Abstract
Enterobacteriaceae can contaminate meat during various processing stages, including slaughter, evisceration, and b utchering, potentially causing foodborne illnesses. The purpose of this study was to investigate the prevalence of Enterobacteriaceae in carcass samples obtained from slaughterhouses and meat cuts collected from butcher shops. A total of 120 samples of camel, cattle, and sheep meat were analyzed for microbial contamination and isolates were identified using the PCR test. Total viable count ranged from 4.91 to 5.37 Log10 CFU/g in slaughterhouses and butcher shops. E. coli dominated, with 84 out of the 120 samples (70%) contaminated, where contamination was highest in camel meat and lowest in sheep meat with 100% and 30% of contaminated samples, respectively. Salmonella was confirmed in 40% of camel, 47.5% of cattle, and 32.5% of sheep samples. In addition, twenty-five Enterobacteriaceae strains belonging to 19 different genera were detected in the meat samples. The highest occurrence was in the sheep samples with 15 different genera followed by the camels and the cattle samples with 14 different genera each. The presence of Enterobacteriaceae in camel, cattle, and sheep carcasses raises significant concerns regarding food safety. Adherence to good hygiene practices throughout animal slaughtering is crucial to minimize the risk of infection and transmission and ensure food safety.
Keywords:
Enterobacteriaceae; camels; cattle; sheep; slaughterhouses; butcher shops; Salmonella; Al-Ahsa 1. Introduction
Foodborne diseases pose a significant public health concern worldwide, leading to substantial morbidity and mortality (WHO, 2017) [1]. Every year, millions of people suffer from foodborne diseases that are globally important because of their high incidence and the costs that they impose on society. Meat, an excellent protein source for human beings, is a perishable food that is easily contaminated with microorganisms, resulting in potential economic losses and health hazards. The contamination of meat with Enterobacteriaceae is a significant public health concern with far-reaching consequences for both consumers and the food industry. These pathogens, including Escherichia coli (especially E. coli O157:H7), Salmonella spp., Campylobacter jejuni, Yersinia enterocolitica, and Klebsiella spp., cause severe illnesses such as salmonellosis, hemolytic uremic syndrome, and hemorrhagic colitis [2]. In animals, Enterobacteriaceae are predominant in the gastrointestinal tract and can contaminate meat during slaughter [3]. Therefore, their prevalence in livestock warrants comprehensive investigation [4]. In most countries, including Saudi Arabia, camels, cattle, and sheep are primary meat sources, and the microbiological safety of meat is crucial, given the scale of their consumption. Slaughterhouses are critical points in the meat production process where many studies have shown the prevalence of E. coli and Salmonella spp. on carcasses is mainly due to improper slaughter practices and poor fed-animal hygiene [3]. Along the same lines, outlets have been documented as a source of Enterobacteriaceae prevalence in raw meats [5,6,7,8]. The contamination of Enterobacteriaceae can occur during slaughtering and handling practices, causing health risks. The health risks that E. coli and Salmonella spp. pose are not uniform and depend on the specific strains involved. Some E. coli strains, such as enterohemorrhagic E. coli (EHEC), can cause severe bloody diarrhea, kidney failure, and even death, while Salmonella Enteritidis and S. Typhimurium are responsible for the majority of human salmonellosis cases [9].
According to the Centers for Disease Control and Prevention (CDCP), Salmonella spp. cause approximately 1.35 million infections, 26,500 hospitalizations, and 420 deaths in the United States annually, while in Europe, Salmonella spp. were reported as the second most frequent causative agent of foodborne illness and the second cause of bacterial inflammation of the small intestine in Germany [10,11]. Another area of concern is the potential for cross-contamination in slaughterhouses and butcher shops. Despite the ability to eliminate pathogens during cooking, there is a risk posed by cross-contamination with other foods, such as ready-to-eat food [12]. A study carried out by Brichta-Harhay et al. (2008) [3] found that the prevalence of E. coli O157:H7 on cattle carcasses ranged from 0% to 3.6%, while Salmonella spp. prevalence ranged from 0% to 1.8% on carcasses. In retail outlets, the presence of E. coli and Salmonella spp. can be influenced by cross-contamination, temperature control, and the overall quality of the meat [6]. Understanding the prevalence and load of Enterobacteriaceae contamination present on the hides and carcasses of animals during processing is a significant prerequisite for risk assessment and management. However, comprehensive research into the prevalence and diversity of genera of Enterobacteriaceae, particularly in camels, remains limited. This study, therefore, aims to elucidate the prevalence of Enterobacteriaceae in camels, cattle, and sheep during slaughtering and presentation in butcher shops in Al-Ahsa Governorate.
