Microbial Peptidases: Key Players in Reducing Gluten Immunogenicity Through Peptide Degradation
Abstract
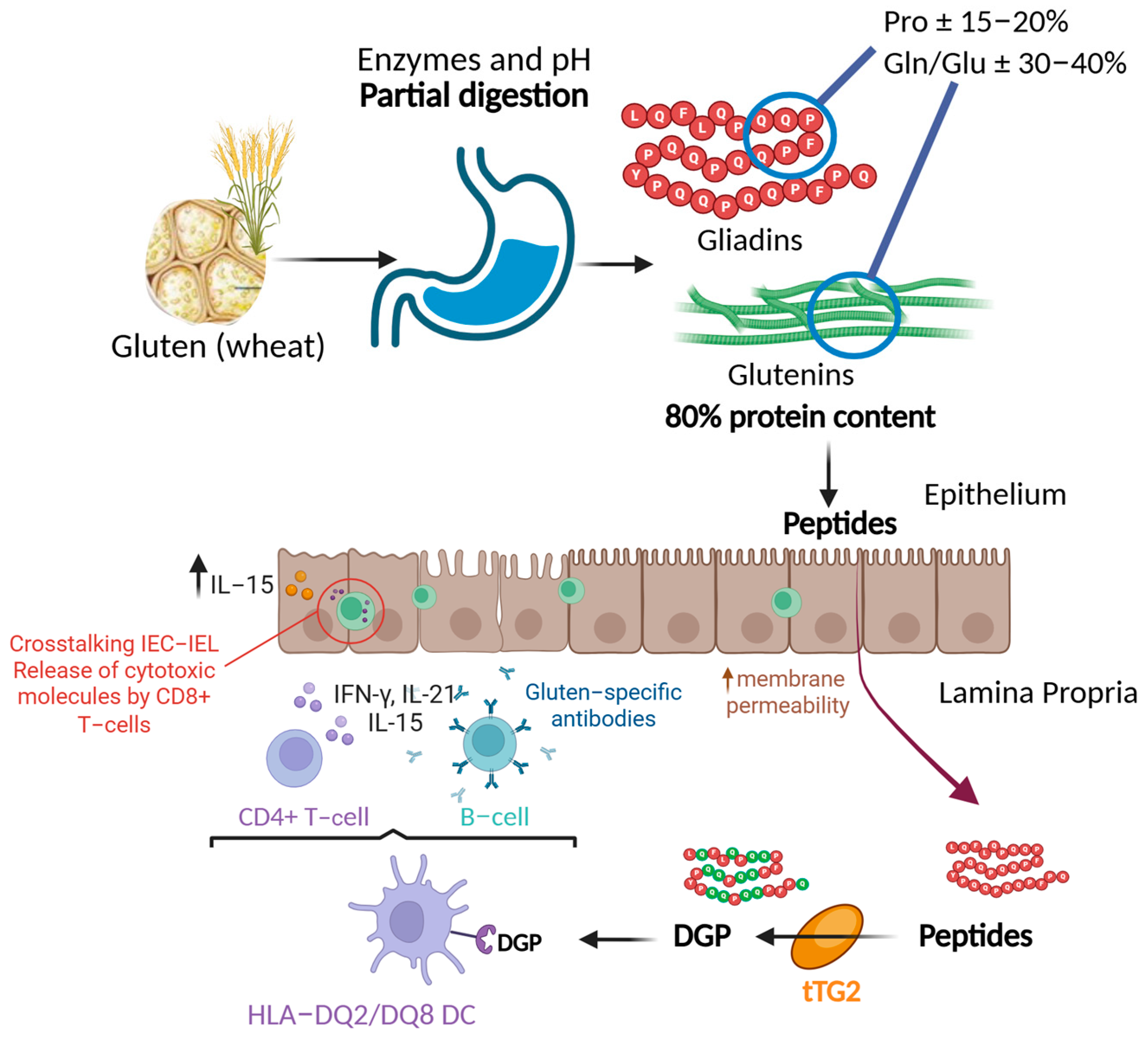
1. Introduction to Celiac Disease and Non-Celiac Gluten Sensitivity
2. Gluten, Gliadin, and Immunogenic Peptides: Characteristics
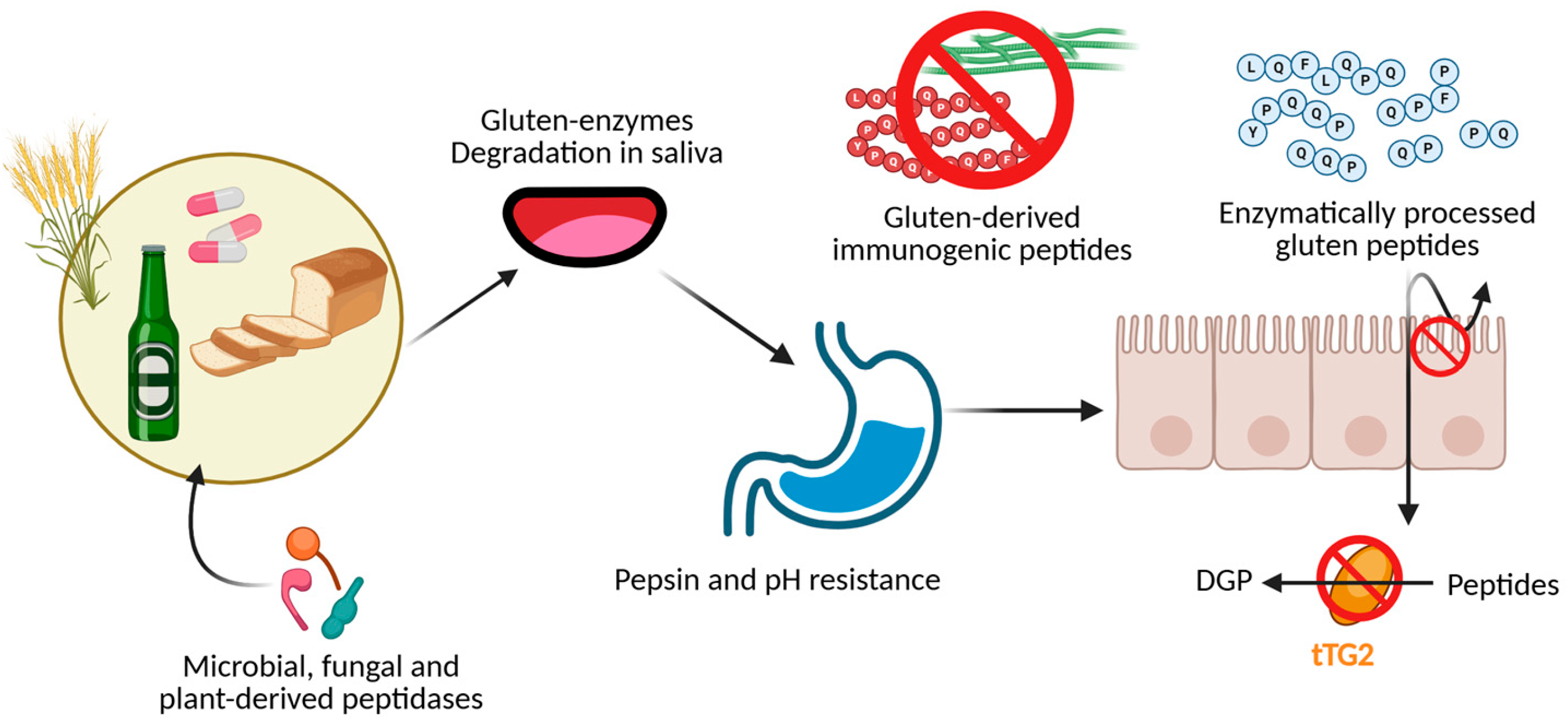
