Effects of Saccharomyces Cerevisiae Fermentation Products on the Microbial Community throughout the Gastrointestinal Tract of Calves
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Animal Trials
2.3. Sample Collections
2.3.1. Rumen Fluid Collection on Day 28
2.3.2. Collection of Rumen and Intestine Contents on Day 56
2.4. DNA Isolation and Illumina Hiseq Sequencing
2.5. Data Processing and Analysis
2.5.1. Quality Control and Paired-End Reads Assemblies
2.5.2. OTU Cluster and Species Annotation
2.5.3. Data Analysis
2.6. Nucleotide Sequence Accession Numbers
3. Results
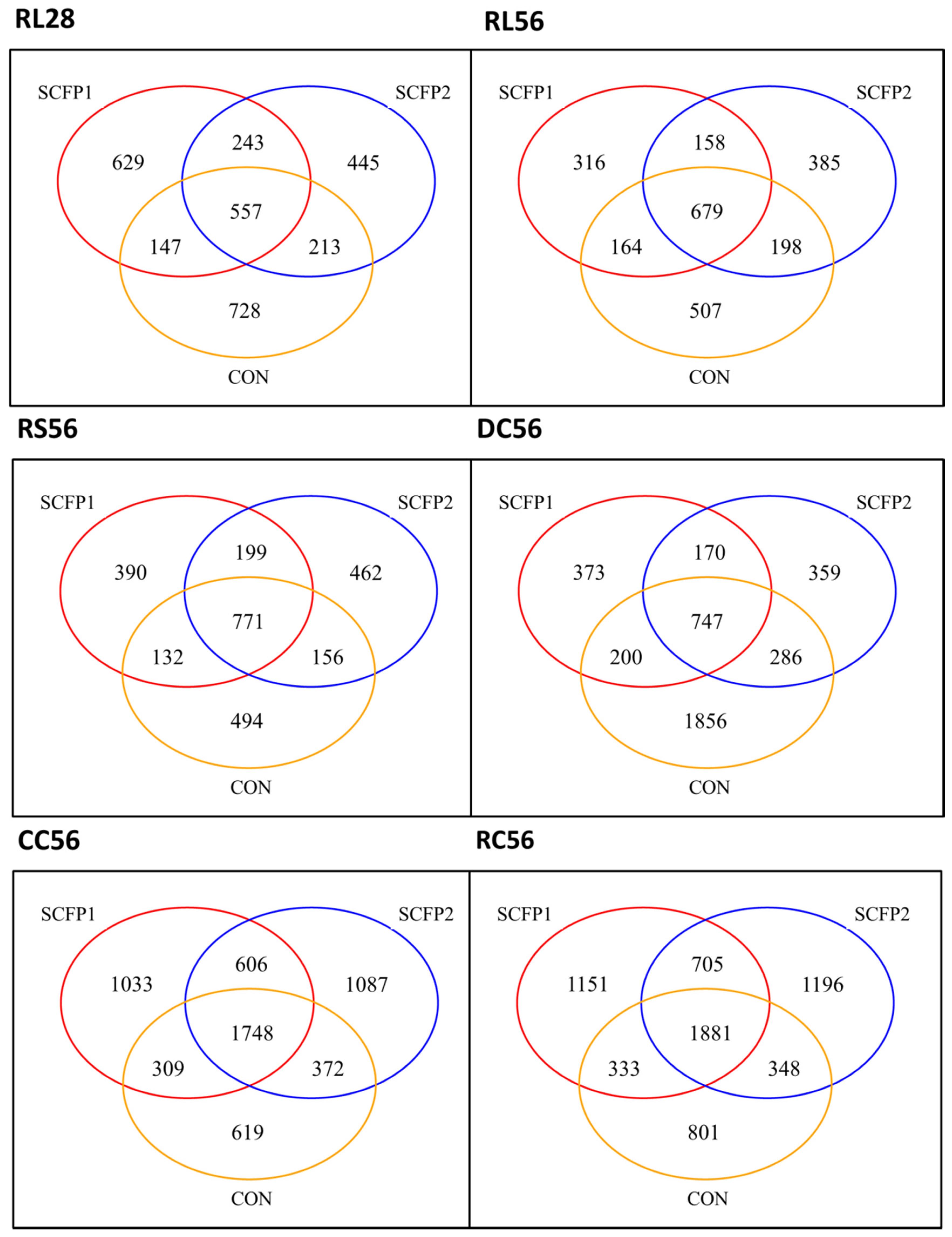
3.1. Sequences and OTUs
3.2. Impact of SCFP on Microbiota
3.2.1. Microbial Richness and Diversity
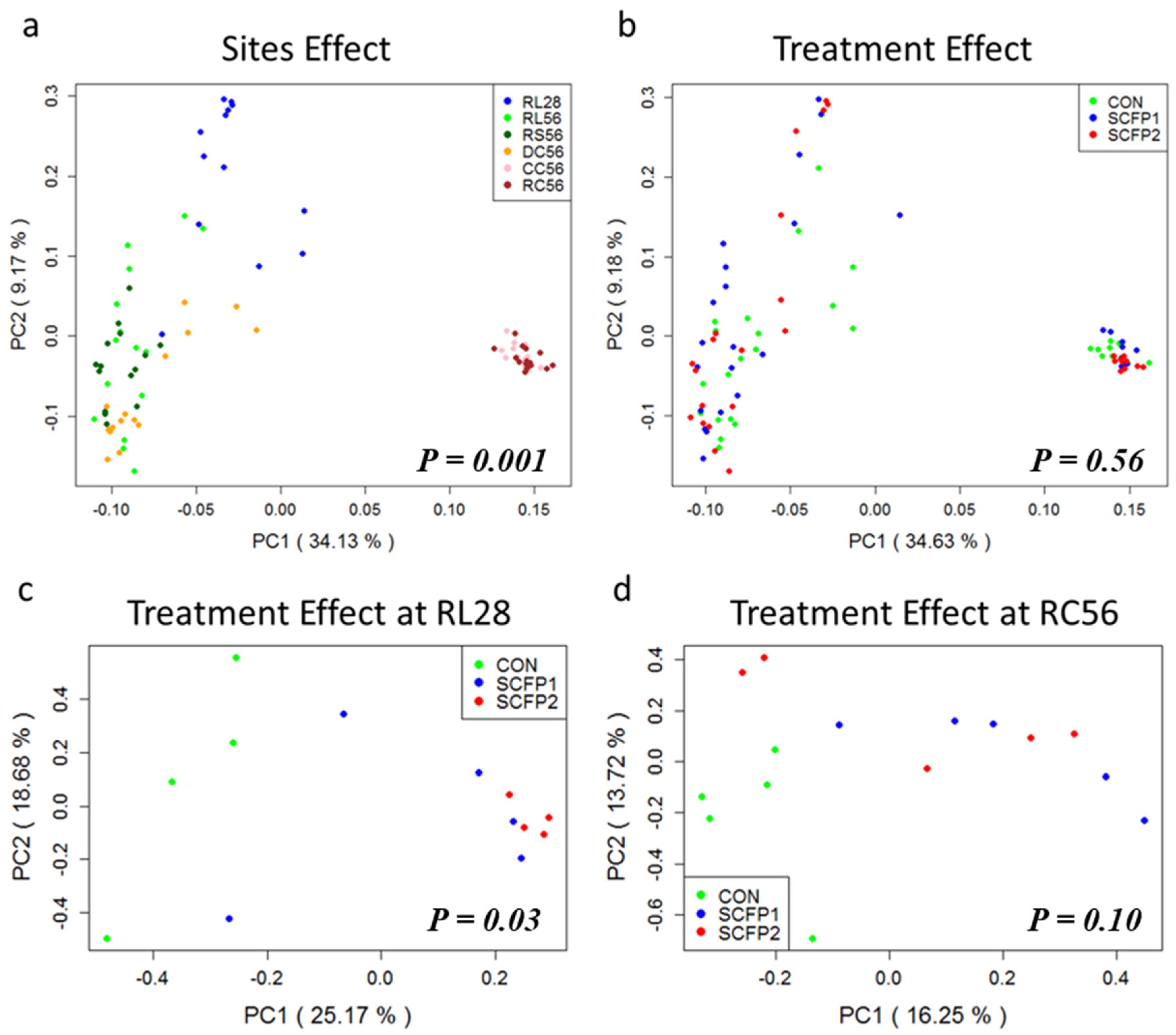
3.2.2. Sample Clustering
3.2.3. Taxonomic Characteristics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
References
- Rey, M.; Enjalbert, F.; Combes, S.; Cauquil, L.; Bouchez, O.; Monteils, V. Establishment of ruminal bacterial community in dairy calves from birth to weaning is sequential. J. Appl. Microbiol. 2014, 116, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.W. The development of the flora of the alimentary tract in young animals. J. Pathol. Bacteriol. 1965, 90, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Vlková, E.; Trojanová, I.; Rada, V. Distribution of bifidobacteria in the gastrointestinal tract of calves. Folia Microbiol. 2006, 51, 325–328. [Google Scholar] [CrossRef]
- Guzman, C.E.; Bereza-Malcolm, L.T.; De, G.B.; Franks, A.E. Presence of Selected Methanogens, Fibrolytic Bacteria, and Proteobacteria in the Gastrointestinal Tract of Neonatal Dairy Calves from Birth to 72 Hours. PLoS ONE 2015, 10, e0133048. [Google Scholar] [CrossRef] [PubMed]