2. Materials and Methods
2.1. Sample Design
A random sampling technique was employed to ensure the unbiased selection of sampling units. This method facilitated the random selection of samples based on the number of animals slaughtered on sampling days. Data were gathered from two high-throughput municipality slaughterhouses (located in the north and the south) and six butcher shops located in Al-Ahsa governorate (Eastern province, Saudi Arabia) between August and October 2022. To comprehensively understand microbial diversity among consumed meat, this study focused on three distinct animal types, camels, cattle, and sheep. All animal intents to slaughter were subjected to comprehensive veterinary examination.
2.2. Sample Collection
A comprehensive collection of 120 samples from camels, cattle, and sheep was undertaken from two distinct municipal slaughterhouses and six butcher shops. From each animal variety, 40 samples (20 specimens from slaughterhouse, and the same from butcher shops) were analyzed. All animals were subjected to comprehensive veterinary inspection before and after the slaughtering phase. The samples at the slaughterhouses were obtained during slaughtering phase, after evisceration process, whereas in butcher shops, they were collected from the display refrigerators. Approximately 250 g of each carcass were aseptically taken from different parts of the carcass including the neck, chest, backchain, belly, and legs. The same weight was collected from butcher shops from different parts of the carcasses. All samples were placed in sterile plastic bags and stored in an ice box to avoid microbial development. The samples were then transferred to the laboratories within 4–8 h for microbiological analyses.
2.3. Microbiological Analyses
2.3.1. Enumeration of Total Viable Count
The Total Viable Count (TVC) of all samples was enumerated using standard microbiological techniques. Briefly, 25 g of sample was aseptically placed into sterile stomacher bag containing 225 mL of Buffered Peptone Water (BPW) (Oxoid, UK), and completely homogenized using stomacher (Seward Medical Ltd., London, UK) at 200 rpm for 3 min. Serial decimal dilutions of the samples (up to 10−6) were prepared. The samples were then cultured in plate count agar (PCA) (Oxoid, UK) in three replicates. The plates were incubated at 37 °C for 48 h, allowing bacterial growth. Only plates with 30–300 colonies were considered for TVC enumeration. The results were presented in log10 CFU/g.
2.3.2. Presumptive Testing for Enterobacteriaceae
All meat samples were analyzed for detection of Enterobacteriaceae. Aseptically, 25 g of sample was placed into sterile stomacher bag containing 225 mL of sterile 0.1% buffered peptone water (BPW) (Oxoid, UK and homogenized [13]. For Salmonella spp., pre-enrichment was carried out by incubating the mixture at 37 °C for 24 h. Aliquots of 100 µL were transferred into 100 mL. Tetrathionate Broth (TTB) (Oxoid, UK) tubes containing potassium iodide and iodine solution, as recommended by the manufacturer, were then incubated again at 37 °C for 24 h for enrichment. Aliquots of 1 mL from each final dilution were inoculated into Petri dishes containing different agars of MacConkey (MCA) (Oxoid, UK), Eosin Methylene Blue (EMB) (Oxoid, UK), and Salmonella-Shigella (SS) (Oxoid, UK), and incubated for a minimum of 24 h until visible colonies were observed. All suspected colonies were subcultured based on their phenotypic appearances as follows: colonies that appeared on MacConkey agar (MCA) as lactose and non-lactose fermenters were subculture separately using different MacConkey agar and Salmonella-Shigella (SS) agar, while colonies with dark centers and colonies with green metallic sheen were sub-cultured on Salmonella-Shigella (SS) agar and Eosin methylene blue (EMB) agar, respectively and subsequently screened on sorbitol MacConkey agar (SMAC) as described by Cox et al., (2010) [13].
2.3.3. Confirmation of Identification
Biochemical Testing
All bacterial isolates were subjected to preliminary standard biochemical test for identification. Presumptively identified members of Enterobacteriaceae were further screened using Analytical Profile Index API® 20E (BioMérieux®, Inc., Paris, France), following the manufacturer’s instructions.