3. Treatment for Gluten Elimination: Pros and Cons
4. Enzymes Capable of Degrading Immunogenic Peptides and Their Industrial Applications from Microorganisms
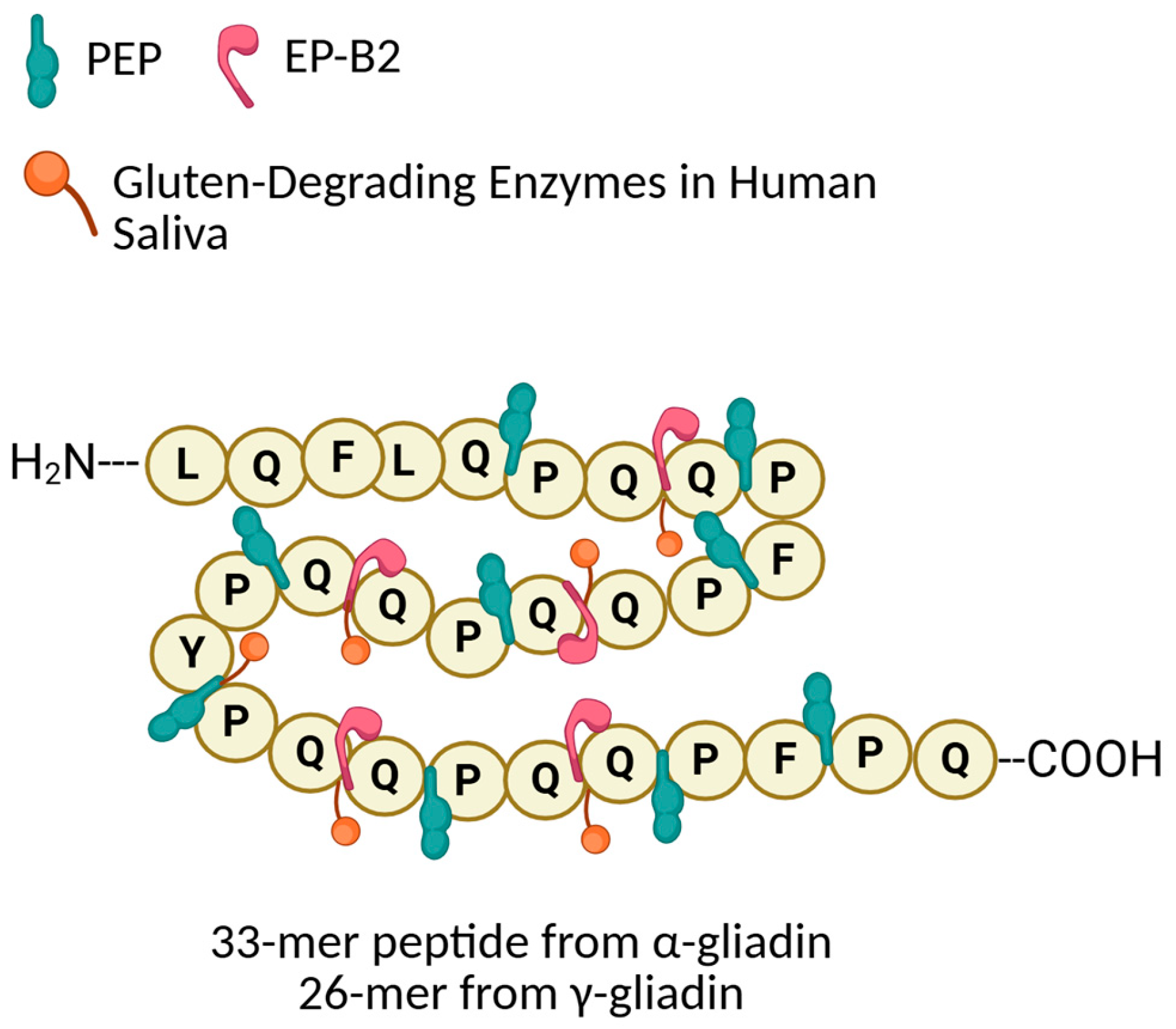
- Prolyl Endopeptidases (PEP):
- 2.
- Glutamine-Specific Cysteine Endoprotease (EP-B2):
- 3.
