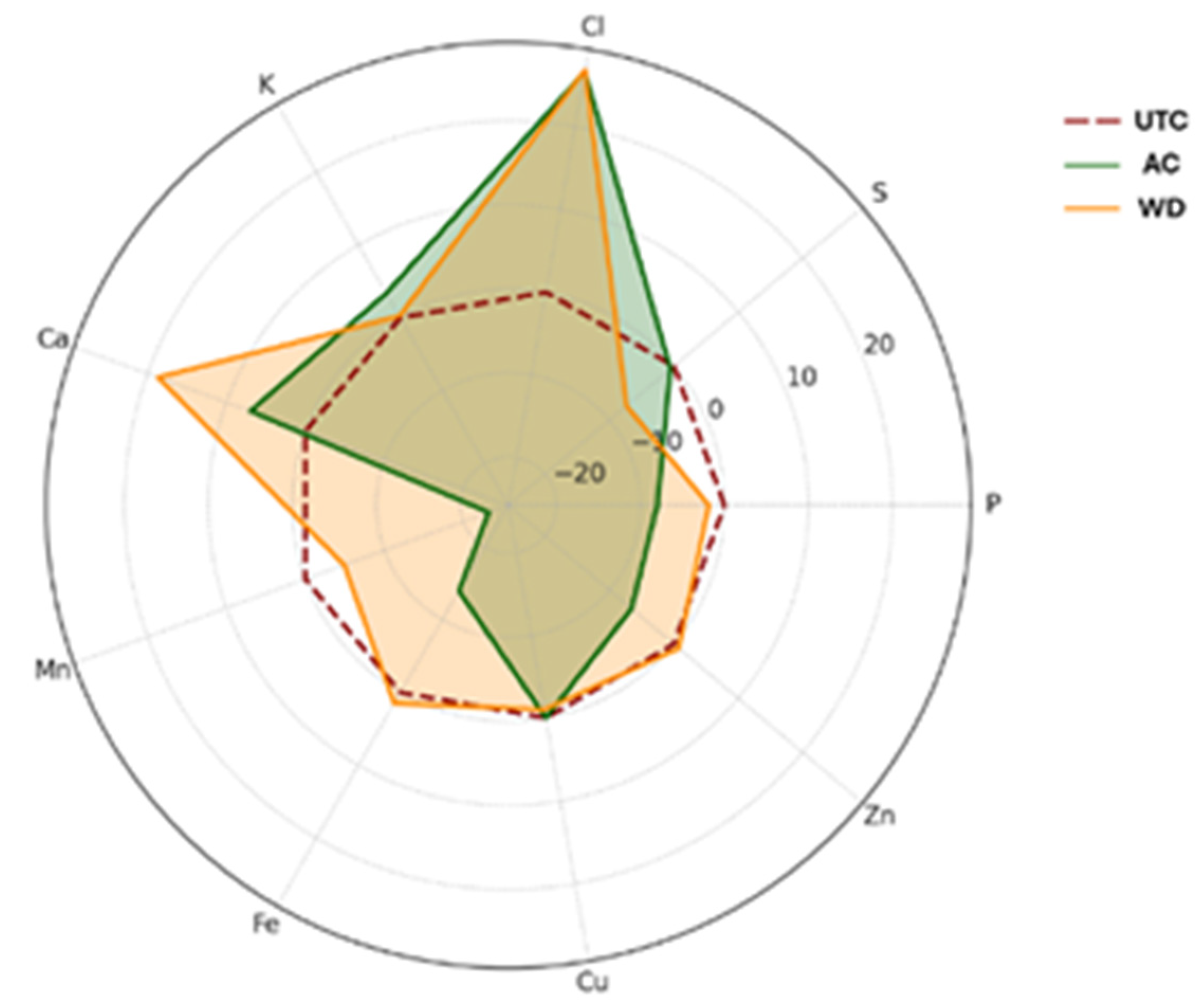
2.5. Mineral Element Content
2.6. Green Extraction of Walnut Samples
2.7. UHPLC-PDA-ESI-Orbitrap-MS/MS Analyses
2.8. Semi-Quantitative Analysis
2.9. 1H Nuclear Magnetic Resonance (NMR) Investigation
2.10. Cell Culture
2.11. Scratch Wound Assay and Analysis
2.12. Quantitative Reverse Transcriptase PCR (qRT-PCR)
2.13. Statistical Analysis
3. Results and Discussion
3.1. Nutritional Profile of Walnuts
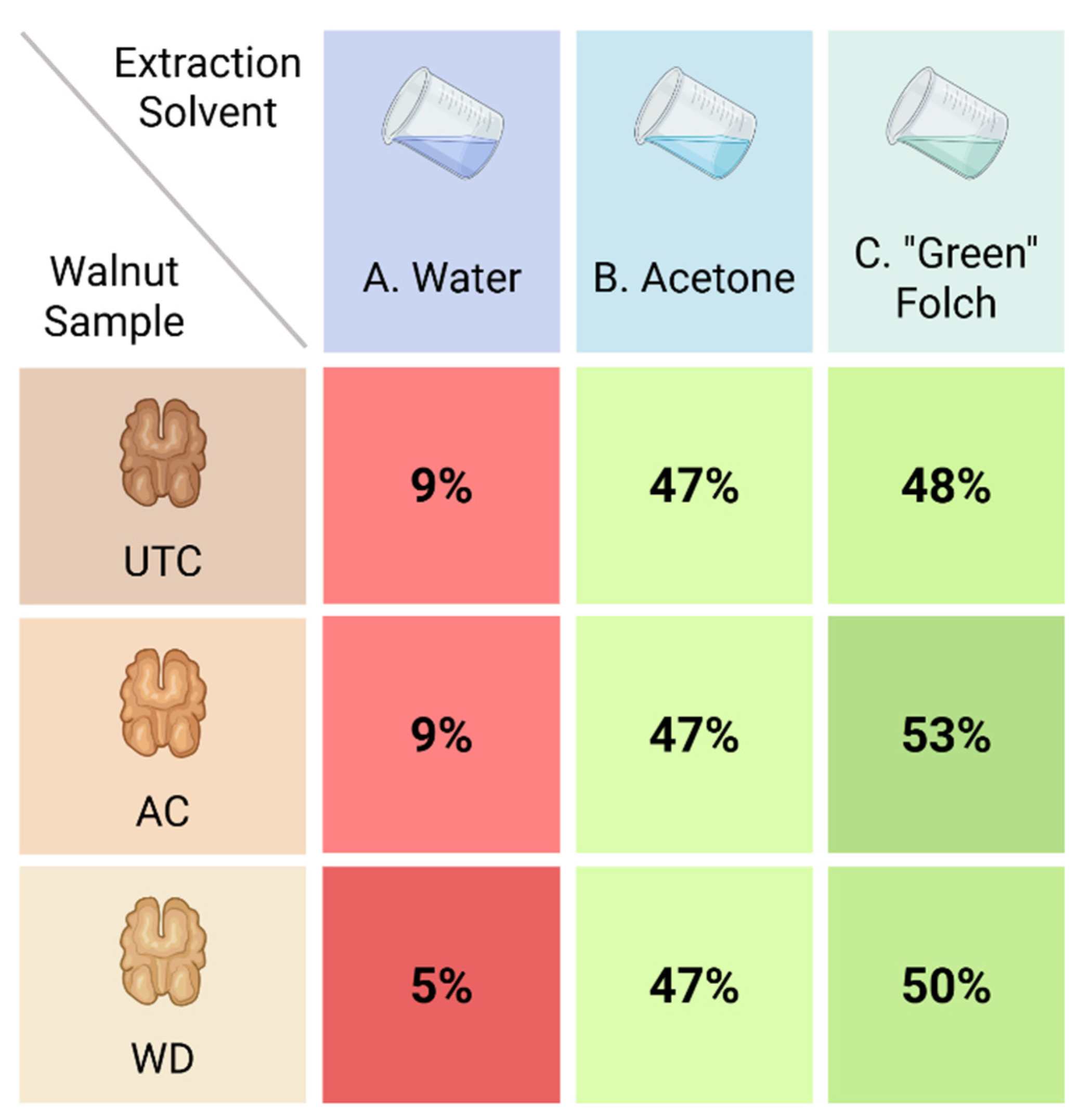
3.2. Preparation of Walnut Phytocomplexes: A Green Approach to Extraction
3.3. LC-MS/MS Identification and Quantification
3.3.1. Sinapic and p-Coumaric Acid Derivatives
3.3.2. Gallic Acid Derivatives and Tannins
3.3.3. Ellagic Acid Derivatives and Glansreginins
3.3.4. Lipid-Derived Hydroxy Fatty Acids
3.4. 1H NMR Analysis
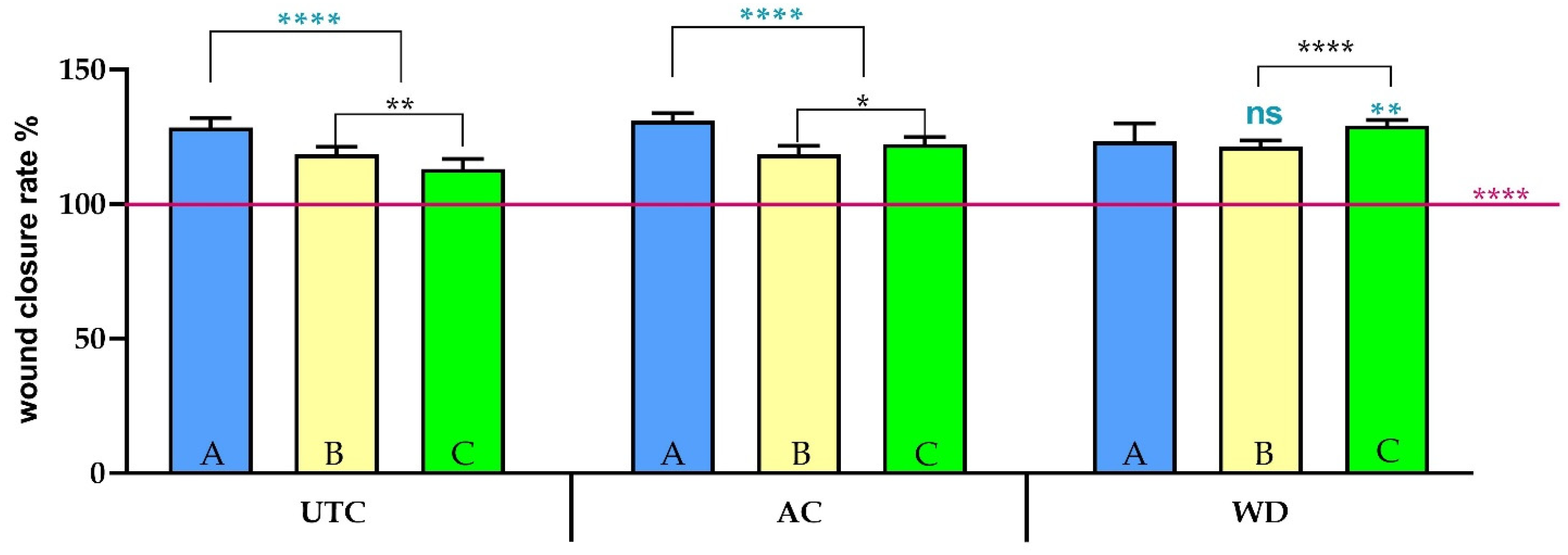
3.5. Scratch Assay Analysis of Different Phytocomplexes—Wound Closure and EMT Activation
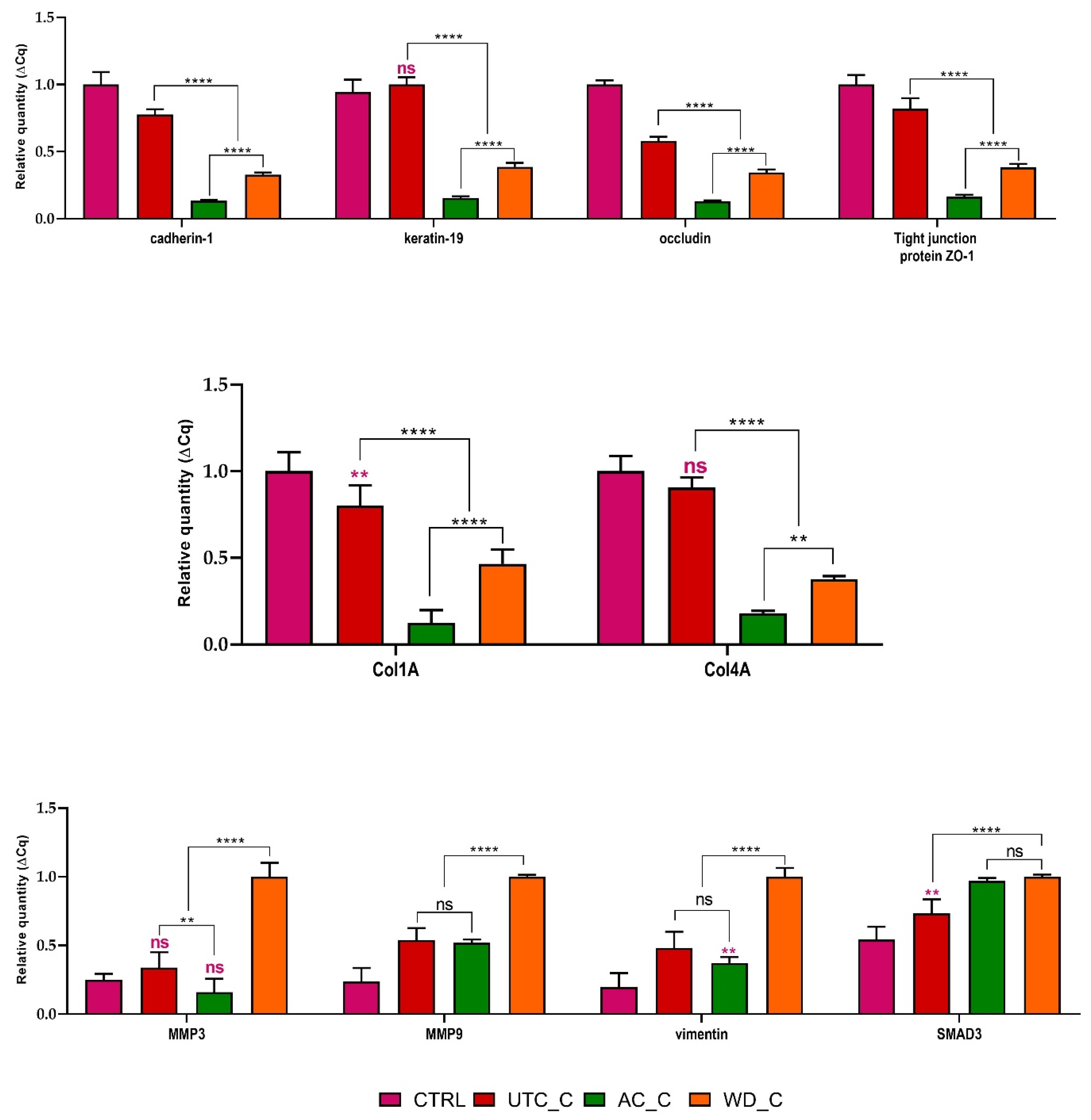
3.6. Molecular Analysis of Epithelial–Mesenchymal Transition and Extracellular Matrix Remodeling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Mukarram, S.A.; Wandhekar, S.S.; Ahmed, A.E.M.; Pandey, V.K.; Csaba, O.; Lajos, D.; József, P.; Harsányi, E.; Bela, K. Exploring the Ecological Implications, Gastronomic Applications, and Nutritional and Therapeutic Potential of Juglans regia L. (Green Walnut): A Comprehensive Review. Nutrients 2024, 16, 1183. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Lin, L.; Zhang, Y.; Li, C.; Chen, B.; Shen, Y. Effects of Walnut Kernel Pellicle on the Composition and Properties of Enzymatic Hydrolysates of Walnut Meal by Peptidomics and Bioinformatics. J. Food Sci. 2025, 90, e17604. [Google Scholar] [CrossRef]
- Ni, Z.J.; Zhang, Y.G.; Chen, S.X.; Thakur, K.; Wang, S.; Zhang, J.G.; Shang, Y.F.; Wei, Z.J. Exploration of Walnut Components and Their Association with Health Effects. Crit. Rev. Food Sci. Nutr. 2022, 62, 5113–5129. [Google Scholar] [CrossRef]
- Ros, E. Contribution of Nuts to the Mediterranean Diet. In The Mediterranean Diet: An Evidence-Based Approach; Academic Press: Cambridge, MA, USA, 2015; pp. 175–184. [Google Scholar] [CrossRef]
- Wen, C.; Zhang, Z.; Cao, L.; Liu, G.; Liang, L.; Liu, X.; Zhang, J.; Li, Y.; Yang, X.; Li, S.; et al. Walnut Protein: A Rising Source of High-Quality Protein and Its Updated Comprehensive Review. J. Agric. Food Chem. 2023, 71, 10525–10542. [Google Scholar] [CrossRef]