2.3.4. Molecular Testing
The identification of isolated bacterial strains was performed using Extract-N-Amp™ Tissue PCR Kit (XNAT2R, Sigma-Aldrich Pty Ltd., Darmstadt, Germany) according to the manufacturer’s instruction [14]. For identification, 16SRNA sequences were used. For amplification, universal primers NS1 (forward 5′-AGA GTT TGA TCM TGG CTC AG-3′) and NS2 (reverse 5′-ACGGYTACCTTGTTACGACTT-3′) were used [15]. The polymerase chain reaction (PCR) was carried out using a Rotor-Gene 6000 thermocycler (Corbett Life Science, Qiagen, Australia) as the following procedure: 3 min initial denaturation at 95 °C, 35 cycles of denaturation (30 s at 95 °C), annealing (30 s at 55 °C), extension (1.5 min at 72 °C), and a final extension at 72 °C for 7 min. Each 25 µL PCR reaction mixture contained 12.5 µL of No-ROX Kit, 3.5 µL deionized water, 2 µL each of 10 µM forward and reverse primer, and 5 µL of the extracted DNA. The sequencing was performed by Macrogen Inc. (Seoul, Republic of Korea). All sequences were assembled using the Seqman program of DNASTAR 7.1 software (DNASTAR Inc., Madison, WI, USA).
2.4. Statistical Analysis
The data were analyzed using the Statistical Package for the Social Science (SPSS) version 26 (IBM Corporation, Armonk, NY, USA). Descriptive statistics were used to describe the frequency of Enterobacteriaceae along the camel, cattle, and sheep production chain. The statistical significance of the differences in counts and Enterobacteriaceae prevalence between different sources was determined. A p-value < 0.05 was considered statistically significant.
3. Results and Discussion
3.1. Total Viable Count (TVC)
The total viable count of bacteria (TVC) in samples gathered from two slaughterhouses and six butcher shops was done using culture methods and listed in Table 1 and Figure 1. The mean TVC of camel samples ranged from log10 3.3 to log10 6.2 CFU/g in slaughterhouses and log10 2.8 to log10 6.9 CFU/g in butcher shops. Ten of 20 camel samples collected from the slaughterhouses fell within the critical limits of the microbiological criteria for foodstuffs created by G.C.C Standardization Organization (GSO 1016:2015) [16]. while the remaining samples were within standard limits. Three out of the 20 butcher shops samples exceeded the limits with counts ≥log10 6 CFU/g, 12 were within critical limits, and the remaining five were within standard limits. Similarly, the average TVC of cattle samples varied from log10 3.8 to log10 5.7 CFU/g in slaughterhouses and from log10 4.2 to log10 6.7 CFU/g in butcher shops. Half of the 20 cattle samples from slaughterhouses were within critical limits and the remaining were within standard limits while 3 cattle samples from butcher shops surpassed the standard limits and 8 were within standard limits.

Table 1.
Prevalence of total viable bacteia and total coliforms in camel, cattle, and sheep samples from slaughterhouses and butcher shops.
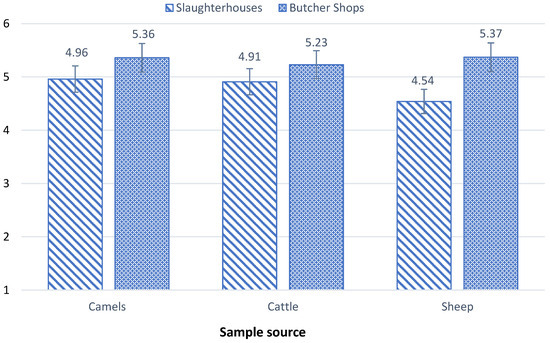
Figure 1.
Contamination level of TVC in camel, cattle, and sheep samples collected from slaughterhouses and butcher shops.
Sheep samples, in general, demonstrated lower bacterial contamination levels, with TVC means ranging from log10 3.4 to log10 6.6 CFU/g in slaughterhouses and from log10 3 to log10 6.9 CFU/g in butcher shops. Two sheep samples from slaughterhouses were within the critical limits with counts of log10 5 CFU/g, and only one sample exceeded standard limits with log10 6.6 CFU/g and 17 samples were within standard limits. With regard to butcher shop samples, 5 samples showed TVC values exceeding standard limits with ≥log10 6 CFU/g, 9 samples were within the critical limits with log10 5 CFU/g, while the remaining samples were within standard limits. These results are in alignment with findings reported in many researches [17,18].
The TVC levels found in this study were significantly higher in butcher shops than in slaughterhouses. This heightened contamination in butcher shops can be attributed to the fact that different types of meat are often handled side by side in butcher shops, allowing bacteria to transfer from one product to another (cross-contamination). In addition, more hands touching meat products in butcher shops is a potential for contamination from humans, improper utensil sanitation is another source of contamination, and the long exposure time in dis-playing meats allows bacteria more time to multiply [19].
