Chronic Chemogenetic Stimulation of the Nucleus Accumbens Produces Lasting Reductions in Binge Drinking and Ameliorates Alcohol-Related Morphological and Transcriptional Changes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Drinking in the Dark (DID)
2.3. Drugs
2.4. Experiment 1
2.5. Experiment 2
2.6. Experiment 3
2.7. Data Analysis and Statistics
2.7.1. Behavior
2.7.2. Morphology
2.7.3. Transcriptomic Analysis
3. Results
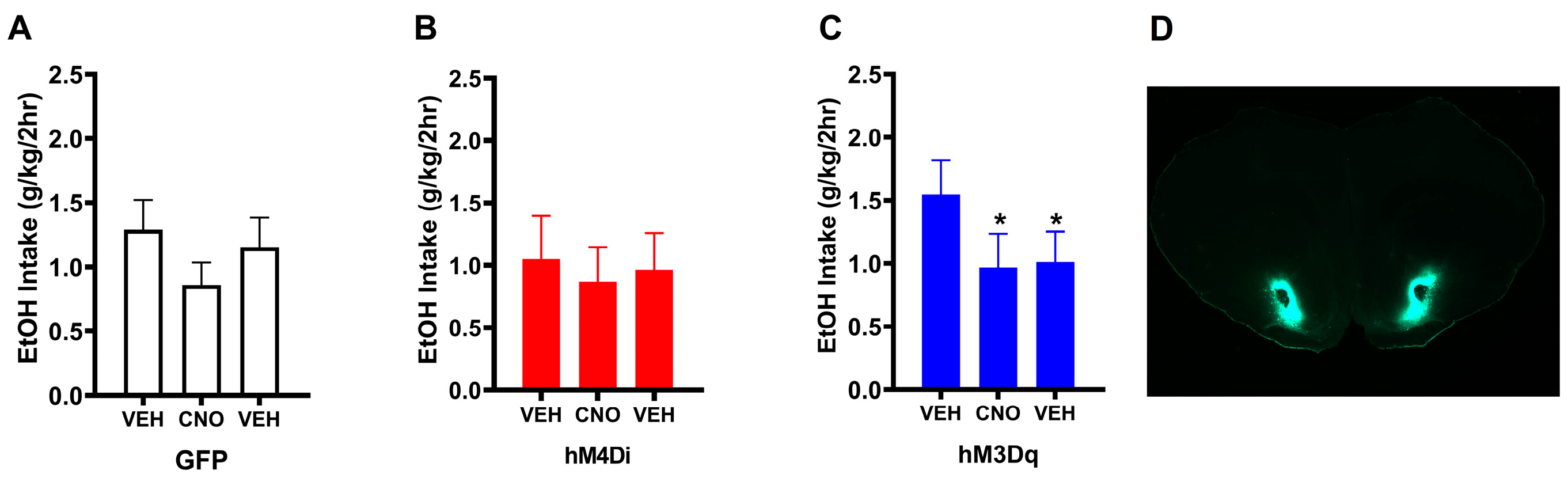
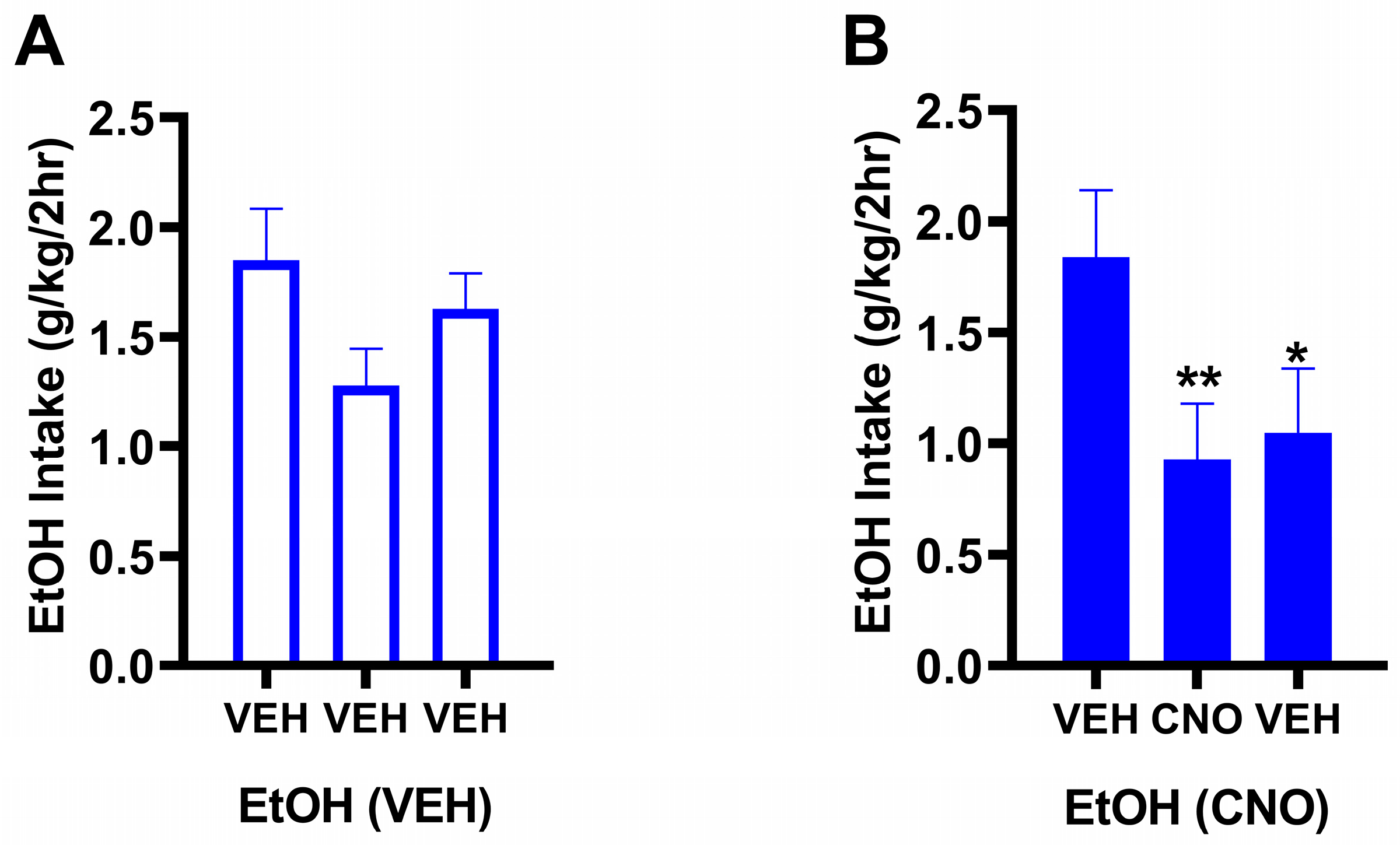
3.1. Experiment 1: Chronically Increasing Gq Signaling in the NAc Produces Lasting Reductions in Binge-Like Drinking
3.2. Experiment 2: Chronically Increasing Gq Signaling in the NAc Produces Lasting Reductions in Binge-Like Drinking, the Transcriptome, and Neuronal Morphology
3.2.1. Behavioral Results
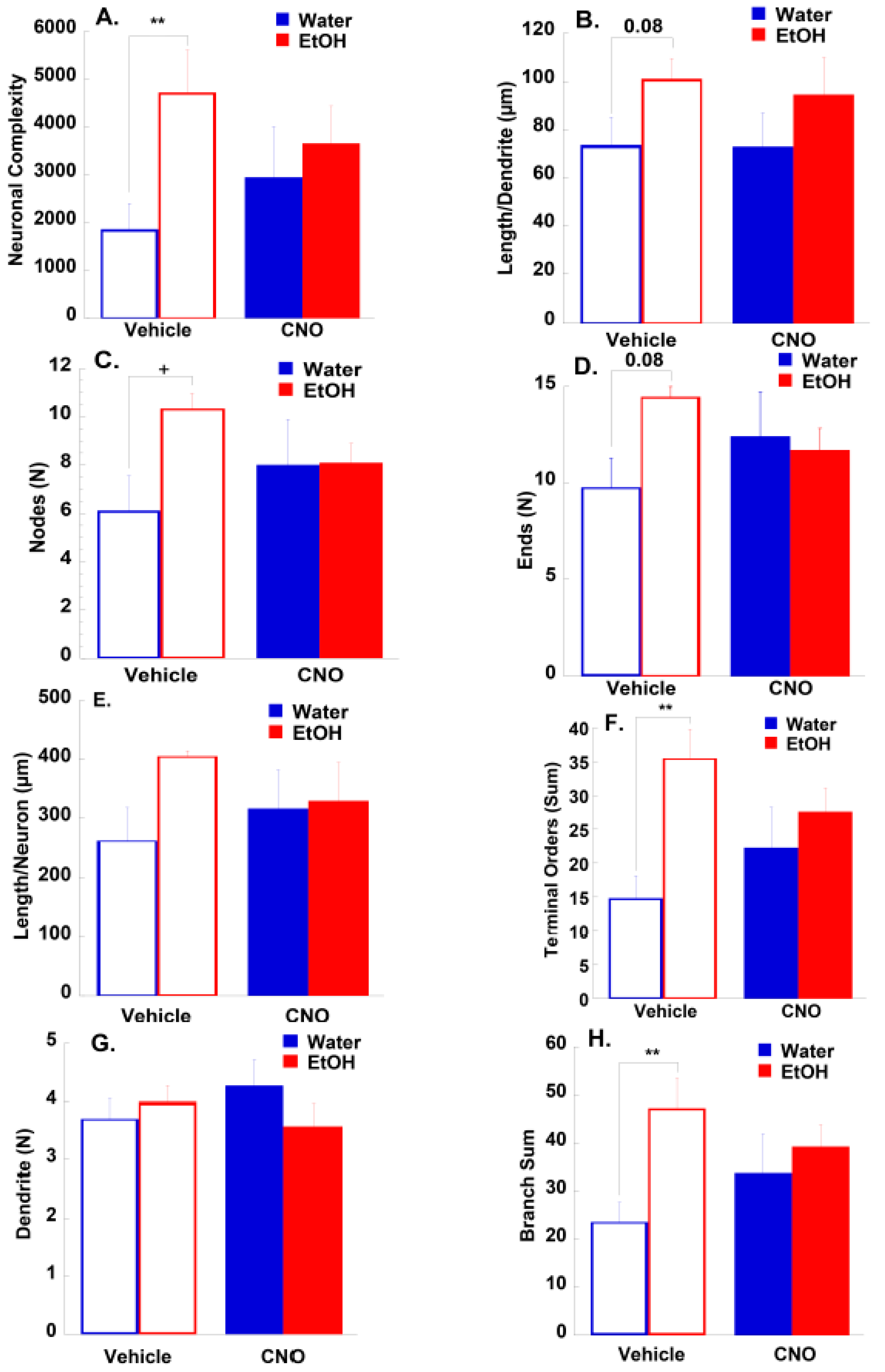
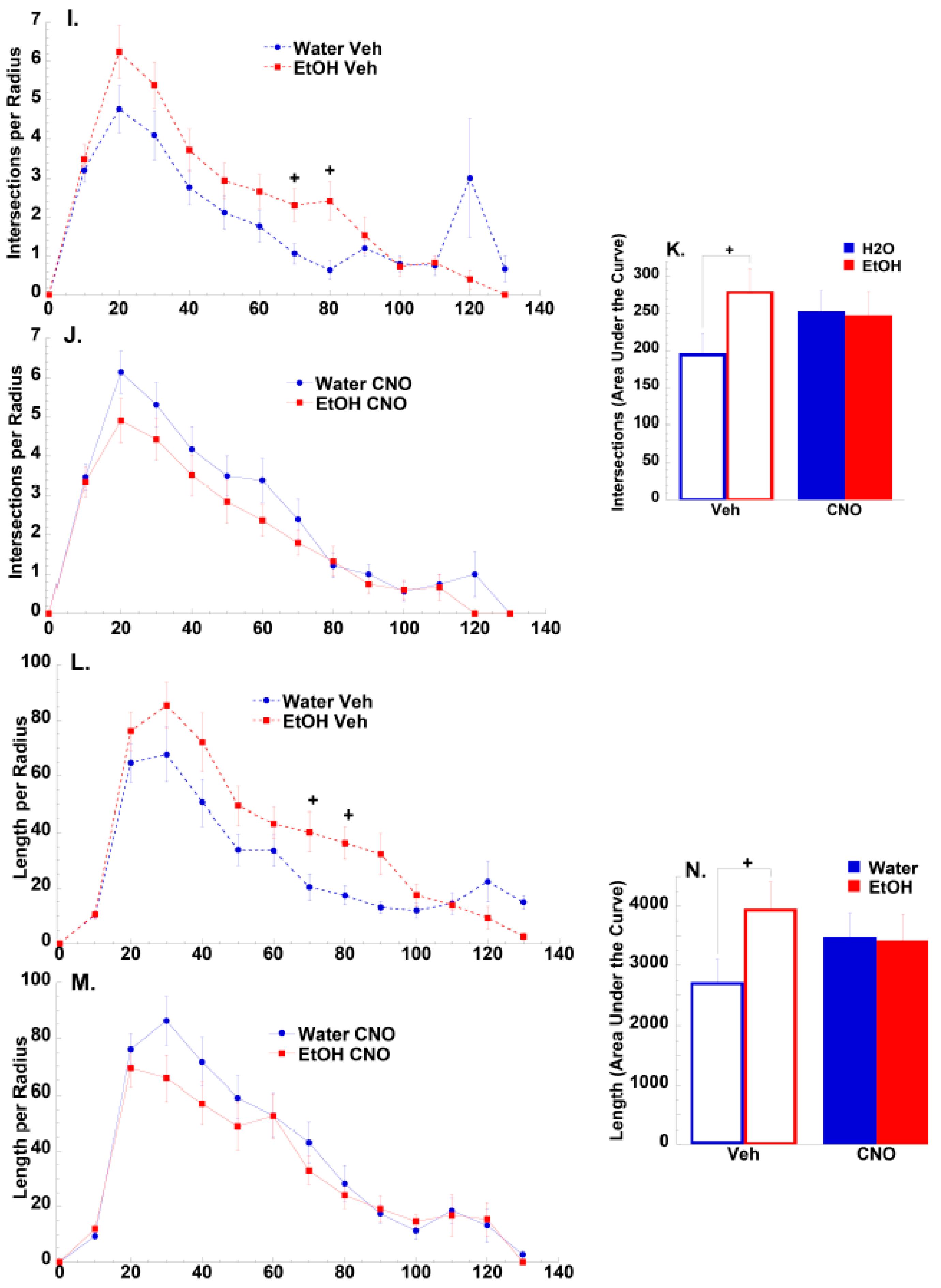
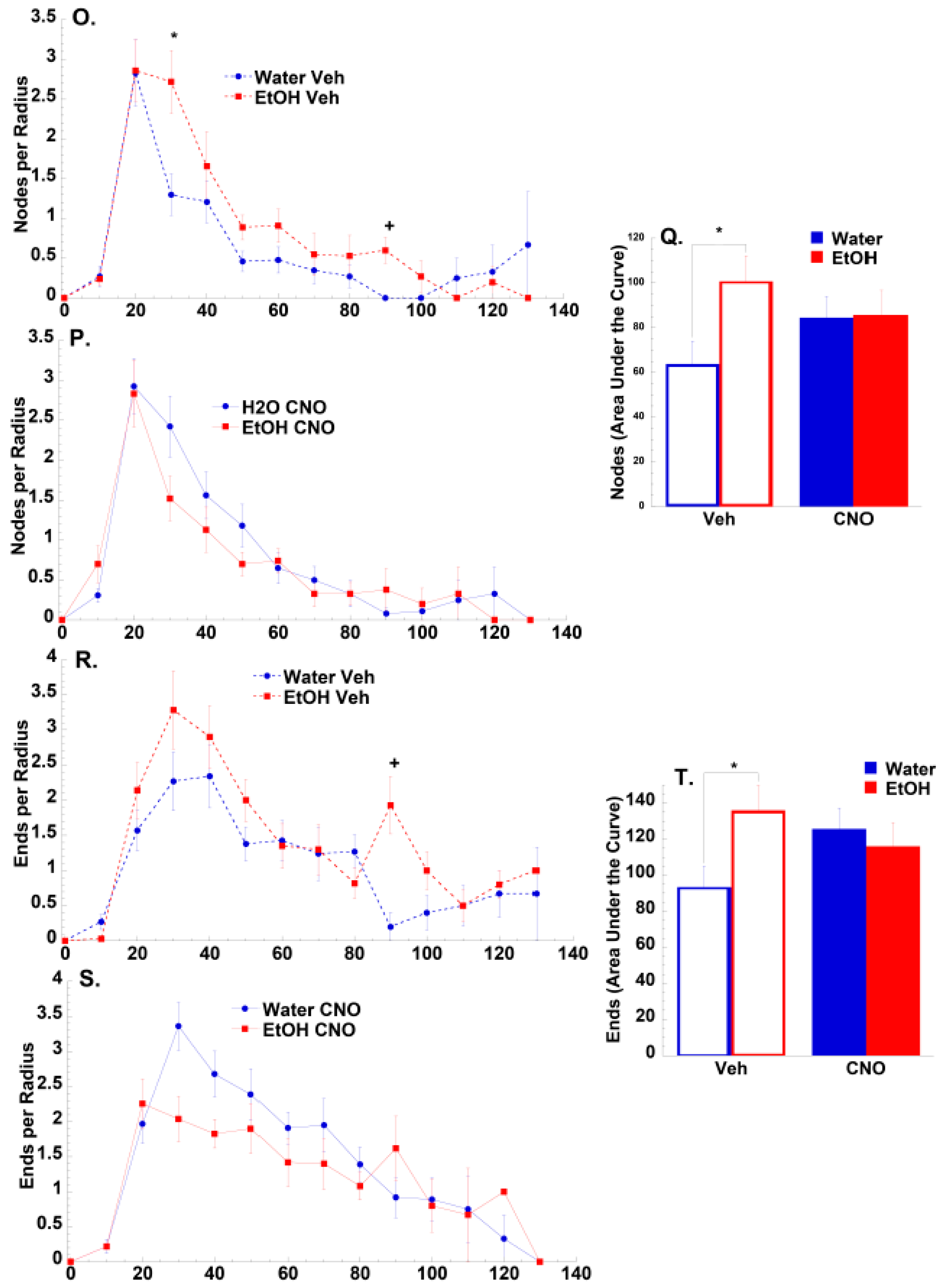
3.2.2. Morphological Results
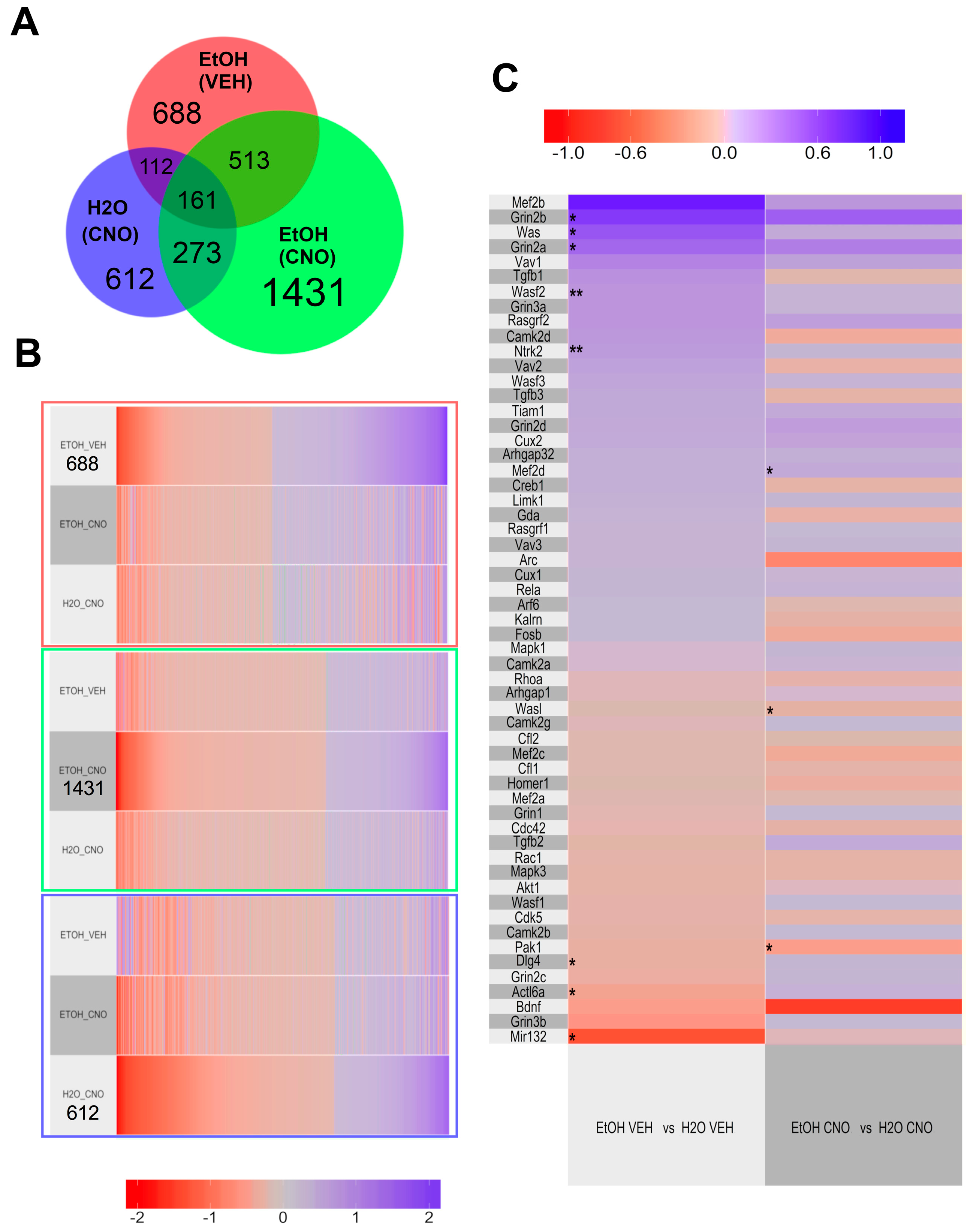
3.2.3. Transcriptomic Results
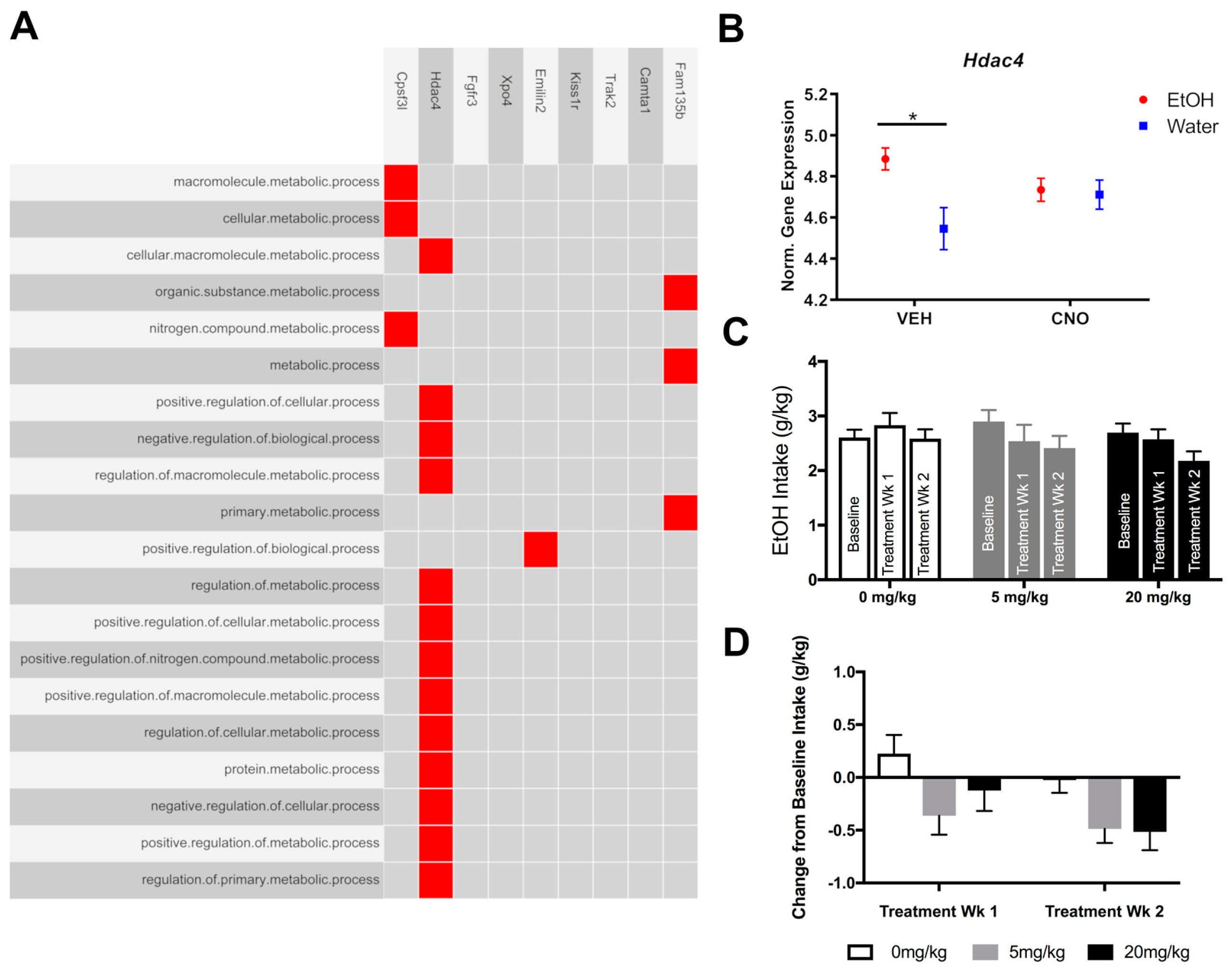
3.3. Experiment 3: Pharmacologically Testing the Role of the Overrepresented Gene, Hdac4, in Binge-Like Drinking
4. Discussion
4.1. Animal Models of Binge Drinking and DREADDs
4.2. Effects of Binge-Drinking and DREADDs on NAc Neuronal Morphology
4.3. Effects of Chronic Binge-Drinking and DREADDs on the NAc Transcriptome
4.4. Pharmacologically Targeting HDAC4 to Reduce Binge-Like Drinking
5. Summary
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Rhodes, J.S.; Best, K.; Belknap, J.K.; Finn, D.A.; Crabbe, J.C. Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol. Behav. 2005, 84, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.S.; Ford, M.M.; Yu, C.H.; Brown, L.L.; Finn, D.A.; Garland, T.; Crabbe, J.C. Mouse inbred strain differences in ethanol drinking to intoxication. Genes, Brain Behav. 2007, 6, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Crabbe, J.C.; Spence, S.E.; Brown, L.L.; Metten, P. Alcohol preference drinking in a mouse line selectively bred for high drinking in the dark. Alcohol. 2011, 45, 427–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crabbe, J.C.; Metten, P.; Rhodes, J.S.; Yu, C.-H.; Brown, L.L.; Phillips, T.J.; Finn, D.A. A line of mice selected for high blood ethanol concentrations shows drinking in the dark to intoxication. Biol. Psychiatry. 2009, 65, 662–670. [Google Scholar] [CrossRef] [Green Version]