- Wang, W.; Li, Q.; Cui, M.; Shen, D.; Mo, R.; Tang, F.; Liu, Y. Integrating Widely Targeted Metabolomics and Network Pharmacology to Provide Insights into the Mechanism of Nutrition Changes in Walnut Kernel under Different Drying Methods. Food Chem. 2025, 485, 144498. [Google Scholar] [CrossRef]
- Colaric, M.; Veberic, R.; Solar, A.; Hudina, M.; Stampar, F. Phenolic Acids, Syringaldehyde, and Juglone in Fruits of Different Cultivars of Juglans regia L. J. Agric. Food Chem. 2005, 53, 6390–6396. [Google Scholar] [CrossRef]
- Amaral, J.S.; Casal, S.; Pereira, J.A.; Seabra, R.M.; Oliveira, B.P.P. Determination of Sterol and Fatty Acid Compositions, Oxidative Stability, and Nutritional Value of Six Walnut (Juglans regia L.) Cultivars Grown in Portugal. J. Agric. Food Chem. 2003, 51, 7698–7702. [Google Scholar] [CrossRef]
- Calcabrini, C.; De Bellis, R.; Mancini, U.; Cucchiarini, L.; Stocchi, V.; Potenza, L. Protective Effect of Juglans regia L. Walnut Extract Against Oxidative DNA Damage. Plant Foods Hum. Nutr. 2017, 72, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Zhao, Z.; Hua, Z.; Fan, J.; Du, J.; Guo, B. Evaluation of Juglans regia L., Root for Wound Healing via Antioxidant, Antimicrobial and Anti-Inflammatory Activity. Indian J. Biochem. Biophys. 2020, 57, 304–311. [Google Scholar] [CrossRef]
- Hoh, J.F.Y.; Finsterer, J.; Finsterer, J.; Taheri, A.; Mirghazanfari, S.M.; Dadpay, M. European Journal of Translational Myology. Eur. J. Transl. Myol. 2025, 30, 210–218. [Google Scholar] [CrossRef]
- Nasiry, D.; Khalatbary, A.R.; Ghaemi, A.; Ebrahimzadeh, M.A.; Hosseinzadeh, M.H. Topical Administration of Juglans regia L. Leaf Extract Accelerates Diabetic Wound Healing. BMC Complement. Med. Ther. 2022, 22, 255. [Google Scholar] [CrossRef]
- Al-Nadaf, A.H.; Awadallah, A.; Thiab, S. Superior Rat Wound-Healing Activity of Green Synthesized Silver Nanoparticles from Acetonitrile Extract of Juglans regia L.: Pellicle and Leaves. Heliyon 2024, 10, e24473. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pizá, M.C.; Sautua, F.J.; Kocira, S.; Bohatá, A.; Bedrníček, J.; Sozoniuk, M.; Carmona, M.A. Quick and Effective Evaluation Methods for Biocontrol Agents and Biostimulants Against Phytopathogenic Fungi Relevant for Various Cropping Systems. Plant Pathol. 2025, 74, 641–668. [Google Scholar] [CrossRef]
- Fedeli, R.; Dichiara, M.; Carullo, G.; Tudino, V.; Gemma, S.; Butini, S.; Campiani, G.; Loppi, S. Unlocking the Potential of Biostimulants in Sustainable Agriculture: Effect of Wood Distillate on the Nutritional Profiling of Apples. Heliyon 2024, 10, e37599. [Google Scholar] [CrossRef]