3.2. Prevalence of Coliform and E. coli
Table 1 summarizes the presence of coliforms and E. coli in the evaluated samples. The assessment aimed to explore the overall hygiene quality and safety practices when handling carcasses in both slaughterhouses and butcher shops. In this study, coliforms were found in all of the 60 samples collected from slaughterhouses. The mean value of camel samples was 7.5 × 102 MPN/g. cattle samples 8.1 × 102 MPN/g and sheep samples was 4.5 × 102 MPN/g. The results from butcher shops were slightly lower than those found in butcher shops, except the sample of cattle that showed higher than those obtained from slaughterhouses. The high prevalence of coliforms indicates inadequate sanitary conditions and poor general hygiene during the slaughtering and handling practices of carcasses. In addition, coliforms can proliferate at temperatures from −2 to 37 °C [20], which allows bacteria to multiply during display in shops.
Detection of E. coli was carried out irrespective of whether the strain is pathogenic or non-pathogenic to estimate the level of hygiene. Among isolated Enterobacteriaceae genera, E. coli was the most predominant species as shown in Table 2. E. coli. was found in 84 (70%) out of the 120 camels, cattle, and sheep samples investigated, with 35.8% slaughterhouses and 34.1% butcher shops samples having levels of contamination higher than limits established in guidelines [21]. In detail, E. coli was detected in all camel carcasses (100%), 17 (85%) of cattle carcasses, and 6 (30%) of sheep carcasses obtained from slaughterhouses. While in butcher shops, the raw meat-cut samples contaminated with E. coli were 14 (70%) camel samples, 12 (60%) cattle samples, and 15 (75%) sheep samples. There were no significant differences (p > 0.05) in the occurrence of E. coli between the two slaughterhouses, nor between butcher shops.

Table 2.
Presence of Enterobacteriaceae isolated from camels, cattle, and sheep in slaughterhouses and butcher shops (Identified using PCR-test).
The presence of E. coli in food is a significant concern for public health and food safety. The bacteria of E. coli are typically found in the lower intestine of warm-blooded organisms. While many strains of E. coli are harmless, certain serotypes, including E. coli O157:H7, Shiga-toxin-producing E. coli (STEC), and enterohemorrhagic E. coli (EHEC), pose serious illnesses threats [22]. These pathogens carry several different virulence factors, controlled by genes located on chromosomes, plasmids, or phages [23]. Animals are considered the main source of E. coli found in fresh meat. This is mainly due to the abundance and natural presence of this bacterium in the digestive system of many animals [23]. The presence of E. coli in carcasses and raw meat typically indicates fecal contamination, which can occur during slaughtering, handling, packaging, or from cross-contamination with equipment, surfaces, or other foods [24]. This presents a significant risk to public health and can result in foodborne illnesses [25].
The high level of E. coli contamination in the samples tested indicates unhygienic practices, which is also an indication of the potential presence of unacceptable levels of other pathogens. It is worth mentioning that the samples of carcasses in this study were obtained after the evisceration phase. Thus, it is likely that the worker hands and utensils used during the slaughter process are some of the major causes of the E. coli prevalence in carcasses and meat cut samples. Similar results have been reported for abattoirs, and retail outlets in Lahore, Pakistan, where it was found that 63 (45%) out of 140 samples were contaminated with E. coli [5]. Previous studies have documented the prevalence of E. coli in meats. A study carried out in Nigeria revealed that the presence of E. coli in different types of meat including beef, pork, chicken, and mutton was 23.6%. Similarly, E. coli was the most frequently isolated bacterium with 45.4% contamination rate among all tested samples collected from pig slaughterhouse in South Africa [23]. The current study revealed a higher level of contamination in comparison with the study conducted in Ethiopia by Mohammed, Shimelis, Admasu, and Feyera (2014) [26] who found that the prevalence of E. coli in meat samples collected from abattoirs at 15.89%.
3.3. Detection of Salmonella spp.
The second most predominant isolated genera were Salmonella spp. Out of 120 different samples obtained from slaughterhouses and butcher shops, 48 (40%) tested positive for Salmonella spp. distributed on 16 (13.3%) camels, 19 (15.8%) cattle, and 13 (10.8%) sheep samples (Table 2). A total of 48 different isolates of Salmonella spp. were recovered from the positive sample, 47 (39%) of them identified as S. enterica serotype Paratyphi A and only 1 (0.8%) as S. enterica serotype Arizonae as illustrated in Table 2. The total prevalence of the most predominant bacteria in camel, cattle and sheep samples collected from slaughterhouses and butcher shops is shown in Figure 2. In general, the samples of butcher shops showed more contamination with Salmonella spp. (25%), compared with slaughterhouses (15%). S. Paratyphi A is causal agent for serious disease called paratyphoid fever, causing an esti-mated 5.4 million illnesses worldwide [27]. As per the GSO 1016:2015, FAO/WHO (2005) and Health Protection Agency (2009), Salmonella spp. must not be detected in meat and meat product intended to be consumed by humans [16].