- Crabbe, J.C.; Metten, P.; Belknap, J.K.; Spence, S.E.; Cameron, A.J.; Schlumbohm, J.P.; Huang, L.C.; Barkley-Levenson, A.M.; Ford, M.M.; Phillips, T.J. Progress in a replicated selection for elevated blood ethanol concentrations in HDID mice. Genes, Brain Behav. 2014, 13, 236–246. [Google Scholar] [CrossRef] [Green Version]
- Barkley-Levenson, A.M.; Crabbe, J.C. High Drinking in the Dark Mice: A genetic model of drinking to intoxication. Alcohol 2014, 48, 217–223. [Google Scholar] [CrossRef] [Green Version]
- Crabbe, J.C.; Metten, P.; Savarese, A.M.; Ozburn, A.R.; Schlumbohm, J.P.; Spence, S.E.; Hack, W.R. Ethanol Conditioned Taste Aversion in High Drinking in the Dark Mice. Brain Sci. 2019, 9, 2. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, L.B.; Ozburn, A.R.; Ponomarev, I.; Metten, P.; Reilly, M.; Crabbe, J.C.; Harris, R.A.; Mayfield, R.D. Genome-Wide Expression Profiles Drive Discovery of Novel Compounds that Reduce Binge Drinking in Mice. Neuropsychopharmacology 2018, 43, 1257–1266. [Google Scholar] [CrossRef]
- Iancu, O.D.; Oberbeck, D.; Darakjian, P.; Metten, P.; McWeeney, S.; Crabbe, J.C.; Hitzemann, R. Selection for Drinking in the Dark Alters Brain Gene Coexpression Networks. Alcohol. Clin. Exp. Res. 2013, 37, 1295–1303. [Google Scholar] [CrossRef] [Green Version]
- Hitzemann, R.; Oberbeck, D.; Iancu, O.; Darakjian, P.; McWeeney, S.; Spence, S.; Schlumbohm, J.; Metten, P.; Crabbe, J. Alignment of the transcriptome with individual variation in animals selectively bred for High Drinking-In-the-Dark (HDID). Alcohol 2017, 60, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Barkley-Levenson, A.M.; Crabbe, J.C. Distinct ethanol drinking microstructures in two replicate lines of mice selected for drinking to intoxication. Genes, Brain Behav. 