- Kleinjöbstl, D.; Schornsteiner, E.; Mann, E.; Wagner, M.; Drillich, M.; Schmitzesser, S. Pyrosequencing reveals diverse fecal microbiota in Simmental calves during early development. Front. Microbiol. 2014, 5, 622. [Google Scholar]
- Castro, J.J.; Gomez, A.; White, B.; Loften, J.R.; Drackley, J.K. Changes in the intestinal bacterial community, short-chain fatty acid profile, and intestinal development of preweaned Holstein calves. 2. Effects of gastrointestinal site and age. J. Dairy Sci. 2016, 99, 9703–9715. [Google Scholar] [CrossRef]
- Dawson, K.; Newman, K.; Boling, J. Effects of microbial supplements containing yeast and lactobacilli on roughage-fed ruminal microbial activities. J. Anim. Sci. 1990, 68, 3392–3398. [Google Scholar] [CrossRef]
- Chaucheyras-Durand, F.; Fonty, G. Establishment of cellulolytic bacteria and development of fermentative activities in the rumen of gnotobiotically-reared lambs receiving the microbial additive Saccharomyces cerevisiae CNCM I-1077. Reprod. Nutr. Dev. 2001, 41, 57–68. [Google Scholar] [CrossRef]
- Hristov, A.N.; Varga, G.; Cassidy, T.; Long, M.; Heyler, K.; Karnati, S.K.R.; Corl, B.; Hovde, C.J.; Yoon, I. Effect of Saccharomyces cerevisiae fermentation product on ruminal fermentation and nutrient utilization in dairy cows. J. Dairy Sci. 2010, 93, 682–692. [Google Scholar] [CrossRef]
- Magalhães, V.J.; Susca, F.; Lima, F.S.; Branco, A.F.; Yoon, I.; Santos, J.E. Effect of feeding yeast culture on performance, health, and immunocompetence of dairy calves. J. Dairy Sci. 2008, 91, 1497–1509. [Google Scholar] [CrossRef] [PubMed]
- Lesmeister, K.E.; Heinrichs, A.J.; Gabler, M.T. Effects of supplemental yeast (Saccharomyces cerevisiae) culture on rumen development, growth characteristics, and blood parameters in neonatal dairy calves. J. Dairy Sci. 2004, 87, 1832–1839. [Google Scholar] [CrossRef]