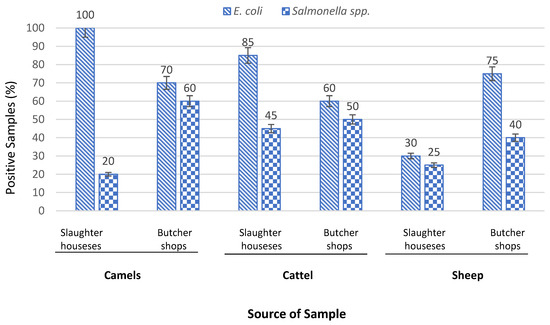
Figure 2.
Prevalence of E. coli and Salmonella spp. In camel, cattle, and sheep samples collected from slaughterhouses and butcher shops.
Salmonella spp., a member of the Enterobacteriaceae family, are the second most common cause of global foodborne infections. Animal products, especially meat, are recognized as primary vectors transmitting Salmonella spp. to humans. In this study, the prevalence of Salmonella spp. in carcasses and meat cuts of camels, cattle, and sheep sourced from both slaughterhouses and butcher shops was investigated. According to our knowledge, a single research study was carried out by Mandour and Altabary (2014) [28] on the microbial quality of camel and mutton carcasses at Al-Ahsa abattoirs. The findings revealed 40% prevalence of E. coli in animal carcasses and meat cuts, aligning with several studies [29,30,31]. However, with respect to Salmonella spp. this study detects no Salmonella spp. [28].
The elevated levels of Salmonella contamination in the samples tested in this study underscore the suboptimal hygiene standards and practices during the slaughtering process. Additionally, exposing carcasses and meat cuts to high temperatures prior to refrigeration could markedly lead to acceleration of the growth of Salmonella and other food-borne microorganisms. The prevalence of Salmonella spp. in this study was higher than some previous studies [5,32,33]. However, some other research have reported more than 60% of prevalence in raw meat samples [34,35]. Based on the current results, S. enterica Paratyphi A, was the predominant serovar, it was found in 47 (39%) of the 120 meat samples studied, while the serovar S. arizonae was found in only one sample. The distribution of Salmonella serovars in raw meat including beef, lambs and poultry can vary considerably among different regions of the world [36]. The difference could be influenced by local environmental factors. For instance, S. enteritidis was identified as the most prevalent contaminant in another study, with a rate of 37.5% contamination [31].
The rising incidence of Salmonella-induced foodborne illnesses underscores the urgency of addressing this public health concern. This study indicates that camel, cattle, and sheep carcasses and meat cuts obtained from slaughterhouses and butcher shops in Al-Ahsa are heavily contaminated with Salmonella spp., and this level of contamination in beef suggests poor sanitary conditions of raw meat handling where it is being produced. Such extensive contamination points to inadequate sanitation in meat production facilities. Potential sources of contamination include fecal matter near butchering sites, direct contact during skinning, and contaminated water used for rinsing meat [37]. Designing slaughtering lines to facilitate hygienic operations is critical. Moreover, effectively enforcing sanitary practices, including the regular disinfection of working tools, is crucial in mitigating the risk of microbiological contamination of carcasses.
3.4. Prevalence of Other Enterobacteriaceae Genera
Twenty-five different strains belonging to nineteen Enterobacteriaceae genera were isolated from camel, cattle, and sheep samples obtained from slaughterhouses and butcher shops, as illustrated in Table 2. Figure 3 shows the most predominant bacterial genera occurring in the samples, where E. coli was highest with 84 (70%) followed by Salmonella spp. with 48 (40%), Proteus spp. with 15 (12%), and Citrobacter spp. with 8 (7%) among all samples. Other genera including Raoultella spp. and Serratia spp. were also identified in four (3.2%) samples. Klebsiella spp., Shigella spp., and Yersinia spp. were also confirmed in meat samples at 1.6%, 0.83%, and 0.83% respectively. The occurrence of pathogens belonging to the Enterobacteriaceae family including Salmonella spp., Shigella spp., and Yersinia spp. in meat and meat products may pose significant risks to human health. These bacteria cause foodborne illnesses, leading to severe complications and even death, particularly in vulnerable populations like young children, the elderly, pregnant women, and immunocompromised individuals [38]. Despite the common practice in Saudi Arabia of cooking meat at high temperatures considered sufficient to eliminate any present pathogens, it does not guarantee a reduction in the risk associated with cross-contamination. This is particularly concerning in the context of other types of food, such as fruits and vegetables, which are often consumed raw. Thus, the potential for cross-contamination presents a significant risk to other foods, underscoring the need for attention and precautionary measures [7].