2015, 14, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Barkley-Levenson, A.M.; Crabbe, J.C. Genotypic and sex differences in anxiety-like behavior and alcohol-induced anxiolysis in High Drinking in the Dark selected mice. Alcohol 2015, 49, 29–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barkley-Levenson, A.M.; Cunningham, C.L.; Smitasin, P.J.; Crabbe, J.C. Rewarding and aversive effects of ethanol in High Drinking in the Dark selectively bred mice. Addict. Biol. 2015, 20, 80–90. [Google Scholar] [CrossRef] [Green Version]
- McCulley, W.D.; Ascheid, S.; Crabbe, J.C.; Rosenwasser, A.M. Selective breeding for ethanol-related traits alters circadian phenotype. Alcohol 2013, 47, 187–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamdar, N.K.; Miller, S.A.; Syed, Y.M.; Bhayana, R.; Gupta, T.; Rhodes, J.S. Acute effects of naltrexone and GBR 12909 on ethanol drinking-in-the-dark in C57BL/6J mice. Psychopharmacology (Berl.) 2007, 192, 207–217. [Google Scholar] [CrossRef]
- Ozburn, A.R.; Harris, R.A.; Blednov, Y.A. Chronic voluntary alcohol consumption results in tolerance to sedative/hypnotic and hypothermic effects of alcohol in hybrid mice. Pharmacol. Biochem. Behav. 2013, 104, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Kasten, C.R.; Blasingame, S.N.; Boehm, S.L. Bidirectional enantioselective effects of the GABAB receptor agonist baclofen in two mouse models of excessive ethanol consumption. Alcohol 2015, 49, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Navarro, M.; Carvajal, F.; Lerma-Cabrera, J.M.; Cubero, I.; Picker, M.J.; Thiele, T.E. Evidence that Melanocortin Receptor Agonist Melanotan-II Synergistically Augments the Ability of Naltrexone to Blunt Binge-Like Ethanol Intake in Male C57BL/6J Mice. Alcohol. Clin. Exp. Res. 2015, 39, 1425–1433. [Google Scholar] [CrossRef] [Green Version]
- Crabbe, J.C.; Ozburn, A.R.; Metten, P.; Barkley-Levenson, A.; Schlumbohm, J.P.; Spence, S.E.; Hack, W.R.; Huang, L.C. High Drinking in the Dark (HDID) mice are sensitive to the effects of some clinically relevant drugs to reduce binge-like drinking. Pharmacol. Biochem. Behav. 2017, 160, 55–62. [Google Scholar] [CrossRef]