- Fedeli, R.; Marotta, L.; Frattaruolo, L.; Panti, A.; Carullo, G.; Fusi, F.; Saponara, S.; Gemma, S.; Butini, S.; Cappello, A.R.; et al. Nutritionally enriched tomatoes (Solanum lycopersicum L.) grown with wood distillate: Chemical and biological characterization for quality assessment. J. Food Sci. 2023, 88, 5324–5338. [Google Scholar] [CrossRef]
- Akley, E.K.; Ampim, P.A.Y.; Obeng, E.; Sanyare, S.; Yevu, M.; Owusu Danquah, E.; Asirifi Amoako, O.; Tengey, T.K.; Avedzi, J.K.; Avornyo, V.K.; et al. Wood vinegar promotes soil health and the productivity of cowpea. Agronomy 2023, 13, 2497. [Google Scholar] [CrossRef]
- Haensel, D.; Dai, X. Epithelial-to-Mesenchymal Transition in Cutaneous Wound Healing: Where We Are and Where We Are Heading. Dev. Dyn. 2018, 247, 473–480. [Google Scholar] [CrossRef]
- Bio-Dea. Distillato di Legno BioDea®: Estratto Vegetale Biodegradabile Ricco in Polifenoli, Tannini e Acido Acetico; Bio-Esperia Srl: Umbertide/Perugia, Italy, 2025; Available online: https://www.biodea.bio (accessed on 27 April 2025).
- Wakeel, A.; Jan, S.A.; Ullah, I.; Shinwari, Z.K.; Xu, M. Solvent Polarity Mediates Phytochemical Yield and Antioxidant Capacity of Isatis Tinctoria. PeerJ 2019, 2019, e7857. [Google Scholar] [CrossRef]
- Aparna, B.; Hema, B.P. Preliminary screening and quantification of flavonoids in selected seeds of Apiaceae by UV-visible spectrophotometry with evaluation study on different aluminium chloride complexation reaction. Indian J. Sci. Technol. 2022, 15, 857–868. [Google Scholar] [CrossRef]
- Fedeli, R.; Mazza, I.; Perini, C.; Salerni, E.; Loppi, S. New Frontiers in the Cultivation of Edible Fungi: The Application of Biostimulants Enhances the Nutritional Characteristics of Pleurotus Eryngii (DC). Quél. Agric. 2024, 14, 1012. [Google Scholar] [CrossRef]
- Fedeli, R.; Vannini, A.; Grattacaso, M.; Loppi, S. Wood Distillate (Pyroligneous Acid) Boosts Nutritional Traits of Potato Tubers. Ann. Appl. Biol. 2023, 183, 135–140. [Google Scholar] [CrossRef]
- Bogo, G.; Bortolotti, L.; Sagona, S.; Felicioli, A.; Galloni, M.; Barberis, M.; Nepi, M. Effects of non-protein amino acids in nectar on bee survival and behavior. J. Chem. Ecol. 2019, 45, 278–285. [Google Scholar] [CrossRef]
- Olympus. Vanta M Series Handheld XRF Analyzer; Olympus Scientific Solutions: Waltham, MA, USA. Available online: https://www.olympus-ims.com/en/products/xrf-analyzers/vanta (accessed on 27 April 2025).