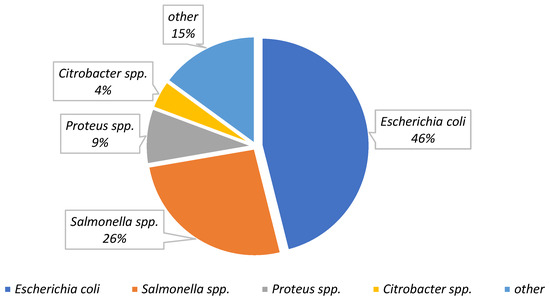
Figure 3.
The most predominant bacterial genera detected in camel, cattle, and sheep carcasses and meat cuts. All values are based on the total samples (n = 120).
4. Conclusions
This study was carried out in two municipal slaughterhouses and six butcher shops in Al-Ahsa. The findings evidenced a considerable prevalence of well-known pathogens, including E. coli, Salmonella spp., Shigella spp., and Klebsilla spp. across different types of livestock including camels, cattle, and sheep in both slaughterhouses and butcher shops. The data indicate that animal carcasses exhibited high contamination levels of E. coli and Salmonella spp., with a rate of 70% and 40%, respectively. E. coli was predominant among all isolates with 70% presence in samples of carcasses and meat cuts. E. coli was found in 100% of camel carcasses, 58% of cattle carcasses, and 30% of sheep carcasses. While in butcher shops, E. coli was detected in 70%, 60%, and 75% in camel, cattle, and sheep meat samples, respectively. On the other hand, Salmonella was positive in 40% of camel 47.5% cattle and 32.5% sheep samples, collected from both slaughterhouses and butcher shops. Twenty-five Enterobacteriaceae species belonging to 19 bacterial genera were isolated and confirmed using PCR-test. The samples of sheep had the highest occurrence of Enterobacteriaceae with 15 different genera followed by camels and cattle samples with 14 different genera for both. In conclusion, the profound prevalence of Enterobacteriaceae among camel, cattle, and sheep carcasses collected from slaughterhouses and meat cuts obtained from butcher shops raises significant concerns regarding food safety. These findings underscore the need to enhance hygiene practices and implement stringent microbial monitoring procedures in both slaughterhouses and butcher shops. It highlights the potential risks to public health since these pathogens have historical of human foodborne illnesses outbreaks. More research is needed to identify the main reasons of the high occurrence of contamination and to help to design and implement an action plan to minimize or prevent foodborne illnesses. Designing slaughtering lines to facilitate hygienic operations is evidently critical. Moreover, the effective enforcement of sanitary practices, including the regular disinfection of working tools, plays a crucial role in mitigating the risk of microbiological contamination of carcasses.
Author Contributions
F.A.-A.: writing—review and editing, supervision, project administration, funding acquisition; S.H.H.: data curation, writing—original draft, preparation; S.A.A.H.: data curation, writing—original draft, preparation. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work (Project number INST114).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Drug-Resistant Salmonella. Fact Sheet N°139. 2005. Available online: http://www.who.int/mediacentre/factsheets/fs139/en/ (accessed on 30 September 2023).