- Zhou, F.C.; Anthony, B.; Dunn, K.W.; Lindquist, W.B.; Xu, Z.C.; Deng, P. Chronic alcohol drinking alters neuronal dendritic spines in the brain reward center nucleus accumbens. Brain Res. 2007, 1134, 148–161. [Google Scholar] [CrossRef] [Green Version]
- Koob, G.F.; Volkow, N.D. Neurocircuitry of Addiction. Neuropsychopharmacology 2010, 35, 217–238. [Google Scholar] [CrossRef] [Green Version]
- Heinze, H.-J.; Heldmann, M.; Voges, J.; Hinrichs, H.; Marco-Pallares, J.; Hopf, J.-M.; Müller, U.J.; Galazky, I.; Sturm, V.; Bogerts, B.; et al. Counteracting incentive sensitization in severe alcohol dependence using deep brain stimulation of the Nucleus accumbens: clinical and basic science aspects. Front. Hum. Neurosci. 2009, 3, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, J.; Gründler, T.O.J.; Bauer, R.; Huff, W.; Fischer, A.G.; Lenartz, D.; Maarouf, M.; Bührle, C.; Klosterkötter, J.; Ullsperger, M.; et al. Successful deep brain stimulation of the nucleus accumbens in severe alcohol dependence is associated with changed performance monitoring. Addict. Biol. 2011, 16, 620–623. [Google Scholar] [CrossRef] [PubMed]
- Voges, J.; Müller, U.; Bogerts, B.; Münte, T.; Heinze, H.-J. Deep Brain Stimulation Surgery for Alcohol Addiction. World Neurosurg. 2013, 80, S28.e21–S28.e31. [Google Scholar] [CrossRef] [PubMed]
- Knapp, C.M.; Tozier, L.; Pak, A.; Ciraulo, D.A.; Kornetsky, C. Deep brain stimulation of the nucleus accumbens reduces ethanol consumption in rats. Pharmacol. Biochem. Behav. 2009, 92, 474–479. [Google Scholar] [CrossRef] [Green Version]
- Henderson, M.B.; Green, A.I.; Bradford, P.S.; Chau, D.T.; Roberts, D.W.; Leiter, J.C. Deep brain stimulation of the nucleus accumbens reduces alcohol intake in alcohol-preferring rats. Neurosurg. Focus 2010, 29, E12. [Google Scholar] [CrossRef]
- Ho, A.L.; Salib, A.-M.N.; Pendharkar, A.V.; Sussman, E.S.; Giardino, W.J.; Halpern, C.H. The nucleus accumbens and alcoholism: a target for deep brain stimulation. Neurosurg. Focus 2018, 45, E12. [Google Scholar] [CrossRef] [Green Version]
- Roth, B.L. DREADDs for Neuroscientists. Neuron 2016, 89, 683–694. [Google Scholar] [CrossRef] [Green Version]