- Fedeli, R.; Di Lella, L.A.; Loppi, S. Suitability of XRF for Routine Analysis of Multi-Elemental Composition: A Multi-Standard Verification. Methods Protoc. 2024, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Breil, C.; Abert Vian, M.; Zemb, T.; Kunz, W.; Chemat, F. “Bligh and Dyer” and Folch Methods for Solid–Liquid–Liquid Extraction of Lipids from Microorganisms. Comprehension of Solvatation Mechanisms and towards Substitution with Alternative Solvents. Int. J. Mol. Sci. 2017, 18, 708. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Carullo, G.; Sciubba, F.; Governa, P.; Mazzotta, S.; Frattaruolo, L.; Grillo, G.; Cappello, A.R.; Cravotto, G.; Di Cocco, M.E.; Aiello, F. Mantonico and Pecorello Grape Seed Extracts: Chemical Characterization and Evaluation of in Vitro Wound-Healing and Anti-Inflammatory Activities. Pharmaceuticals 2020, 13, 97. [Google Scholar] [CrossRef] [PubMed]
- Krunić, A.; Orjala, J. Application of high-field NMR spectroscopy for characterization and quantitation of submilligram quantities of isolated natural products. Magn. Reson. Chem. 2015, 53, 1043–1050. [Google Scholar] [CrossRef]
- Carullo, G.; Ahmed, A.; Fusi, F.; Sciubba, F.; Di Cocco, M.E.; Restuccia, D.; Spizzirri, U.G.; Saponara, S.; Aiello, F. Vasorelaxant Effects Induced by Red Wine and Pomace Extracts of Magliocco Dolce Cv. Pharmaceuticals 2020, 13, 87. [Google Scholar] [CrossRef]
- Farooq, H.; Courtier-Murias, D.; Soong, R.; Masoom, H.; Maas, W.; Fey, M.; Kumar, R.; Monette, M.; Stronks, H.; Simpson, M.J.; et al. Rapid parameter optimization of low signal-to-noise samples in NMR spectroscopy using rapid CPMG pulsing during acquisition: Application to recycle delays. Magn. Reson. Chem. 2013, 51, 129–135. [Google Scholar] [CrossRef]
- Ranzato, E.; Martinotti, S.; Burlando, B. Epithelial Mesenchymal Transition Traits in Honey-Driven Keratinocyte Wound Healing: Comparison among Different Honeys. Wound Repair Regen. 2012, 20, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Governa, P.; Carullo, G.; Biagi, M.; Rago, V.; Aiello, F. Evaluation of the in Vitro Wound-Healing Activity of Calabrian Honeys. Antioxidants 2019, 8, 36. [Google Scholar] [CrossRef]
- Dontchos, B.N.; Coyle, C.H.; Izzo, N.J.; Didiano, D.M.; Karpie, J.C.; Logar, A.; Chu, C.R. Optimizing CO2 normalizes pH and enhances chondrocyte viability during cold storage. J. Orthop. Res. 2008, 26, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Mazzotta, S.; Governa, P.; Borgonetti, V.; Marcolongo, P.; Nanni, C.; Gamberucci, A.; Manetti, F.; Pessina, F.; Carullo, G.; Brizzi, A.; et al. Pinocembrin and Its Linolenoyl Ester Derivative Induce Wound Healing Activity in HaCaT Cell Line Potentially Involving a GPR120/FFA4 Mediated Pathway. Bioorg. Chem. 2021, 108, 104657. [Google Scholar] [CrossRef]
- Martinotti, S.; Calabrese, G.; Ranzato, E. Honeydew Honey: Biological Effects on Skin Cells. Mol. Cell. Biochem. 2017, 435, 185–192. [Google Scholar] [CrossRef]
- Ranzato, E.; Bonsignore, G.; Martinotti, S. ER Stress Response and Induction of Apoptosis in Malignant Pleural Mesothelioma: The Achilles Heel Targeted by the Anticancer Ruthenium Drug BOLD-100. Cancers 2022, 14, 4126. [Google Scholar] [CrossRef] [PubMed]
- GraphPad Software, GraphPad Prism version 10.0.0 for Windows; GraphPad Software: Boston, MA, USA, 2023. Available online: https://www.graphpad.com (accessed on 27 April 2025).
- MetaboAnalyst 6.0. Available online: https://www.metaboanalyst.ca/ (accessed on 27 April 2025).
- Cheynier, V.; Comte, G.; Davies, K.M.; Lattanzio, V.; Martens, S. Plant Phenolics: Recent Advances on Their Biosynthesis, Genetics, and Ecophysiology. Plant Physiol. Biochem. 2013, 72, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Gharibi, S.; Tabatabaei, B.E.S.; Saeidi, G.; Goli, S.A.H. Effect of Drought Stress on Total Phenolic, Lipid Peroxidation, and Antioxidant Activity of Achillea Species. Appl. Biochem. Biotechnol. 2016, 178, 796–809. [Google Scholar] [CrossRef]
- Smirnoff, N. Ascorbic Acid Metabolism and Functions: A Comparison of Plants and Mammals. Free. Radic. Biol. Med. 2018, 122, 116–129. [Google Scholar] [CrossRef]
- Gu, S.; Zhu, W.; Ren, L.; Sun, B.; Ren, Y.; Niu, Y.; Li, X.; He, Q. New Insights into the Impact of Wood Vinegar on the Growth and Rhizosphere Microorganisms of Cherry Radish (Raphanus sativus L.). PeerJ 2024, 12, e18505. [Google Scholar] [CrossRef]
- Calvo, P.; Nelson, L.; Kloepper, J.W. Agricultural Uses of Plant Biostimulants. Plant Soil 2014, 383, 3–41. [Google Scholar] [CrossRef]
- du Jardin, P. Plant Biostimulants: Definition, Concept, Main Categories and Regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef]
- Lin, Z.H.; Chen, C.S.; Zhao, S.Q.; Liu, Y.; Zhong, Q.S.; Ruan, Q.C.; Chen, Z.H.; You, X.M.; Shan, R.Y.; Li, X.L.; et al. Molecular and Physiological Mechanisms of Tea (Camellia sinensis (L.) O. Kuntze) Leaf and Root in Response to Nitrogen Deficiency. BMC Genom. 2023, 24, 27. [Google Scholar] [CrossRef] [PubMed]
- Wormit, A.; Usadel, B. The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs). Int. J. Mol. Sci. 2018, 19, 2878. [Google Scholar] [CrossRef]
- Malhotra, H.; Vandana; Sharma, S.; Pandey, R. Phosphorus nutrition: Plant growth in response to deficiency and excess. In Plant Nutrients and Abiotic Stress Tolerance; Springer: Singapore, 2018; pp. 171–190. [Google Scholar] [CrossRef]
- Sharma, A. A Review on the Effect of Organic and Chemical Fertilizers on Plants. Int. J. Res. Appl. Sci. Eng. Technol. 2017, 5, 677–680. [Google Scholar] [CrossRef]