- Quigley, J.D.; Boehms, S.I.; Steen, T.M.; Heitmann, R.N. Effects of lasalocid on selected ruminal and blood metabolites in young calves. J. Dairy Sci. 1992, 75, 2235–2241. [Google Scholar] [CrossRef]
- Brewer, M.T.; Anderson, K.L.; Yoon, I.; Scott, M.F.; Carlson, S.A. Amelioration of salmonellosis in pre-weaned dairy calves fed Saccharomyces cerevisiae fermentation products in feed and milk replacer. Vet. Microbiol. 2014, 172, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E.S.; Martin, S.A. Effects of a Saccharomyces cerevisiae Culture on Ruminal Bacteria that Utilize Lactate and Digest Cellulose. J. Dairy Sci. 1997, 80, 2035–2044. [Google Scholar] [CrossRef]
- He, S.; Zhou, Z.; Meng, K.; Zhao, H.; Yao, B.; Ringø, E.; Yoon, I. Effects of dietary antibiotic growth promoter and Saccharomyces cerevisiae fermentation product on production, intestinal bacterial community, and nonspecific immunity of hybrid tilapia (Oreochromis niloticus female x Oreochromis aureus male). J. Anim. Sci. 2011, 89, 84–92. [Google Scholar] [CrossRef]
- Zhu, W.; Wei, Z.; Xu, N.; Yang, F.; Yoon, I.; Chung, Y.; Liu, J.; Wang, J. Effects of Saccharomyces cerevisiae fermentation products on performance and rumen fermentation and microbiota in dairy cows fed a diet containing low quality forage. J. Anim. Sci. Biotechnol. 2017, 8, 677–685. [Google Scholar] [CrossRef]
- Xiao, J.; Alugongo, G.; Chung, R.; Dong, S.; Li, S.; Yoon, I.; Wu, Z.; Cao, Z. Effects of Saccharomyces cerevisiae fermentation products on dairy calves: Ruminal fermentation, gastrointestinal morphology, and microbial community. J. Dairy Sci. 2016. [Google Scholar] [CrossRef]