- Purohit, K.; Parekh, P.K.; Kern, J.; Logan, R.W.; Liu, Z.; Huang, Y.; McClung, C.A.; Crabbe, J.C.; Ozburn, A.R. Pharmacogenetic Manipulation of the Nucleus Accumbens Alters Binge-Like Alcohol Drinking in Mice. Alcohol. Clin. Exp. Res. 2018, 42, 879–888. [Google Scholar] [CrossRef]
- Bayram-Weston, Z.; Olsen, E.; Harrison, D.J.; Dunnett, S.B.; Brooks, S.P. Optimising Golgi-Cox staining for use with perfusion-fixed brain tissue validated in the zQ175 mouse model of Huntington’s disease. J. Neurosci. Methods 2016, 265, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Aarts, E.; Verhage, M.; Veenvliet, J.V.; Dolan, C.V.; van der Sluis, S. A solution to dependency: using multilevel analysis to accommodate nested data. Nat. Neurosci. 2014, 17, 491–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Software 2015. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk?/projects/fastqc/ (accessed on 10 February 2020).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Development Core Team, R. R: A Language and Environment for Statistical Computing. Available online: http://www.r-project.org (accessed on 10 February 2020).
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [Green Version]
- Hulsen, T.; de Vlieg, J.; Alkema, W. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 2008, 9, 488. [Google Scholar] [CrossRef] [Green Version]
- Russo, S.J.; Dietz, D.M.; Dumitriu, D.; Morrison, J.H.; Malenka, R.C.; Nestler, E.J. The addicted synapse: Mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 2010. [Google Scholar] [CrossRef] [Green Version]
- Arikkath, J. Molecular mechanisms of dendrite morphogenesis. Front. Cell. Neurosci. 2012. [Google Scholar] [CrossRef] [Green Version]
- Uys, J.D.; McGuier, N.S.; Gass, J.T.; Griffin, W.C.; Ball, L.E.; Mulholland, P.J. Chronic intermittent ethanol exposure and withdrawal leads to adaptations in nucleus accumbens core postsynaptic density proteome and dendritic spines. Addict. Biol. 2016. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tritchler, D.; Parkhomenko, E.; Beyene, J. Filtering Genes for Cluster and Network Analysis. BMC Bioinform. 2009, 10, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Akbar, M.; Egli, M.; Cho, Y.-E.; Song, B.-J.; Noronha, A. Medications for alcohol use disorders: An overview. Pharmacol. Ther. 2018, 185, 64–85. [Google Scholar] [CrossRef]