- He, L.; Geng, K.; Li, B.; Li, S.; Gustave, W.; Wang, J.; Jeyakumar, P.; Zhang, X.; Wang, H. Enhancement of Nutrient Use Efficiency with Biochar and Wood Vinegar: A Promising Strategy for Improving Soil Productivity. J. Sci. Food Agric. 2025, 105, 465–472. [Google Scholar] [CrossRef]
- Phibunwatthanawong, T.; Riddech, N. Liquid Organic Fertilizer Production for Growing Vegetables under Hydroponic Condition. Int. J. Recycl. Org. Waste Agric. 2019, 8, 369–380. [Google Scholar] [CrossRef]
- Hänsch, R.; Mendel, R.R. Physiological Functions of Mineral Micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 2009, 12, 259–266. [Google Scholar] [CrossRef]
- Jindo, K.; Goron, T.L.; Pizarro-Tobías, P.; Sánchez-Monedero, M.Á.; Audette, Y.; Deolu-Ajayi, A.O.; van der Werf, A.; Goitom Teklu, M.; Shenker, M.; Pombo Sudré, C.; et al. Application of Biostimulant Products and Biological Control Agents in Sustainable Viticulture: A Review. Front. Plant Sci. 2022, 13, 932311. [Google Scholar] [CrossRef]
- Baccelli, I.; Mauch-Mani, B. Beta-Aminobutyric Acid Priming of Plant Defense: The Role of ABA and Other Hormones. Plant Mol. Biol. 2016, 91, 703–711. [Google Scholar] [CrossRef]
- Netto, L.E.S.; de Oliveira, M.A.; Monteiro, G.; Demasi, A.P.D.; Cussiol, J.R.R.; Discola, K.F.; Demasi, M.; Silva, G.M.; Alves, S.V.; Faria, V.G.; et al. Reactive Cysteine in Proteins: Protein Folding, Antioxidant Defense, Redox Signaling and More. Comp. Biochem. Physiol.—C Toxicol. Pharmacol. 2007, 146, 180–193. [Google Scholar] [CrossRef]
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine Salvage and S-Adenosylmethionine: Essential Links between Sulfur, Ethylene and Polyamine Biosynthesis. Biochem. J. 2013, 451, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid Biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Chemat, F.; Vian, M.A.; Cravotto, G. Green Extraction of Natural Products: Concept and Principles. Int. J. Mol. Sci. 2012, 13, 8615–8627. [Google Scholar] [CrossRef]
- Adamovic, M.; Adamovic, A.; Andjic, M.; Dimitrijevic, J.; Zdravkovic, N.; Kostic, O.; Pecarski, D.; Pecarski, T.; Obradovic, D.; Tomovic, M. The Botany, Phytochemistry and the Effects of the Juglans regia on Healthy and Diseased Skin. Cosmetics 2024, 11, 163. [Google Scholar] [CrossRef]
- Ferreres, F.; Sousa, C.; Vrchovská, V.; Valentão, P.; Pereira, J.A.; Seabra, R.M.; Andrade, P.B. Chemical Composition and Antioxidant Activity of Tronchuda Cabbage Internal Leaves. Eur. Food Res. Technol. 2006, 222, 88–98. [Google Scholar] [CrossRef]
- Moco, S.; Bino, R.J.; Vorst, O.; Verhoeven, H.A.; De Groot, J.; Van Beek, T.A.; Vervoort, J.; Ric De Vos, C.H. A Liquid Chromatography-Mass Spectrometry-Based Metabolome Database for Tomato. Plant Physiol. 2006, 141, 1205–1218. [Google Scholar] [CrossRef]
- Bystrom, L.M.; Lewis, B.A.; Brown, D.L.; Rodriguez, E.; Obendorf, R.L. Characterisation of Phenolics by LC–UV/Vis, LC–MS/MS and Sugars by GC in Melicoccus bijugatus Jacq. ‘Montgomery’ Fruits. Food Chem. 2008, 111, 1017–1024. [Google Scholar] [CrossRef]
- Medic, A.; Kunc, P.; Zamljen, T.; Hudina, M.; Veberic, R.; Solar, A. Identification and Quantification of the Major Phenolic Constituents in Castanea Sativa and Commercial Interspecific Hybrids (C. sativa x C. crenata) Chestnuts Using HPLC–MS/MS. Int. J. Mol. Sci. 2023, 24, 13086. [Google Scholar] [CrossRef]
- Medic, A.; Jakopic, J.; Solar, A.; Hudina, M.; Veberic, R. Walnut (J. regia) Agro-Residues as a Rich Source of Phenolic Compounds. Biology 2021, 10, 535. [Google Scholar] [CrossRef]
- Slatnar, A.; Mikulic-Petkovsek, M.; Stampar, F.; Veberic, R.; Solar, A. Identification and Quantification of Phenolic Compounds in Kernels, Oil and Bagasse Pellets of Common Walnut (Juglans regia L.). Food Res. Int. 2015, 67, 255–263. [Google Scholar] [CrossRef]
- Formato, M.; Piccolella, S.; Zidorn, C.; Vastolo, A.; Calabrò, S.; Cutrignelli, M.I.; Pacifico, S. UHPLC-ESI-QqTOF Analysis and In Vitro Rumen Fermentation for Exploiting Fagus Sylvatica Leaf in Ruminant Diet. Molecules 2022, 27, 2217. [Google Scholar] [CrossRef]
- Hama, J.R.; Fitzsimmons-Thoss, V. Determination of Unsaturated Fatty Acids Composition in Walnut (Juglans regia L.) Oil Using NMR Spectroscopy. Food Anal. Methods 2022, 15, 1226–1236. [Google Scholar] [CrossRef]
- Schmitt, C.; Bastek, T.; Stelzer, A.; Schneider, T.; Fischer, M.; Hackl, T. Detection of Peanut Adulteration in Food Samples by Nuclear Magnetic Resonance Spectroscopy. J. Agric. Food Chem. 2020, 68, 14364–14373. [Google Scholar] [CrossRef]
- Vitale, S.; Colanero, S.; Placidi, M.; Di Emidio, G.; Tatone, C.; Amicarelli, F.; D’Alessandro, A.M. Phytochemistry and biological activity of medicinal plants in wound healing: An overview of current research. Molecules 2022, 27, 3566. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.S.; de Melo Bisneto, A.V.; Silva, L.S.; Bailão, E.F.L.C.; Cardoso, C.G.; Carneiro, C.C.; da Costa Santos, S.; Chen-Chen, L. Pedunculagin and Tellimagrandin-I Stimulate Inflammation and Angiogenesis and Upregulate Vascular Endothelial Growth Factor and Tumor Necrosis Factor-Alpha In Vivo. Microvasc. Res. 2024, 151, 104615. [Google Scholar] [CrossRef] [PubMed]
- Yeni, Y.; Adresi, Y. Investigation of the İn Vitro Effect of Vanillic Acid on Wound Healing via FN1 and COL1α1 Genes. Dicle Med. J. 2024, 51, 233–240. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, C.; Lu, W.; Li, S.; Chen, Z.; Xia, J.; Tian, Y.; Ma, A.; Jia, Y. Transcriptomics Reveals the Potential Mechanism of Ellagic Acid Extract from Raspberry on Wound Healing. Food Agric. Immunol. 2023, 34, 2214709. [Google Scholar] [CrossRef]
- Ranzato, E.; Patrone, M.; Mazzucco, L.; Burlando, B. Platelet Lysate Stimulates Wound Repair of HaCaT Keratinocytes. Br. J. Dermatol. 2008, 159, 537–545. [Google Scholar] [CrossRef]