- Gluten-Degrading Enzymes in Human Saliva:
| Enzyme | Organism | Degraded Substrate | Probiotic | Degradation Efficiency | pH Stability Range | Commercial or Clinical Use | Reference |
|---|---|---|---|---|---|---|---|
| Prolyloligopeptidase (POP) | Myxococcus xanthus | Proline-rich peptides | No | High activity against 33-mer; no reported kinetic constants | Neutral to slightly alkaline | Research stage | [81] |
| Prolyloligopeptidase (POP) | Sphingomonas capsulata | Proline-rich peptides | No | Km ≈ 82 µM; effective in combination therapy (ALV003) | pH 6–8 | Approved for clinical testing | |
| Prolyloligopeptidase (POP) | Flavobacterium meningosepticum | Immunogenic gliadin peptides | No | Effective in simulated digestion; structural features described | Neutral | Research stage | [43] |
| Dipeptidylpeptidase IV (DPP IV) | Lactobacillus casei | Proline-rich peptides | Yes | Moderate cleavage activity; kinetics not reported | ~7 | Over the counter supplement | [43] |
| Prolylendopeptidase (PEP) | Aspergillus niger | Immunogenic gluten peptides | No | >80% 33-mer degradation in vitro; Km ≈ 170 µM | pH 3–5 | Available as Tolerase G | [48,82] |
| Prolylendopeptidase (PEP) | Flammulina velutipes | Immunogenic gliadin peptides | No | Specific activity = 56.7 U/mg (Z-Gly-Pro-pNA) | Neutral | Used in industrial gluten removal | [43] |
| Glutamine-specific cysteine endoprotease EP-B2 | Hordeum vulgare | Immunogenic gliadin peptides | No | Km = 30–100 µM; kcat ≈ 3.2–5.4 s−1 | pH 3–5 | Combined with SC-PEP in ALV003 | [48,64] |
| Prolyloligopeptidase | Tenebrio molitor | Proline-rich peptides | No | In vitro activity confirmed; no kinetic data | Acidic to neutral | Potential for food processing | [48] |
| Postglutamine peptidase | Tribolium castaneum | Glutamine-rich peptides | No | Km ≈ 25 µM (QPQLPYPQPQ); effective on immunogenic targets | Acid stable | Research stage | [48] |
| Prolyloligopeptidase (POP) | Rothia sp. | Immunogenic gliadin peptides | Yes | Comparable to AN-PEP in oral gluten hydrolysis | pH 5–7 | Experimental probiotic use | [70,83,84] |
| Peptidase | Actinomyces odontolyticus | 33-mer peptide | Yes | Selective degradation shown in vitro | Neutral | Investigational oral application | [84,85] |
5. Future Perspectives on the Application of Enzymes for Celiac Disease Treatment
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scherf, K.A. Gluten and Wheat-Related Disorders. In Handbook of Food Allergen Detection and Control; Elsevier: Amsterdam, The Netherlands, 2025; pp. 369–387. [Google Scholar]
- Catassi, G.; Marmo, C.; Gasbarrini, A.; Riccioni, M.E. Role of Device-Assisted Enteroscopy in Crohn’s Disease. J. Clin. Med. 2024, 13, 3919. [Google Scholar] [CrossRef]
- Rahmani, S.; Galipeau, H.J.; Clarizio, A.V.; Wang, X.; Hann, A.; Rueda, G.H.; Kirtikar, U.N.; Constante, M.; Wulczynski, M.; Su, H.-M.; et al. Gluten-Dependent Activation of CD4+ T Cells by MHC Class II–Expressing Epithelium. Gastroenterology 2024, 167, 1113–1128. [Google Scholar] [CrossRef]
- San-Martin, M.I.; Chamizo-Ampudia, A.; Sanchiz, Á.; Ferrero, M.Á.; Martínez-Blanco, H.; Rodríguez-Aparicio, L.B.; Navasa, N. Microbiome Markers in Gastrointestinal Disorders: Inflammatory Bowel Disease, Colorectal Cancer, and Celiac Disease. Int. J. Mol. Sci. 2025, 26, 4818. [Google Scholar] [CrossRef]
- Caio, G.; Volta, U.; Sapone, A.; Leffler, D.A.; De Giorgio, R.; Catassi, C.; Fasano, A. Celiac Disease: A Comprehensive Current Review. BMC Med. 2019, 17, 142. [Google Scholar] [CrossRef] [PubMed]
- Poddighe, D.; Abdukhakimova, D. Celiac Disease in Asia beyond the Middle East and Indian Subcontinent: Epidemiological Burden and Diagnostic Barriers. World J. Gastroenterol. 2021, 27, 2251–2256. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Alaedini, A.; Bojarski, C.; Bonaz, B.; Bouma, G.; Carroccio, A.; Castillejo, G.; De Magistris, L.; Dieterich, W.; Di Liberto, D.; et al. The Overlapping Area of Non-Celiac Gluten Sensitivity (NCGS) and Wheat-Sensitive Irritable Bowel Syndrome (IBS): An Update. Nutrients 2017, 9, 1268. [Google Scholar] [CrossRef] [PubMed]
- Rubin, E.B.; Viscuso, M.R.; Moleski, S.M. Maintaining a Balanced Diet While Gluten-Free: Treatment Options. Curr. Treat. Options Gastroenterol. 2020, 18, 699–717. [Google Scholar] [CrossRef]
- Wieser, H.; Segura, V.; Ruiz-Carnicer, Á.; Sousa, C.; Comino, I. Food Safety and Cross-Contamination of Gluten-Free Products: A Narrative Review. Nutrients 2021, 13, 2244. [Google Scholar] [CrossRef]
- Liu, S.; El Khoury, D.; Joye, I.J. Gluten-Free Product Recalls and Their Impact on Consumer Trust. Nutrients 2023, 15, 4170. [Google Scholar] [CrossRef]