- Vogt, R.L.; Dippold, L. Escherichia coli O157: H7 outbreak associated with consumption of ground beef, June–July 2002. Public Health Rep. 2005, 120, 174–178. [Google Scholar] [CrossRef]
- Brichta-Harhay, D.M.; Guerini, M.N.; Arthur, T.M.; Bosilevac, J.M.; Kalchayanand, N.; Shackelford, S.D.; Koohmaraie, M. Salmonella and Escherichia coli O157: H7 contamination on hides and carcasses of cull cattle presented for slaughter in the United States: An evaluation of prevalence and bacterial loads by immunomagnetic separation and direct plating methods. Appl. Environ. Microbiol. 2008, 74, 6289–6297. [Google Scholar] [CrossRef]
- Bintsis, T.; Litopoulou-Tzanetaki, E.; Robinson, R.K. Existing and potential applications of ultraviolet light in the food industry—A critical review. J. Sci. Food Agric. 2000, 80, 637–645. [Google Scholar] [CrossRef]
- Ahmad, M.; Sarwar, A.; Najeeb, M.; Nawaz, M.; Anjum, A.; Ali, M.; Mansur, N. Assessment of microbial load of raw meat at abattoirs and retail outlets. J. Anim. Plant Sci. 2013, 23, 745–748. [Google Scholar]
- Bohaychuk, V.; Gensler, G.; King, R.; Manninen, K.; Sorensen, O.; Wu, J.; McMullen, L. Occurrence of pathogens in raw and ready-to-eat meat and poultry products collected from the retail marketplace in Edmonton, Alberta, Canada. J. Food Prot. 2006, 69, 2176–2182. [Google Scholar] [CrossRef]
- Bosilevac, J.M.; Gassem, M.A.; Al Sheddy, I.A.; Almaiman, S.A.; Al-Mohizea, I.S.; Alowaimer, A.; Koohmaraie, M. Prevalence of Escherichia coli O157: H7 and Salmonella in camels, cattle, goats, and sheep harvested for meat in Riyadh. J. Food Prot. 2015, 78, 89–96. [Google Scholar] [CrossRef]
- Jaja, I.F.; Green, E.; Muchenje, V. Aerobic mesophilic, coliform, Escherichia coli, and Staphylococcus aureus counts of raw meat from the formal and informal meat sectors in South Africa. Int. J. Environ. Res. Public Health 2018, 15, 819. [Google Scholar] [CrossRef]
- CDC. What Is Salmonellosis. 2010. Available online: http:www.cdc.gov/Salmonella/-general/index.html (accessed on 30 September 2023).
- Meyer, C.; Thiel, S.; Ullrich, U.; Stolle, A. Salmonella in raw meat and by-products from pork and beef. J. Food Prot. 2010, 73, 1780–1784. [Google Scholar] [CrossRef]
- Terentjeva, M.; Avsejenko, J.; Streikiša, M.; Utināne, A.; Kovaļenko, K.; Bērziņš, A. Prevalence and antimicrobial resistance of Salmonella in meat and meat products in Latvia. Ann. Agric. Environ. Med. 2017, 24, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Todd, E.C.; Greig, J.D.; Bartleson, C.A.; Michaels, B.S. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 6. Transmission and survival of pathogens in the food processing and preparation environment. J. Food Prot. 2009, 72, 202–219. [Google Scholar] [CrossRef]
- Cox, N.; Richardson, L.; Cason, J.; Buhr, R.; Vizzier-Thaxton, Y.; Smith, D.; Doyle, M. Comparison of neck skin excision and whole carcass rinse sampling methods for microbiological evaluation of broiler carcasses before and after immersion chilling. J. Food Prot. 2010, 73, 976–980. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Przemieniecki, S.W.; Kurowski, T.P.; Kotlarz, K.; Krawczyk, K.; Damszel, M.; Karwowska, A. Plant growth promoting properties of Serratia fonticola ART-8 and Pseudomonas putida ART-9 and their effect on the growth of spring wheat (Triticum aestivum L.). Environ. Biotechnol. 2016, 12, 35–39. [Google Scholar] [CrossRef]
- GSO/FDS 1016/2014; Microbiological Criteria for Foodstuffs. Standardization Organization for G.C.C (GSO): Riyadh, Saudi Arabia, 2014; p. 4.
- Ali, N.H.; Farooqui, A.; Khan, A.; Khan, A.Y.; Kazmi, S.U. Microbial contamination of raw meat and its environment in retail shops in Karachi, Pakistan. J. Infect. Dev. Ctries. 2010, 4, 382–388. [Google Scholar]
- Bhandare, S.G.; Sherikar, A.; Paturkar, A.; Waskar, V.; Zende, R. A comparison of microbial contamination on sheep/goat carcasses in a modern Indian abattoir and traditional meat shops. Food Control 2007, 18, 854–858. [Google Scholar] [CrossRef]
- Nychas, G.-J.E.; Skandamis, P.N.; Tassou, C.C.; Koutsoumanis, K.P. Meat spoilage during distribution. Meat Sci. 2008, 78, 77–89. [Google Scholar] [CrossRef]
- Jay, J.M.; Loessner, M.J.; Golden, D.A. The HACCP and FSO systems for food safety. Mod. Food Microbiol. 2005, 6, 497–515. [Google Scholar]