- Yao, W.; Wang, Z.; Ma, H.; Lin, Y.; Liu, X.; Li, P.; He, X. Epithelial-Mesenchymal Plasticity (EMP) in Wound Healing: Exploring EMT Mechanisms, Regulatory Network, and Therapeutic Opportunities. Heliyon 2024, 10, e34269. [Google Scholar] [CrossRef]
- Gu, Y.; Huang, Q.; Wang, Y.; Wang, H.; Xiang, Z.; Xu, Y.; Wang, X.; Liu, W.; Wang, A. The Vasculogenic Mimicry Related Signature Predicts the Prognosis and Immunotherapy Response in Renal Clear Cell Carcinoma. BMC Cancer 2024, 24, 420. [Google Scholar] [CrossRef]
- Li, W.; Li, P.; Li, X.; Hou, H.; Lin, H.; Jin, M.; Liu, K.; Zhang, X.; Sheng, W. Metabolomics-Based Profiling of Anti-Inflammatory Compounds from Mentha Spicata in Shanghe, China. Heliyon 2024, 10, e35974. [Google Scholar] [CrossRef] [PubMed]
- Stone, R.C.; Pastar, I.; Ojeh, N.; Chen, V.; Liu, S.; Garzon, K.I.; Tomic-Canic, M. Epithelial-Mesenchymal Transition in Tissue Repair and Fibrosis. Cell Tissue Res. 2016, 365, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Desai, N.; Sahel, D.; Kubal, B.; Postwala, H.; Shah, Y.; Chavda, V.P.; Fernandes, C.; Khatri, D.K.; Vora, L.K. Role of the extracellular matrix in cancer: Insights into tumor progression and therapy. Adv. Ther. 2025, 8, 2400370. [Google Scholar] [CrossRef]
- Ilmer, M.; Vykoukal, J.; Boiles, A.R.; Coleman, M.; Alt, E. Two sides of the same coin: Stem cells in cancer and regenerative medicine. FASEB J. 2014, 28, 2748–2761. [Google Scholar] [CrossRef]



| Gene | Sequence |
|---|---|
| cadherin-1 | CCGAGAGCTACACGTTCTCTTCAAAATTCACTCTGCC |
| Keratin-19 | AACCATGAGGAGGAAATCAGCATGACCTCATATTGGCTTC |
| occludin | GGACTGGATCAGGGAATATCATTCTTTATCCAAACGGGAG |
| ZO-1 | TTGTCTTCAAAAACTCCCACGACTCACAGGAATAGCTTTAG |
| Col1A | GCTATGATGAGAAATCAACCGTCATCTCCATTCTTTCCAGG |
| Col4A | AAAGGGAGATCAAGGGATAGTCACCTTTTTCTCCAGGTAG |
| MMP3 | GCAGTTAGAGAACATGGAGACGAGAAATAAATTGGTCCC |
| MMP9 | AAGGATGGGAAGTACTGGGCCCAGAGAAGAAGAAAAG |
| vimentin | GGAAACTAATCTGGATTCACTCCATCTCTAGTTTCAACCGTC |
| SMAD3 | CTACCAGAGAGTAGAGACACTCTCTGGAATATTGCTCTGG |
| β-actin | TCCCTGGAGAAGAGCTACGAAGCACTGTGTTGGCGTACAG |
| Parameter | UTC | AC | WD |
|---|---|---|---|
| P | 2545 ± 33 a | 2342 ± 19 b | 2498 ± 27 a |
| S | 585 ± 3 a | 582 ± 4 a | 542 ± 3 b |
| K | 2467 ± 5 b | 2550 ± 29 a | 2473 ± 7 ab |
| Ca | 530 ± 12 b | 567 ± 12 b | 629 ± 3 a |
| Mn | 22.4 ± 0.2 a | 17.2 ± 0.2 b | 21.3 ± 0.4 a |
| Fe | 20.1 ± 0.2 a | 17.3 ± 0.3 b | 20.4 ± 0.4 a |
| Cu | 9.7 ± 0.3 | 9.7 ± 0.1 | 9.6 ± 0.2 |
| Zn | 25.9 ± 0.2 a | 24.2 ± 0.1 b | 26.1 ± 0.2 a |
| Parameter | UTC | AC | WD |
|---|---|---|---|
| Total soluble protein content | 8.7 ± 0.2 b | 11.3 ± 0.1 a | 13.6 ± 0.1 a |
| Aspartic acid | 84.7 ± 8.1 | 80.2 ± 10.8 | 94.1 ± 9.1 |
| Serine | 35.1 ± 6.6 | 26.9 ± 4.4 | 40.1 ± 7.6 |
| Glutamic acid | 248 ± 11 | 262 ± 28 | 272 ± 17 |
| Glycine + Histidine | 34.6 ± 10.3 | 23.2 ± 2.1 | 38.3 ± 9.3 |
| Arginine + Threonine | 104 ± 8 | 108 ± 16 | 124 ± 9 |
| Taurine | 27.2 ± 5.5 | 19.5 ± 3.6 | 31.6 ± 6.6 |
| β-alanine | 40.6 ± 28.2 | 9.1 ± 3.9 | 16.3 ± 3.3 |
| Alanine | 58.9 ± 11.1 | 48.1 ± 6.3 | 56.8 ± 7.3 |
| Proline | 69.4 ± 9.6 | 92.5 ± 27.7 | 93.9 ± 18.3 |
| γ-aminobutyric acid | 64.1 ± 9.2 | 58.2 ± 8.9 | 53.7 ± 9.1 |
| β- and α- aminobutyric acid | 8.2 ± 3.6 a | 2.6 ± 1.6 b | 9.1 ± 1.4 a |
| Cysteine | 100 ± 12 a | 47.8 ± 5.1 b | 12.5 ± 4.4 c |
| Tyrosine | 39.1 ± 11.2 | 26.4 ± 4.6 | 44.7 ± 9.9 |
| Valine | 35.3 ± 8.6 | 22.6 ± 3.7 | 40.4 ± 7.5 |
| Methionine | 9.7 ± 3.6 a | 3.9 ± 1.8 b | 12.8 ± 4.2 a |
| Ornithine | 21.3 ± 4.8 | 13.5 ± 2.3 | 17.1 ± 3.9 |
| Lysine | 49.9 ± 10.3 | 31.6 ± 7.1 | 57.1 ± 9.1 |
| Isoleucine | 29.9 ± 10.8 | 15.2 ± 3.1 | 26.9 ± 5.5 |
| Leucine | 33.1 ± 5.4 a | 18.2 ± 3.8 b | 38.7 ± 4.1 a |
| Phenylalanine | 24.7 ± 6.0 a | 14.5 ± 3.4 b | 29.6 ± 6.4 a |
| Peak | Name | Equivalents Expressed | UTC_A | AC_A | WD_A | |||
| Concentration * µg g−1 dw | Precision # (RSD, %) | Concentration * µg g−1 dw | Precision # (RSD, %) | Concentration * µg g−1 dw | Precision # (RSD, %) | |||
| 2 | Monogalloyl-glucose | Gallic acid | 2.53 ± 0.05 b | 1.81% | 4.79 ± 0.07 a | 1.37% | 2.62 ± 0.13 b | 4.88% |
| 3 | Pedunculagin (bis-HHDP-glucose) | Gallic acid | 1.34 ± 0.01 c | 0.87% | 4.46 ± 0.11 a | 2.54% | 1.64 ± 0.05 b | 3.19% |
| 5 | Pedunculagin (bis-HHDP-glucose) | Gallic acid | 1.41 ± 0.07 b | 4.71% | 4.07 ± 0.15 a | 3.63% | 1.65 ± 0.08 b | 4.67% |
| 6 | Gallic acid methylester | Gallic acid | 4.16 ± 0.03 b | 0.70% | 11.81 ± 0.09 a | 0.77% | 2.85 ± 0.07 c | 2.29% |
| 8 | (+)Catechin | Catechin | 0.84 ± 0.01 b | 0.87% | 10.9 ± 0.04 a | 0.34% | 0.89 ± 0.01 b | 0.83% |
| 9 | Dicarboxylic acid derivative 2 | Ellagic acid | 9.89 ± 0.6 b | 6.08% | 15.11 ± 0.14 a | 0.91% | 5.05 ± 0.05 c | 0.92% |
| 10 | Dicarboxylic acid derivative 3 | Ellagic acid | 15.05 ± 0.26 b | 1.76% | 20.33 ± 0.5 a | 2.48% | 7.35 ± 0.17 c | 2.35% |
| 11 | Dihydroxytetralone hexoside | Chlorogenic acid | 14.62 ± 0.3 b | 2.05% | 25.46 ± 0.36 a | 1.43% | 8.24 ± 0.36 c | 4.40% |
| 14 | HHDP digalloyl glucose | Gallic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 16 | Sinapic acid hexose | Sinapic acid | 43.31 ± 0.43 c | 0.98% | 101.52 ± 0.65 a | 0.64% | 65.64 ± 0.89 b | 1.35% |
| 17 | p-Coumaric acid derivative 1 | 4-Coumaric acid | 42.58 ± 1.31 c | 3.08% | 111.93 ± 0.32 a | 0.28% | 67.32 ± 0.62 b | 0.91% |
| 19 | Gallic acid derivative 4 | Gallic acid | 48.06 ± 1.72 b | 3.57% | 135.88 ± 2.64 a | 1.95% | 42.24 ± 0.77 c | 1.83% |
| 20 | Glansreginin B | Ellagic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 21 | Dicarboxylic acid derivative 3 | Ellagic acid | 2.74 ± 0.05 b | 1.76% | 14.4 ± 0.08 a | 0.57% | 2.55 ± 0.06 c | 2.31% |