- Catassi, C.; Monachesi, C.; Catassi, G.N.; Lionetti, E. Lessons from Low-Gluten Challenge Studies in Celiac Disease: Is It Time to Reconsider the Gluten Threshold in Gluten-Free Food? Dig. Liver Dis. 2025, 57, 1105–1106. [Google Scholar] [CrossRef]
- Mandal, S.; Verma, A.K. Wheat Breeding, Fertilizers, and Pesticides: Do They Contribute to the Increasing Immunogenic Properties of Modern Wheat? Gastrointest. Disord. 2021, 3, 247–264. [Google Scholar] [CrossRef]
- Ali, A.; AlHussaini, K.I. Pesticides: Unintended Impact on the Hidden World of Gut Microbiota. Metabolites 2024, 14, 155. [Google Scholar] [CrossRef]
- Sawant, S.S.; Park, H.-Y.; Sim, E.-Y.; Kim, H.-S.; Choi, H.-S. Microbial Fermentation in Food: Impact on Functional Properties and Nutritional Enhancement—A Review of Recent Developments. Fermentation 2025, 11, 15. [Google Scholar] [CrossRef]
- Fasano, A.; Catassi, C. Current Approaches to Diagnosis and Treatment of Celiac Disease: An Evolving Spectrum. Gastroenterology 2001, 120, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, H.; Huilian, C.; Fu, L. Detoxification Mechanism and the Impact of Transamidation-Modified Gliadin on Celiac-Based Gluten Sensitivity: The Potential of Unlocking Gluten Tolerance in Functional Food. J. Agric. Food Chem. 2025, 73, 1682–1693. [Google Scholar] [CrossRef] [PubMed]
- Gélinas, P.; Théolier, J. How to Reduce Gluten in Foods: A Critical Review of Patents. Int. J. Food Sci. Technol. 2024, 59, 8069–8081. [Google Scholar] [CrossRef]
- Mamone, G.; Di Stasio, L.; Vitale, S.; Picascia, S.; Gianfrani, C. Analytical and Functional Approaches to Assess the Immunogenicity of Gluten Proteins. Front. Nutr. 2023, 9, 1049623. [Google Scholar] [CrossRef]
- Pourmohammadi, K.; Abedi, E. Hydrolytic Enzymes and Their Directly and Indirectly Effects on Gluten and Dough Properties: An Extensive Review. Food Sci. Nutr. 2021, 9, 3988–4006. [Google Scholar] [CrossRef]
- Scherf, K.A.; Catassi, C.; Chirdo, F.; Ciclitira, P.J.; Feighery, C.; Gianfrani, C.; Koning, F.; Lundin, K.E.A.; Schuppan, D.; Smulders, M.J.M.; et al. Recent Progress and Recommendations on Celiac Disease From the Working Group on Prolamin Analysis and Toxicity. Front. Nutr. 2020, 7, 29. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, H.; Fu, L. A Review of Gluten Detoxification in Wheat for Food Applications: Approaches, Mechanisms, and Implications. Crit. Rev. Food Sci. Nutr. 2025, 65, 2100–2116. [Google Scholar] [CrossRef]
- Xu, K.; Kuang, J. Effects of Thermal Treatment on the Physicochemical and Structural Properties of Wheat Gluten Proteins: Insights from Gluten, Glutenin, and Gliadin Fractions. Int. J. Food Sci. Technol. 2024, 59, 2275–2285. [Google Scholar] [CrossRef]
- Ogilvie, O.; Roberts, S.; Sutton, K.; Gerrard, J.; Larsen, N.; Domigan, L. The Effect of Baking Time and Temperature on Gluten Protein Structure and Celiac Peptide Digestibility. Food Res. Int. 2021, 140, 109988. [Google Scholar] [CrossRef] [PubMed]
- Frick, R.; Høydahl, L.S.; Petersen, J.; du Pré, M.F.; Kumari, S.; Berntsen, G.; Dewan, A.E.; Jeliazkov, J.R.; Gunnarsen, K.S.; Frigstad, T.; et al. A High-Affinity Human TCR-like Antibody Detects Celiac Disease Gluten Peptide–MHC Complexes and Inhibits T Cell Activation. Sci. Immunol. 2021, 6, eabg4925. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, B. Gluten-Related Disorders: Celiac Disease, Wheat Allergy, and Nonceliac Gluten Sensitivity. Crit. Rev. Food Sci. Nutr. 2020, 60, 2606–2621. [Google Scholar] [CrossRef]
- Abbasi, A.; Bazzaz, S.; Ibrahim, S.A.; Hekmatdoost, A.; Hosseini, H.; Sabahi, S.; Sheykhsaran, E.; Rahbar Saadat, Y.; Asghari Ozma, M.; Lahouty, M. A Critical Review on the Gluten-Induced Enteropathy/Celiac Disease: Gluten-Targeted Dietary and Non-Dietary Therapeutic Approaches. Food Rev. Int. 2024, 40, 883–923. [Google Scholar] [CrossRef]
- Baharin, A.; Ting, T.-Y.; Goh, H.-H. Post-Proline Cleaving Enzymes (PPCEs): Classification, Structure, Molecular Properties, and Applications. Plants 2022, 11, 1330. [Google Scholar] [CrossRef]
- Pinto-Sanchez, M.I.; Silvester, J.A.; Lebwohl, B.; Leffler, D.A.; Anderson, R.P.; Therrien, A.; Kelly, C.P.; Verdu, E.F. Society for the Study of Celiac Disease Position Statement on Gaps and Opportunities in Coeliac Disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 875–884. [Google Scholar] [CrossRef]
- de Lourdes Moreno, M.; Segura, V.; Ruiz-Carnicer, Á.; Nájar, A.M.; Comino, I.; Sousa, C. Oral Enzyme Strategy in Celiac Disease. In Biotechnological Strategies for the Treatment of Gluten Intolerance; Elsevier: Amsterdam, The Netherlands, 2021; pp. 201–220. [Google Scholar]