- Williams, E.C.; Matson, T.E.; Harris, A.H.S. Strategies to increase implementation of pharmacotherapy for alcohol use disorders: a structured review of care delivery and implementation interventions. Addict. Sci. Clin. Pract. 2019, 14, 6. [Google Scholar] [CrossRef] [Green Version]
- Cassataro, D.; Bergfeldt, D.; Malekian, C.; Van Snellenberg, J.X.; Thanos, P.K.; Fishell, G.; Sjulson, L. Reverse pharmacogenetic modulation of the nucleus accumbens reduces ethanol consumption in a limited access paradigm. Neuropsychopharmacology 2014, 39, 283–290. [Google Scholar] [CrossRef]
- DePoy, L.; Daut, R.; Brigman, J.L.; MacPherson, K.; Crowley, N.; Gunduz-Cinar, O.; Pickens, C.L.; Cinar, R.; Saksida, L.M.; Kunos, G.; et al. Chronic alcohol produces neuroadaptations to prime dorsal striatal learning. Proc. Natl. Acad. Sci. USA 2013, 110, 14783–14788. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Cheng, Y.; Wang, X.; Roltsch Hellard, E.; Ma, T.; Gil, H.; Ben Hamida, S.; Ron, D. Alcohol Elicits Functional and Structural Plasticity Selectively in Dopamine D1 Receptor-Expressing Neurons of the Dorsomedial Striatum. J. Neurosci. 2015, 35, 11634–11643. [Google Scholar] [CrossRef] [Green Version]
- Kupchik, Y.M.; Kalivas, P.W. The Direct and Indirect Pathways of the Nucleus Accumbens are not What You Think. Neuropsychopharmacology 2017, 42, 369–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moonat, S.; Starkman, B.G.; Sakharkar, A.; Pandey, S.C. Neuroscience of alcoholism: molecular and cellular mechanisms. Cell. Mol. Life Sci. 2010, 67, 73–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spijker, S.; Houtzager, S.W.J.; De Gunst, M.C.M.; De Boer, W.P.H.; Schoffelmeer, A.N.M.; Smit, A.B. Morphine exposure and abstinence define specific stages of gene expression in the rat nucleus accumbens. FASEB J. 2004, 18, 848–850. [Google Scholar] [CrossRef] [PubMed]
- Roche, K.W. The expanding role of PSD-95: a new link to addiction. Trends Neurosci. 2004, 27, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.-D.; Gainetdinov, R.R.; Arbuckle, M.I.; Sotnikova, T.D.; Cyr, M.; Beaulieu, J.-M.; Torres, G.E.; Grant, S.G.N.; Caron, M.G. Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron 2004, 41, 625–638. [Google Scholar] [CrossRef] [Green Version]