| 22 | Dicarboxylic acid derivative 3 | Ellagic acid | 2.65 ± 0.07 b | 2.68% | 15.67 ± 0.56 a | 3.56% | 2.86 ± 0.1 b | 3.39% |
| 23 | Ellagic acid pentoside | Ellagic acid | 2.35 ± 0.14 b | 6.16% | 5.27 ± 0.04 a | 0.68% | 1.13 ± 0.07 c | 6.11% |
| 24 | Dicarboxylic acid derivative 3 | Ellagic acid | 50.09 ± 0.46 c | 0.92% | 122.64 ± 0.31 a | 0.26% | 51.5 ± 0.41 b | 0.79% |
| 25 | Ellagic acid | Ellagic acid | 48.6 ± 1.2 b | 2.46% | 88.24 ± 1.83 a | 2.07% | 40.31 ± 0.95 c | 2.36% |
| 27 | Glansreginin A | Ellagic acid | 194.89 ± 4.7 b | 2.41% | 281.88 ± 4.04 a | 1.43% | 202.25 ± 1.31 b | 0.65% |
| 29 | Glansreginin A | Ellagic acid | 12.22 ± 0.11 c | 0.94% | 69.18 ± 0.15 a | 0.22% | 14.5 ± 0.15 b | 1.02% |
| 30 | Glansreginin A | Ellagic acid | 8.25 ± 0.18 b | 2.14% | 11.66 ± 0.03 a | 0.24% | 7.74 ± 0.06 c | 0.71% |
| 35 | 3-p-Coumaroylquinic acid | 4-Coumaric acid | 10.7 ± 0.02 b | 0.20% | 66.89 ± 0.7 a | 1.05% | 8.61 ± 0.15 c | 1.70% |
| 36 | 5-O-(3′-O-Glucosylcaffeoyl)quinic acid | Chlorogenic acid | 2.71 ± 0.06 b | 2.08% | 7.21 ± 0.15 a | 2.05% | 2.19 ± 0.05 c | 2.17% |
| 40 | 9,12,13-trihydroxy-10,15-octadecadienoic acid | Oleic acid | 8.03 ± 0.18 c | 2.24% | 47.77 ± 0.13 a | 0.27% | 9.28 ± 0.31 b | 3.31% |
| 42 | 9,12,13-trihydroxy-10-octadecenoic acid | Oleic acid | 92.85 ± 2.1 b | 2.27% | 173.1 ± 2.61 a | 1.51% | <LOQ c | - |
| 43 | 9,12,13-trihydroxy-10-octadecenoic acid | Oleic acid | 2.63 ± 0.17 c | 6.43% | 9.37 ± 0.05 a | 0.52% | 3.12 ± 0.09 b | 2.95% |
| 44 | 9,12,13-trihydroxy-10,15-octadecadienoic acid | Oleic acid | 2.16 ± 0.06 b | 2.90% | 5.52 ± 0.18 a | 3.22% | 1.43 ± 0.04 c | 3.07% |
| Peak | Name | Equivalents Expressed | UTC_B | AC_B | WD_B | |||
| Concentration * µg g−1 dw | Precision # (RSD, %) | Concentration * µg g−1 dw | Precision # (RSD, %) | Concentration * µg g−1 dw | Precision # (RSD, %) | |||
| 2 | Monogalloyl-glucose | Gallic acid | 0.57 ± 0.01 a | 1.72% | 0.51 ± 0.01 b | 2.88% | <LOQ c | - |
| 3 | Pedunculagin (bis-HHDP-glucose) | Gallic acid | 2.66 ± 0.06 a | 2.23% | 0.82 ± 0.07 b | 8.68% | 0.75 ± 0.01 b | 1.25% |
| 5 | Pedunculagin (bis-HHDP-glucose) | Gallic acid | 5.19 ± 0.32 a | 6.21% | 1.08 ± 0.07 c | 6.27% | 2.3 ± 0.03 b | 1.28% |
| 6 | Gallic acid methylester | Gallic acid | 3.13 ± 0.04 a | 1.25% | 0.51 ± 0.01 c | 1.88% | 0.91 ± 0.01 b | 0.85% |
| 8 | (+)Catechin | Catechin | 0.91 ± 0.01 a | 1.58% | 0.36 ± 0.00 b | 1.27% | 0.25 ± 0.00 c | 0.84% |
| 9 | Dicarboxylic acid derivative 2 | Ellagic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 10 | Dicarboxylic acid derivative 3 | Ellagic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 11 | Dihydroxytetralone hexoside | Chlorogenic acid | 1.61 ± 0.01 a | 0.47% | 0.75 ± 0.01 b | 1.63% | 0.35 ± 0.00 c | 0.71% |
| 14 | HHDP digalloyl glucose | Gallic acid | 2 ± 0.09 a | 4.52% | <LOQ c | - | 0.22 ± 0.01 b | 4.30% |
| 16 | Sinapic acid hexose | Sinapic acid | 8.02 ± 0.1 a | 1.28% | 5.61 ± 0.12 b | 2.20% | 3.16 ± 0.19 c | 6.08% |
| 17 | p-Coumaric acid derivative 1 | 4-Coumaric acid | 5.29 ± 0.18 a | 3.38% | 4.8 ± 0.04 b | 0.78% | 3.81 ± 0.22 c | 5.67% |
| 19 | Gallic acid derivative 4 | Gallic acid | 2.27 ± 0.03 b | 1.25% | 2.1 ± 0.04 c | 1.92% | 2.51 ± 0.00 a | 0.17% |
| 20 | Glansreginin B | Ellagic acid | 2.71 ± 0.07 a | 2.55% | 2.59 ± 0.04 b | 1.47% | 0.63 ± 0.01 c | 1.34% |
| 21 | Dicarboxylic acid derivative 3 | Ellagic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 22 | Dicarboxylic acid derivative 3 | Ellagic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 23 | Ellagic acid pentoside | Ellagic acid | 2.24 ± 0.12 a | 5.43% | 0.73 ± 0.01 c | 1.76% | 1.46 ± 0.01 b | 0.89% |
| 24 | Dicarboxylic acid derivative 3 | Ellagic acid | 3.37 ± 0.1 a | 2.95% | 3.5 ± 0.03 a | 0.74% | 0.24 ± 0.01 b | 3.57% |
| 25 | Ellagic acid | Ellagic acid | 9.55 ± 0.04 a | 0.46% | 4.69 ± 0.24 b | 5.19% | 4.02 ± 0.11 c | 2.85% |
| 27 | Glansreginin A | Ellagic acid | 32.44 ± 1.47 a | 4.54% | 30.75 ± 0.45 a | 1.45% | 15.24 ± 0.26 b | 1.70% |
| 29 | Glansreginin A | Ellagic acid | 1.35 ± 0.02 a | 1.67% | 0.88 ± 0.02 b | 2.00% | <LOQ c | - |
| 30 | Glansreginin A | Ellagic acid | 0.94 ± 0.01 a | 0.65% | 0.94 ± 0.02 a | 2.10% | <LOQ b | - |
| 35 | 3-p-Coumaroylquinic acid | 4-Coumaric acid | 0.68 ± 0.01 b | 1.78% | 0.79 ± 0.03 a | 3.71% | <LOQ c | - |
| 36 | 5-O-(3′-O-Glucosylcaffeoyl)quinic acid | Chlorogenic acid | 1.53 ± 0.03 b | 2.04% | 1.68 ± 0.03 a | 1.98% | 0.44 ± 0.00 c | 0.97% |
| 40 | 9,12,13-trihydroxy-10,15-octadecadienoic acid | Oleic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 42 | 9,12,13-trihydroxy-10- octadecenoic acid | Oleic acid | 3.94 ± 0.19 a | 4.79% | 3.43 ± 0.05 b | 1.41% | 1.23 ± 0.05 c | 3.92% |
| 43 | 9,12,13-trihydroxy-10- octadecenoic acid | Oleic acid | 1.34 ± 0.08 a | 6.22% | 1.4 ± 0.07 a | 4.84% | <LOQ b | - |
| 44 | 9,12,13-trihydroxy-10,15-octadecadienoic acid | Oleic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| Peak | Name | Equivalents Expressed | UTC_C | AC_C | WD_C | |||
| Concentration * µg g−1 dw | Precision # (RSD, %) | Concentration * µg g−1 dw | Precision # (RSD, %) | Concentration * µg g−1 dw | Precision # (RSD, %) | |||
| 2 | Monogalloyl-glucose | Gallic acid | <LOQ c | - | 0.43 ± 0.01 b | 3.46% | 0.78 ± 0.03 a | 3.94% |
| 3 | Pedunculagin (bis-HHDP-glucose) | Gallic acid | 1.45 ± 0.05 b | 3.52% | 3.71 ± 0.15 a | 4.18% | 0.96 ± 0.05 c | 5.46% |
| 5 | Pedunculagin (bis-HHDP-glucose) | Gallic acid | 1.11 ± 0.03 b | 2.39% | 10.08 ± 0.56 a | 5.51% | 0.65 ± 0.05 b | 6.99% |