- Cristofori, F.; Francavilla, R.; Capobianco, D.; Dargenio, V.N.; Filardo, S.; Mastromarino, P. Bacterial-Based Strategies to Hydrolyze Gluten Peptides and Protect Intestinal Mucosa. Front. Immunol. 2020, 11, 567801. [Google Scholar] [CrossRef]
- Gasparre, N.; Rosell, C.M. Wheat Gluten: A Functional Protein Still Challenging to Replace in Gluten-free Cereal-based Foods. Cereal Chem. 2023, 100, 243–255. [Google Scholar] [CrossRef]
- Stanciu, D.; Staykov, H.; Dragomanova, S.; Tancheva, L.; Pop, R.; Ielciu, I.; Crișan, G. Gluten Unraveled: Latest Insights on Terminology, Diagnosis, Pathophysiology, Dietary Strategies, and Intestinal Microbiota Modulations—A Decade in Review. Nutrients 2024, 16, 3636. [Google Scholar] [CrossRef]
- Woldemariam, K.Y.; Yuan, J.; Wan, Z.; Yu, Q.; Cao, Y.; Mao, H.; Liu, Y.; Wang, J.; Li, H.; Sun, B. Celiac Disease and Immunogenic Wheat Gluten Peptides and the Association of Gliadin Peptides with HLA DQ2 and HLA DQ8. Food Rev. Int. 2022, 38, 1553–1576. [Google Scholar] [CrossRef]
- Rani, M.; Sogi, D.S.; Gill, B.S. Characterization of Gliadin, Secalin and Hordein Fractions Using Analytical Techniques. Sci. Rep. 2021, 11, 23135. [Google Scholar] [CrossRef]
- Voisine, J.; Abadie, V. Interplay Between Gluten, HLA, Innate and Adaptive Immunity Orchestrates the Development of Coeliac Disease. Front. Immunol. 2021, 12, 674313. [Google Scholar] [CrossRef]
- Lindstad, C.B.; Dewan, A.E.; Stamnaes, J.; Sollid, L.M.; du Pré, M.F. TG2-Gluten Complexes as Antigens for Gluten-Specific and Transglutaminase-2 Specific B Cells in Celiac Disease. PLoS ONE 2021, 16, e0259082. [Google Scholar] [CrossRef] [PubMed]
- Dodero, V.I.; Herrera, M.G. Oligomerization of 33-mer Gliadin Peptides: Supramolecular Assemblies in Celiac Disease. ChemMedChem 2025, 20, e202400789. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.R.; Byrne, G.; Chirdo, F.G.; Feighery, C. Coeliac Disease Pathogenesis: The Uncertainties of a Well-Known Immune Mediated Disorder. Front. Immunol. 2020, 11, 1374. [Google Scholar] [CrossRef] [PubMed]
- Cardo, A.; Churruca, I.; Lasa, A.; Navarro, V.; Vázquez-Polo, M.; Perez-Junkera, G.; Larretxi, I. Nutritional Imbalances in Adult Celiac Patients Following a Gluten-Free Diet. Nutrients 2021, 13, 2877. [Google Scholar] [CrossRef]
- Jabri, B.; Abadie, V. IL-15 Functions as a Danger Signal to Regulate Tissue-Resident T Cells and Tissue Destruction. Nat. Rev. Immunol. 2015, 15, 771–783. [Google Scholar] [CrossRef]
- Kõiv, V.; Tenson, T. Gluten-Degrading Bacteria: Availability and Applications. Appl. Microbiol. Biotechnol. 2021, 105, 3045–3059. [Google Scholar] [CrossRef]
- Shan, L.; Molberg, Ø.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural Basis for Gluten Intolerance in Celiac Sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef]
- Hausch, F.; Shan, L.; Santiago, N.A.; Gray, G.M.; Khosla, C. Intestinal Digestive Resistance of Immunodominant Gliadin Peptides. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 283, G996–G1003. [Google Scholar] [CrossRef]
- Pinto-Sanchez, M.I.; Blom, J.-J.; Gibson, P.R.; Armstrong, D. Nutrition Assessment and Management in Celiac Disease. Gastroenterology 2024, 167, 116–131.e1. [Google Scholar] [CrossRef]
- Wei, G.; Helmerhorst, E.J.; Darwish, G.; Blumenkranz, G.; Schuppan, D. Gluten Degrading Enzymes for Treatment of Celiac Disease. Nutrients 2020, 12, 2095. [Google Scholar] [CrossRef]
- Freitas, D.; Gómez-Mascaraque, L.G.; Brodkorb, A. Digestion of Protein and Toxic Gluten Peptides in Wheat Bread, Pasta and Cereal and the Effect of a Supplemental Enzyme Mix. Front. Nutr. 2022, 9, 986272. [Google Scholar] [CrossRef]
- Segura, V.; Ruiz-Carnicer, Á.; Sousa, C.; Moreno, M.D.L. New Insights into Non-Dietary Treatment in Celiac Disease: Emerging Therapeutic Options. Nutrients 2021, 13, 2146. [Google Scholar] [CrossRef] [PubMed]
- Dunaevsky, Y.E.; Tereshchenkova, V.F.; Belozersky, M.A.; Filippova, I.Y.; Oppert, B.; Elpidina, E.N. Effective Degradation of Gluten and Its Fragments by Gluten-Specific Peptidases: A Review on Application for the Treatment of Patients with Gluten Sensitivity. Pharmaceutics 2021, 13, 1603. [Google Scholar] [CrossRef]
- Vaquero, L.; Rodríguez-Martín, L.; León, F.; Jorquera, F.; Vivas, S. New Coeliac Disease Treatments and Their Complications. Gastroenterol. Hepatol. (Engl. Ed.) 2018, 41, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Palaiodimou, L.; Kokotis, P.; Zompola, C.; Papagiannopoulou, G.; Bakola, E.; Papadopoulou, M.; Zouvelou, V.; Petras, D.; Vlachopoulos, C.; Tsivgoulis, G. Fabry Disease: Current and Novel Therapeutic Strategies. A Narrative Review. Curr. Neuropharmacol. 2023, 21, 440–456. [Google Scholar] [CrossRef]