- Álvarez-Astorga, M.; Capita, R.; Alonso-Calleja, C.; Moreno, B.; Garcı, C. Microbiological quality of retail chicken by-products in Spain. Meat Sci. 2002, 62, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, S. Foodborne Diseases—Disease Burden. In Food Safety in the 21st Century; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1–10. [Google Scholar]
- Abdalla, S.E.; Abia, A.L.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. Food animals as reservoirs and potential sources of multidrug-resistant diarrheagenic E. coli pathotypes: Focus on intensive pig farming in South Africa. Onderstepoort J. Vet. Res. 2022, 89, 1963. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, R.; Satharasinghe, D.; Anwarama, P.; Parakatawella, P.; Jayasooriya, L.; Ranasinghe, R.; Nakaguchi, Y. Prevalence and Antimicrobial Resistance of Escherichia coli in Chicken Meat and Edible Poultry Organs Collected from Retail Shops and Supermarkets of North Western Province in Sri Lanka. J. Food Qual. 2022, 2022, 8962698. [Google Scholar] [CrossRef]
- Zerabruk, K.; Retta, N.; Muleta, D.; Tefera, A.T. Assessment of microbiological safety and quality of minced meat and meat contact surfaces in selected butcher shops of Addis Ababa, Ethiopia. J. Food Qual. 2019, 2019, 3902690. [Google Scholar] [CrossRef]
- Mohammed, O.; Shimelis, D.; Admasu, P.; Feyera, T. Prevalence and antimicrobial susceptibility pattern of E. coli isolates from raw meat samples obtained from abattoirs in Dire Dawa City, eastern Ethiopia. Int. J. Microbiol. Res. 2014, 5, 35–39. [Google Scholar]
- Sanderson, K.E.; Liu, S.-L.; Tang, L.; Johnston, R.N. Salmonella Typhi and Salmonella Paratyphi A. In Molecular Medical Microbiology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 1275–1306. [Google Scholar]
- Mandour, M.A.; Altabary, G.F. Slaughtered at Al-Ahsaa Abattoir, Saudi Arabia. J. Anim. Vet. Adv. 2014, 13, 1179–1184. [Google Scholar]
- Barkocy-Gallagher, G.A.; Arthur, T.M.; Rivera-Betancourt, M.; Nou, X.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M. Seasonal prevalence of Shiga toxin–producing Escherichia coli, including O157: H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. J. Food Prot. 2003, 66, 1978–1986. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkaway, S.; Samaha, I.A.; El-Galil, H. Prevalence of pathogenic microorganisms in raw meat products from retail outlets in Alexandria province. Alex. J. Vet. Sci. 2016, 51, 374–380. [Google Scholar]
- Sallam, K.I.; Mohammed, M.A.; Hassan, M.A.; Tamura, T. Prevalence, molecular identification and antimicrobial resistance profile of Salmonella serovars isolated from retail beef products in Mansoura, Egypt. Food Control 2014, 38, 209–214. [Google Scholar] [CrossRef]
- Hyeon, J.-Y.; Chon, J.-W.; Hwang, I.-G.; Kwak, H.-S.; Kim, M.-S.; Kim, S.-K.; Seo, K.-H. Prevalence, Antibiotic Resistance, and Molecular Characterizatio of Salmonella Serovars in Retail Meat Products. J. Food Prot. 2011, 74, 161–166. [Google Scholar] [CrossRef]
- Nurye, M.; Demlie, M. Assessment of hygienic practices and microbial quality of meat at slaughterhouses and butcher’s shops in West Hararghe Zone, Ethiopia. Abyssinia J. Sci. Technol. 2021, 6, 32–41. [Google Scholar]
- Ekli, R.; Adzitey, F.; Huda, N. Prevalence of resistant Salmonella spp. isolated from raw meat and liver of cattle in the Wa Municipality of Ghana. IOP Conf. Ser. Earth Environ. Sci. 2019, 287, 012006. [Google Scholar] [CrossRef]
- Hathai, T.; Yamaguchi, R. Molecular characterization of antibiotic-resistant Salmonella isolates from retail meat from markets in Northern Vietnam. J. Food Prot. 2012, 75, 1709–1714. [Google Scholar] [CrossRef]
- Altaf Hussain, M.; Wang, W.; Sun, C.; Gu, L.; Liu, Z.; Yu, T.; Hou, J. Molecular Characterization of Pathogenic Salmonella spp. from Raw Beef in Karachi, Pakistan. Antibiotics 2020, 9, 73. [Google Scholar] [CrossRef]
- McEvoy, J.; Doherty, A.; Finnerty, M.; Sheridan, J.; McGuire, L.; Blair, I.; Harrington, D. The relationship between hide cleanliness and bacterial numbers on beef carcasses at a commercial abattoir. Lett. Appl. Microbiol. 2000, 30, 390–395. [Google Scholar] [CrossRef]
- Gwida, M.; Hotzel, H.; Geue, L.; Tomaso, H. Occurrence of Enterobacteriaceae in raw meat and in human samples from Egyptian retail sellers. Int. Sch. Res. Not. 2014, 2014, 6. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).