- Mulligan, M.K.; Rhodes, J.S.; Crabbe, J.C.; Mayfield, R.D.; Adron Harris, R.; Ponomarev, I. Molecular Profiles of Drinking Alcohol to Intoxication in C57BL/6J Mice. Alcohol. Clin. Exp. Res. 2011, 35, 659–670. [Google Scholar] [CrossRef]
- Ferguson, L.B.; Zhang, L.; Kircher, D.; Wang, S.; Mayfield, R.D.; Crabbe, J.C.; Morrisett, R.A.; Harris, R.A.; Ponomarev, I. Dissecting Brain Networks Underlying Alcohol Binge Drinking Using a Systems Genomics Approach. Mol. Neurobiol. 2019, 56, 2791–2810. [Google Scholar]
- Ozburn, A.R.; Falcon, E.; Mukherjee, S.; Gillman, A.; Arey, R.; Spencer, S.; McClung, C.A. The Role of Clock in Ethanol-Related Behaviors. Neuropsychopharmacology 2013, 38, 2393–2400. [Google Scholar] [CrossRef] [Green Version]
- Blednov, Y.A.; Black, M.; Chernis, J.; Da Costa, A.; Mayfield, J.; Harris, R.A. Ethanol Consumption in Mice Lacking CD14, TLR2, TLR4, or MyD88. Alcohol. Clin. Exp. Res. 2017, 41, 516–530. [Google Scholar] [CrossRef]
- Wolstenholme, J.T.; Warner, J.A.; Capparuccini, M.I.; Archer, K.J.; Shelton, K.L.; Miles, M.F. Genomic analysis of individual differences in ethanol drinking: evidence for non-genetic factors in C57BL/6 mice. PLoS ONE 2011, 6, e21100. [Google Scholar] [CrossRef] [Green Version]
- Sakharkar, A.J.; Zhang, H.; Tang, L.; Shi, G.; Pandey, S.C. Histone Deacetylases (HDAC)-Induced Histone Modifications in the Amygdala: A Role in Rapid Tolerance to the Anxiolytic Effects of Ethanol. Alcohol. Clin. Exp. Res. 2012, 36, 61–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon-O’Brien, E.; Alaux-Cantin, S.; Warnault, V.; Buttolo, R.; Naassila, M.; Vilpoux, C. The histone deacetylase inhibitor sodium butyrate decreases excessive ethanol intake in dependent animals. Addict. Biol. 2015, 20, 676–689. [Google Scholar] [CrossRef] [PubMed]