- Hossain, I.; Mitu, I.J.; Hasan, M.R.; Saha, S.R. Industrial Enzyme Production in Bangladesh: Current Landscape, Scope, and Challenges. Asian J. Med. Biol. Res. 2023, 9, 145–159. [Google Scholar] [CrossRef]
- Ianiro, G.; Pecere, S.; Giorgio, V.; Gasbarrini, A.; Cammarota, G. Digestive Enzyme Supplementation in Gastrointestinal Diseases. Curr. Drug Metab. 2016, 17, 187–193. [Google Scholar] [CrossRef]
- Brzozowski, B.; Stasiewicz, K.; Adamczak, M. Biotransformation of Wheat Proteins with Lactobacillus Sp. Following High Hydrostatic Pressure Pre-Treatment to Reduce Gluten Immunoreactivity. J. Cereal Sci. 2023, 114, 103787. [Google Scholar] [CrossRef]
- Gass, J.; Ehren, J.; Strohmeier, G.; Isaacs, I.; Khosla, C. Fermentation, Purification, Formulation, and Pharmacological Evaluation of a Prolyl Endopeptidase from Myxococcus Xanthus: Implications for Celiac Sprue Therapy. Biotechnol. Bioeng. 2005, 92, 674–684. [Google Scholar] [CrossRef]
- Kabashima, T.; Fujii, M.; Meng, Y.; Ito, K.; Yoshimoto, T. Prolyl Endopeptidase FromSphingomonas Capsulata:Isolation and Characterization of the Enzyme and Nucleotide Sequence of the Gene. Arch. Biochem. Biophys. 1998, 358, 141–148. [Google Scholar] [CrossRef]
- Diefenthal, T.; Dargatz, H.; Witte, V.; Reipen, G.; Svendsen, I. Cloning of Proline-Specific Endopeptidase Gene from Flavobacterium Meningosepticum: Expression in Escherichia Coli and Purification of the Heterologous Protein. Appl. Microbiol. Biotechnol. 1993, 40, 90–97. [Google Scholar] [CrossRef]
- Ekici, Ö.D.; Paetzel, M.; Dalbey, R.E. Unconventional Serine Proteases: Variations on the Catalytic Ser/His/Asp Triad Configuration. Protein Sci. 2008, 17, 2023–2037. [Google Scholar] [CrossRef]
- Pijning, T.; Vujičić-Žagar, A.; van der Laan, J.; de Jong, R.M.; Ramirez-Palacios, C.; Vente, A.; Edens, L.; Dijkstra, B.W. Structural and Time-resolved Mechanistic Investigations of Protein Hydrolysis by the Acidic Proline-specific Endoprotease from Aspergillus Niger. Protein Sci. 2024, 33, e4856. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Zhang, C.; Zhou, J.; Wang, S.; Meng, H.; Wu, M.; Zheng, Y.; Yu, R. Design of SC PEP with Enhanced Stability against Pepsin Digestion and Increased Activity by Machine Learning and Structural Parameters Modeling. Int. J. Biol. Macromol. 2023, 250, 125933. [Google Scholar] [CrossRef] [PubMed]
- Stefanolo, J.P.; Segura, V.; Grizzuti, M.; Heredia, A.; Comino, I.; Costa, A.F.; Puebla, R.; Temprano, M.P.; Niveloni, S.I.; de Diego, G.; et al. Effect of Aspergillus Niger Prolyl Endopeptidase in Patients with Celiac Disease on a Long-Term Gluten-Free Diet. World J. Gastroenterol. 2024, 30, 1545–1555. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Holster, S.; Bruins, M.J.; Brummer, R.J. Randomized Clinical Trial: Effective Gluten Degradation by Aspergillus Niger-Derived Enzyme in a Complex Meal Setting. Sci. Rep. 2017, 7, 13100. [Google Scholar] [CrossRef]
- Shukla, M.; Gharote, A.; Muchahary, S. Industrial Scope of Cysteine Protease and Its Anti-Gluten Property: A Review. Food Nutr. 2025, 1, 100006. [Google Scholar] [CrossRef]
- Gass, J.; Vora, H.; Bethune, M.T.; Gray, G.M.; Khosla, C. Effect of Barley Endoprotease EP-B2 on Gluten Digestion in the Intact Rat. J. Pharmacol. Exp. Ther. 2006, 318, 1178–1186. [Google Scholar] [CrossRef]
- Bethune, M.T.; Strop, P.; Tang, Y.; Sollid, L.M.; Khosla, C. Heterologous Expression, Purification, Refolding, and Structural-Functional Characterization of EP-B2, a Self-Activating Barley Cysteine Endoprotease. Chem. Biol. 2006, 13, 637–647. [Google Scholar] [CrossRef]
- Denessiouk, K.; Uversky, V.N.; Permyakov, S.E.; Permyakov, E.A.; Johnson, M.S.; Denesyuk, A.I. Papain-like Cysteine Proteinase Zone (PCP-Zone) and PCP Structural Catalytic Core (PCP-SCC) of Enzymes with Cysteine Proteinase Fold. Int. J. Biol. Macromol. 2020, 165, 1438–1446. [Google Scholar] [CrossRef]
- Bethune, M.T.; Ribka, E.; Khosla, C.; Sestak, K. Transepithelial Transport and Enzymatic Detoxification of Gluten in Gluten-Sensitive Rhesus Macaques. PLoS ONE 2008, 3, e1857. [Google Scholar] [CrossRef]
- Vora, H.; McIntire, J.; Kumar, P.; Deshpande, M.; Khosla, C. A Scaleable Manufacturing Process for Pro-EP-B2, a Cysteine Protease from Barley Indicated for Celiac Sprue. Biotechnol. Bioeng. 2007, 98, 177–185. [Google Scholar] [CrossRef]
- Yoosuf, S.; Makharia, G.K. Evolving Therapy for Celiac Disease. Front. Pediatr. 2019, 7, 193. [Google Scholar] [CrossRef]