| 6 | Gallic acid methylester | Gallic acid | 1.37 ± 0.04 b | 2.59% | 3.75 ± 0.03 a | 0.86% | 0.34 ± 0.01 c | 4.14% |
| 8 | (+)Catechin | Catechin | 0.47 ± 0.01 b | 1.60% | 0.66 ± 0.01 a | 2.18% | 0.3 ± 0.01 c | 1.68% |
| 9 | Dicarboxylic acid derivative 2 | Ellagic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 10 | Dicarboxylic acid derivative 3 | Ellagic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 11 | Dihydroxytetralone hexoside | Chlorogenic acid | 0.68 ± 0.01 a | 0.74% | 0.71 ± 0.01 a | 1.67% | 0.52 ± 0.02 b | 3.16% |
| 14 | HHDP digalloyl glucose | Gallic acid | 0.15 ± 0.01 b | 4.72% | 3.97 ± 0.03 a | 0.81% | 0.08 ± 0.00 c | 0.69% |
| 16 | Sinapic acid hexose | Sinapic acid | 3.95 ± 0.05 c | 1.15% | 25.1 ± 0.03 a | 0.11% | 16.59 ± 0.05 b | 0.27% |
| 17 | p-Coumaric acid derivative 1 | 4-Coumaric acid | 2.4 ± 0.01 c | 0.44% | 30.03 ± 0.1 b | 0.32% | 40.04 ± 0.11 a | 0.27% |
| 19 | Gallic acid derivative 4 | Gallic acid | 2.18 ± 0.01 b | 0.63% | 1.88 ± 0.04 c | 1.89% | 2.44 ± 0.16 a | 6.47% |
| 20 | Glansreginin B | Ellagic acid | 0.68 ± 0.01 c | 1.94% | 1.57 ± 0.02 b | 1.23% | 3.46 ± 0.07 a | 1.99% |
| 21 | Dicarboxylic acid derivative 3 | Ellagic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 22 | Dicarboxylic acid derivative 3 | Ellagic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 23 | Ellagic acid pentoside | Ellagic acid | 0.41 ± 0.01 c | 2.58% | 3.46 ± 0.07 a | 1.97% | 0.65 ± 0.01 b | 1.39% |
| 24 | Dicarboxylic acid derivative 3 | Ellagic acid | 0.72 ± 0.03 c | 3.86% | 1.57 ± 0.02 b | 1.19% | 3.55 ± 0.24 a | 6.66% |
| 25 | Ellagic acid | Ellagic acid | 7.62 ± 0.24 c | 3.15% | 17.78 ± 0.14 a | 0.78% | 10.06 ± 0.35 b | 3.45% |
| 27 | Glansreginin A | Ellagic acid | 34.76 ± 0.33 c | 0.96% | 39.9 ± 0.74 b | 1.86% | 58.22 ± 1.17 a | 2.01% |
| 29 | Glansreginin A | Ellagic acid | <LOQ | - | 0.83 ± 0.00 a | 0.42% | 0.2 ± 0.01 b | 4.58% |
| 30 | Glansreginin A | Ellagic acid | <LOQ | - | 1.02 ± 0.01 a | 0.87% | 0.95 ± 0.07 a | 7.72% |
| 35 | 3-p-Coumaroylquinic acid | 4-Coumaric acid | <LOQ | - | 0.87 ± 0.01 a | 1.00% | 0.87 ± 0.05 a | 5.80% |
| 36 | 5-O-(3′-O-Glucosylcaffeoyl)quinic acid | Chlorogenic acid | 1.07 ± 0.03 a | 3.13% | 0.85 ± 0.02 b | 2.70% | 0.83 ± 0.03 b | 3.82% |
| 40 | 9,12,13-trihydroxy-10,15-octadecadienoic acid | Oleic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| 42 | 9,12,13-trihydroxy-10-octadecenoic acid | Oleic acid | 2.37 ± 0.11 b | 4.66% | 1.52 ± 0.03 c | 1.84% | 2.61 ± 0.08 a | 2.96% |
| 43 | 9,12,13-trihydroxy-10-octadecenoic acid | Oleic acid | <LOQ | - | 0.91 ± 0.01 a | 1.04% | 0.99 ± 0.04 a | 3.77% |
| 44 | 9,12,13-trihydroxy-10,15-octadecadienoic acid | Oleic acid | <LOQ | - | <LOQ | - | <LOQ | - |
| Molecule | 1H Shift | Multiplicity | Assignment | UTC_A | AC_A | WD_A | UTC_B | AC_B | WD_B | UTC_C | AC_C | WD_C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fatty acids (terminal -CH3) | 0.82 | t | -CH3 | - | - | + | + | + | + | + | + | + |
| Isoleucine | 0.94 | t | -CH3 | - | - | - | + | + | + | + | + | + |
| Leucine | 0.97 | t | -CH3 | + | + | + | - | + | + | + | - | + |
| Valine | 1.05 | d | -CH2 | + | + | + | + | + | + | + | + | + |
| Fatty acids (methylene) | 1.23 | -CH2n | - | - | + | + | + | + | + | + | + | |
| Threonine | 1.33 | d | -CH2 | + | + | + | + | + | + | + | + | + |
| Alanine | 1.48 | d | -CH2 | + | + | + | + | + | + | + | + | + |
| Arginine | 1.62–1.78 | m | n.a. | + | + | + | + | + | + | + | + | + |
| Acetate | 1.92 | s | -CH3 | + | + | + | + | + | + | + | + | + |
| Fatty acids (allylic) | 1.97 | -CH2-CH=CH | - | - | + | + | + | + | + | + | + | |
| Fatty acids (acyl chains) | 2.24 | -CH2-COO | - | - | + | + | + | + | + | + | + | |
| Sucrose | 4.07 | dd | -CHOH | - | - | + | - | + | + | + | + | + |
| Malate | 4.22 | dd | -CHOH | + | + | + | + | + | + | + | + | + |
| Fatty acids (olefinic portions) | 5.28 | m | CH=CH | - | - | + | + | + | + | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedeli, R.; Ranzato, E.; Martinotti, S.; Basilicata, M.G.; Marotta, L.; Fava, M.; Cursaro, I.; Tremori, G.; Bonsignore, G.; Carullo, G.; et al. Unveiling Wound Healing Properties of Biostimulated Walnut Kernel Extracts via Epithelial Mesenchymal Transition: Switching a Nutritional Matrix into a Therapeutic Remedy. Antioxidants 2025, 14, 1079. https://doi.org/10.3390/antiox14091079
Fedeli R, Ranzato E, Martinotti S, Basilicata MG, Marotta L, Fava M, Cursaro I, Tremori G, Bonsignore G, Carullo G, et al. Unveiling Wound Healing Properties of Biostimulated Walnut Kernel Extracts via Epithelial Mesenchymal Transition: Switching a Nutritional Matrix into a Therapeutic Remedy. Antioxidants. 2025; 14(9):1079. https://doi.org/10.3390/antiox14091079
Chicago/Turabian StyleFedeli, Riccardo, Elia Ranzato, Simona Martinotti, Manuela Giovanna Basilicata, Ludovica Marotta, Marianna Fava, Ilaria Cursaro, Giulio Tremori, Gregorio Bonsignore, Gabriele Carullo, and et al. 2025. "Unveiling Wound Healing Properties of Biostimulated Walnut Kernel Extracts via Epithelial Mesenchymal Transition: Switching a Nutritional Matrix into a Therapeutic Remedy" Antioxidants 14, no. 9: 1079. https://doi.org/10.3390/antiox14091079
APA StyleFedeli, R., Ranzato, E., Martinotti, S., Basilicata, M. G., Marotta, L., Fava, M., Cursaro, I., Tremori, G., Bonsignore, G., Carullo, G., Gemma, S., Aquino, G., Campiglia, P., Pepe, G., Butini, S., Loppi, S., & Campiani, G. (2025). Unveiling Wound Healing Properties of Biostimulated Walnut Kernel Extracts via Epithelial Mesenchymal Transition: Switching a Nutritional Matrix into a Therapeutic Remedy. Antioxidants, 14(9), 1079. https://doi.org/10.3390/antiox14091079