- Trazzi, S.; Fuchs, C.; Viggiano, R.; De Franceschi, M.; Valli, E.; Jedynak, P.; Hansen, F.K.; Perini, G.; Rimondini, R.; Kurz, T.; et al. HDAC4: a key factor underlying brain developmental alterations in CDKL5 disorder. Hum. Mol. Genet. 2016, 25, 3887–3907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourgeron, T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 2015, 16, 551–563. [Google Scholar] [CrossRef]
| Number of Animals and Cells Per Group Included for Morphological Analyses | |||||||
|---|---|---|---|---|---|---|---|
| Group | Animal (n) | Cells/Brain#1 | Cells/Brain#2 | Cells/Brain#3 | Cells/Brain#4 | Cells/Brain#5 | Total Cells |
| H2O+Veh | 4 | 9 | 5 | 6 | 9 | 0 | 29 |
| H2O+CNO | 5 | 5 | 7 | 5 | 6 | 13 | 36 |
| EtOH+Veh | 4 | 6 | 10 | 5 | 8 | 0 | 29 |
| EtOH+CNO | 4 | 6 | 5 | 6 | 6 | 0 | 23 |
| Totals | 17 | 117 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pozhidayeva, D.Y.; Farris, S.P.; Goeke, C.M.; Firsick, E.J.; Townsley, K.G.; Guizzetti, M.; Ozburn, A.R. Chronic Chemogenetic Stimulation of the Nucleus Accumbens Produces Lasting Reductions in Binge Drinking and Ameliorates Alcohol-Related Morphological and Transcriptional Changes. Brain Sci. 2020, 10, 109. https://doi.org/10.3390/brainsci10020109
Pozhidayeva DY, Farris SP, Goeke CM, Firsick EJ, Townsley KG, Guizzetti M, Ozburn AR. Chronic Chemogenetic Stimulation of the Nucleus Accumbens Produces Lasting Reductions in Binge Drinking and Ameliorates Alcohol-Related Morphological and Transcriptional Changes. Brain Sciences. 2020; 10(2):109. https://doi.org/10.3390/brainsci10020109
Chicago/Turabian StylePozhidayeva, Dar’ya Y., Sean P. Farris, Calla M. Goeke, Evan J. Firsick, Kayla G. Townsley, Marina Guizzetti, and Angela R. Ozburn. 2020. "Chronic Chemogenetic Stimulation of the Nucleus Accumbens Produces Lasting Reductions in Binge Drinking and Ameliorates Alcohol-Related Morphological and Transcriptional Changes" Brain Sciences 10, no. 2: 109. https://doi.org/10.3390/brainsci10020109
APA StylePozhidayeva, D. Y., Farris, S. P., Goeke, C. M., Firsick, E. J., Townsley, K. G., Guizzetti, M., & Ozburn, A. R. (2020). Chronic Chemogenetic Stimulation of the Nucleus Accumbens Produces Lasting Reductions in Binge Drinking and Ameliorates Alcohol-Related Morphological and Transcriptional Changes. Brain Sciences, 10(2), 109. https://doi.org/10.3390/brainsci10020109