- Darwish, G.; Helmerhorst, E.J.; Schuppan, D.; Oppenheim, F.G.; Wei, G. Pharmaceutically Modified Subtilisins Withstand Acidic Conditions and Effectively Degrade Gluten in Vivo. Sci. Rep. 2019, 9, 7505. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Darwish, G.; Oppenheim, F.G.; Schuppan, D.; Helmerhorst, E.J. Commensal Bacterium Rothia Aeria Degrades and Detoxifies Gluten via a Highly Effective Subtilisin Enzyme. Nutrients 2020, 12, 3724. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Tian, N.; Siezen, R.; Schuppan, D.; Helmerhorst, E.J. Identification of Food-Grade Subtilisins as Gluten-Degrading Enzymes to Treat Celiac Disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 311, G571–G580. [Google Scholar] [CrossRef] [PubMed]
- Zamakhchari, M.; Wei, G.; Dewhirst, F.; Lee, J.; Schuppan, D.; Oppenheim, F.G.; Helmerhorst, E.J. Identification of Rothia Bacteria as Gluten-Degrading Natural Colonizers of the Upper Gastro-Intestinal Tract. PLoS ONE 2011, 6, e24455. [Google Scholar] [CrossRef]
- Tian, N.; Faller, L.; Leffler, D.A.; Kelly, C.P.; Hansen, J.; Bosch, J.A.; Wei, G.; Paster, B.J.; Schuppan, D.; Helmerhorst, E.J. Salivary Gluten Degradation and Oral Microbial Profiles in Healthy Individuals and Celiac Disease Patients. Appl. Environ. Microbiol. 2017, 83, e03330-16. [Google Scholar] [CrossRef]
- Pryor, J.C.; Nieva, C.; Talley, N.J.; Eslick, G.D.; Duncanson, K.; Burns, G.L.; Hoedt, E.C.; Keely, S. Microbial-Derived Peptidases Are Altered in Celiac Disease, Non-Celiac Gluten Sensitivity, and Functional Dyspepsia: A Systematic Review and Re-Analysis of the Duodenal Microbiome. Gut Microbes 2025, 17, 2500063. [Google Scholar] [CrossRef]
- Kriaa, A.; Jablaoui, A.; Rhimi, S.; Soussou, S.; Mkaouar, H.; Mariaule, V.; Gruba, N.; Gargouri, A.; Maguin, E.; Lesner, A.; et al. SP-1, a Serine Protease from the Gut Microbiota, Influences Colitis and Drives Intestinal Dysbiosis in Mice. Cells 2021, 10, 2658. [Google Scholar] [CrossRef] [PubMed]
- Maciejewska, M.; Dąbrowska, A.; Cano-Lamadrid, M. Sustainable Protein Sources: Functional Analysis of Tenebrio Molitor Hydrolysates and Attitudes of Consumers in Poland and Spain Toward Insect-Based Foods. Foods 2025, 14, 333. [Google Scholar] [CrossRef] [PubMed]
- Dvoryakova, E.A.; Klimova, M.A.; Simonyan, T.R.; Dombrovsky, I.A.; Serebryakova, M.V.; Tereshchenkova, V.F.; Dunaevsky, Y.E.; Belozersky, M.A.; Filippova, I.Y.; Elpidina, E.N. Recombinant Cathepsin L of Tribolium Castaneum and Its Potential in the Hydrolysis of Immunogenic Gliadin Peptides. Int. J. Mol. Sci. 2022, 23, 7001. [Google Scholar] [CrossRef]
- Francavilla, R.; De Angelis, M.; Rizzello, C.G.; Cavallo, N.; Dal Bello, F.; Gobbetti, M. Selected Probiotic Lactobacilli Have the Capacity To Hydrolyze Gluten Peptides during Simulated Gastrointestinal Digestion. Appl. Environ. Microbiol. 2017, 83, e00376-17. [Google Scholar] [CrossRef] [PubMed]
- Schulz, K.; Giesler, L.; Linke, D.; Berger, R.G. A Prolyl Endopeptidase from Flammulina Velutipes for the Possible Degradation of Celiac Disease Provoking Toxic Peptides in Cereal Proteins. Process Biochem. 2018, 73, 47–55. [Google Scholar] [CrossRef]
- Bonner, E.R.; Tschollar, W.; Anderson, R.; Mourabit, S. Review Article: Novel Enzyme Therapy Design for Gluten Peptide Digestion Through Exopeptidase Supplementation. Aliment. Pharmacol. Ther. 2025, 61, 1123–1139. [Google Scholar] [CrossRef]
- SHAN, L.; MARTI, T.; SOLLID, L.M.; GRAY, G.M.; KHOSLA, C. Comparative Biochemical Analysis of Three Bacterial Prolyl Endopeptidases: Implications for Coeliac Sprue. Biochem. J. 2004, 383, 311–318. [Google Scholar] [CrossRef]
- Mitea, C.; Havenaar, R.; Drijfhout, J.W.; Edens, L.; Dekking, L.; Koning, F. Efficient Degradation of Gluten by a Prolyl Endoprotease in a Gastrointestinal Model: Implications for Coeliac Disease. Gut 2007, 57, 25–32. [Google Scholar] [CrossRef]
- Helmerhorst, E.J.; Zamakhchari, M.; Schuppan, D.; Oppenheim, F.G. Discovery of a Novel and Rich Source of Gluten-Degrading Microbial Enzymes in the Oral Cavity. PLoS ONE 2010, 5, e13264. [Google Scholar] [CrossRef]
- Fernandez-Feo, M.; Wei, G.; Blumenkranz, G.; Dewhirst, F.E.; Schuppan, D.; Oppenheim, F.G.; Helmerhorst, E.J. The Cultivable Human Oral Gluten-Degrading Microbiome and Its Potential Implications in Coeliac Disease and Gluten Sensitivity. Clin. Microbiol. Infect. 2013, 19, E386–E394. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Zhou, Y.; Wang, C.; Wu, B.; Wan, J. Actinomyces and Alimentary Tract Diseases: A Review of Its Biological Functions and Pathology. Biomed. Res. Int. 2018, 2018, 3820215. [Google Scholar] [CrossRef]
- Krishnareddy, S.; Stier, K.; Recanati, M.; Lebwohl, B.; Green, P.H. Commercially Available Glutenases: A Potential Hazard in Coeliac Disease. Ther. Adv. Gastroenterol. 2017, 10, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, G.; Leroux, J.-C. In Vivo Fluorescence Imaging of Exogenous Enzyme Activity in the Gastrointestinal Tract. Proc. Natl. Acad. Sci. USA 2011, 108, 9032–9037. [Google Scholar] [CrossRef]
- Ma, J.; Yan, L.; Yang, J.; He, Y.; Wu, L. Effect of Modification Strategies on the Biological Activity of Peptides/Proteins. ChemBioChem 2024, 25, e202300481. [Google Scholar] [CrossRef]
- Chugunov, A.O.; Dvoryakova, E.A.; Dyuzheva, M.A.; Simonyan, T.R.; Tereshchenkova, V.F.; Filippova, I.Y.; Efremov, R.G.; Elpidina, E.N. Fighting Celiac Disease: Improvement of PH Stability of Cathepsin L In Vitro by Computational Design. Int. J. Mol. Sci. 2023, 24, 12369. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Ye, R.-L.; Meng, M. Specificity Enhancement of Glutenase Bga1903 toward Celiac Disease-Eliciting Pro-Immunogenic Peptides via Active-Site Modification. Int. J. Mol. Sci. 2023, 25, 505. [Google Scholar] [CrossRef]
- Lähdeaho, M.-L.; Kaukinen, K.; Laurila, K.; Vuotikka, P.; Koivurova, O.-P.; Kärjä-Lahdensuu, T.; Marcantonio, A.; Adelman, D.C.; Mäki, M. Glutenase ALV003 Attenuates Gluten-Induced Mucosal Injury in Patients With Celiac Disease. Gastroenterology 2014, 146, 1649–1658. [Google Scholar] [CrossRef]
- Tye-Din, J.A.; Anderson, R.P.; Ffrench, R.A.; Brown, G.J.; Hodsman, P.; Siegel, M.; Botwick, W.; Shreeniwas, R. The Effects of ALV003 Pre-Digestion of Gluten on Immune Response and Symptoms in Celiac Disease in Vivo. Clin. Immunol. 2010, 134, 289–295. [Google Scholar] [CrossRef]
- Syage, J.A.; Green, P.H.R.; Khosla, C.; Adelman, D.C.; Sealey-Voyksner, J.A.; Murray, J.A. Latiglutenase Treatment for Celiac Disease: Symptom and Quality of Life Improvement for Seropositive Patients on a Gluten-free Diet. GastroHep 2019, 1, 293–301. [Google Scholar] [CrossRef]
- Murray, J.A.; Syage, J.A.; Wu, T.-T.; Dickason, M.A.; Ramos, A.G.; Van Dyke, C.; Horwath, I.; Lavin, P.T.; Mäki, M.; Hujoel, I.; et al. Latiglutenase Protects the Mucosa and Attenuates Symptom Severity in Patients With Celiac Disease Exposed to a Gluten Challenge. Gastroenterology 2022, 163, 1510–1521.e6. [Google Scholar] [CrossRef] [PubMed]
- Pultz, I.S.; Hill, M.; Vitanza, J.M.; Wolf, C.; Saaby, L.; Liu, T.; Winkle, P.; Leffler, D.A. Gluten Degradation, Pharmacokinetics, Safety, and Tolerability of TAK-062, an Engineered Enzyme to Treat Celiac Disease. Gastroenterology 2021, 161, 81–93.e3. [Google Scholar] [CrossRef] [PubMed]
- Ehren, J.; Morón, B.; Martin, E.; Bethune, M.T.; Gray, G.M.; Khosla, C. A Food-Grade Enzyme Preparation with Modest Gluten Detoxification Properties. PLoS ONE 2009, 4, e6313. [Google Scholar] [CrossRef] [PubMed]
- Mulvihill, E.E.; Drucker, D.J. Pharmacology, Physiology, and Mechanisms of Action of Dipeptidyl Peptidase-4 Inhibitors. Endocr. Rev. 2014, 35, 992–1019. [Google Scholar] [CrossRef]
- Gupta, S.; Sen, U. More than Just an Enzyme: Dipeptidyl Peptidase-4 (DPP-4) and Its Association with Diabetic Kidney Remodelling. Pharmacol. Res. 2019, 147, 104391. [Google Scholar] [CrossRef]
- Li, D.; Silvester, J.A.; Crowley, M.J.; Yang, J.Y.; Alexopoulos, A.-S.; Xu, Y.; Zhan, S.; Wang, T. Assessing the Association between Dipeptidyl Peptidase-4 Inhibitors Use and Celiac Disease through Drug Adverse Event Reporting. Ther. Adv. Chronic Dis. 2020, 11, 2040622320904301. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchiz, A.; San-Martín, M.I.; Navasa, N.; Martínez-Blanco, H.; Ferrero, M.Á.; Rodríguez-Aparicio, L.B.; Chamizo-Ampudia, A. Microbial Peptidases: Key Players in Reducing Gluten Immunogenicity Through Peptide Degradation. Appl. Sci. 2025, 15, 8111. https://doi.org/10.3390/app15148111
Sanchiz A, San-Martín MI, Navasa N, Martínez-Blanco H, Ferrero MÁ, Rodríguez-Aparicio LB, Chamizo-Ampudia A. Microbial Peptidases: Key Players in Reducing Gluten Immunogenicity Through Peptide Degradation. Applied Sciences. 2025; 15(14):8111. https://doi.org/10.3390/app15148111
Chicago/Turabian StyleSanchiz, Africa, M. Isabel San-Martín, N. Navasa, Honorina Martínez-Blanco, Miguel Ángel Ferrero, Leandro Benito Rodríguez-Aparicio, and Alejandro Chamizo-Ampudia. 2025. "Microbial Peptidases: Key Players in Reducing Gluten Immunogenicity Through Peptide Degradation" Applied Sciences 15, no. 14: 8111. https://doi.org/10.3390/app15148111
APA StyleSanchiz, A., San-Martín, M. I., Navasa, N., Martínez-Blanco, H., Ferrero, M. Á., Rodríguez-Aparicio, L. B., & Chamizo-Ampudia, A. (2025). Microbial Peptidases: Key Players in Reducing Gluten Immunogenicity Through Peptide Degradation. Applied Sciences, 15(14), 8111. https://doi.org/10.3390/app15148111