- NRC. Nutrient Requirements of Dairy Cattle, 7th ed.; NRC: Rockville, MD, USA, 2001; p. 381.
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berglyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. Multidiscip. J. Microb. Ecol. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Dillies, M.-A.; Rau, A.; Aubert, J.; Hennequet-Antier, C.; Jeanmougin, M.; Servant, N.; Keime, C.; Marot, G.; Castel, D.; Estelle, J. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013, 14, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R. Pheatmap: Pretty heatmaps. In R Package Version 2.4-1; Microsoft: Redmond, WA, USA, 2012. [Google Scholar]
- Stearns, J.C.; Lynch, M.D.; Senadheera, D.B.; Tenenbaum, H.C.; Goldberg, M.B.; Cvitkovitch, D.G.; Croitoru, K.; Moreno-Hagelsieb, G.; Neufeld, J.D. Bacterial biogeography of the human digestive tract. Sci. Rep. 2011, 1, 170. [Google Scholar] [CrossRef] [PubMed]
- Mrazek, J.; Tepšič, K.; Avguštin, G.; Kopečný, J. Diet-dependent shifts in ruminal butyrate-producing bacteria. Folia Microbiol. 2006, 51, 294–298. [Google Scholar] [CrossRef]
- Meehan, C.J.; Beiko, R.G. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Boil. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef]
- Li, R.W.; Wu, S.; Vi, R.L.B.; Li, W.; Li, C. Perturbation dynamics of the rumen microbiota in response to exogenous butyrate. PLoS ONE 2012, 7, e29392. [Google Scholar] [CrossRef]
- Kiarie, E.; Bhandari, S.; Scott, M.; Krause, D.; Nyachoti, C. Growth performance and gastrointestinal microbial ecology responses of piglets receiving Saccharomyces cerevisiae fermentation products after an oral challenge with Escherichia coli (K88). J. Anim. Sci. 2011, 89, 1062–1078. [Google Scholar] [CrossRef]
- Price, K.; Totty, H.; Lee, H.; Utt, M.; Fitzner, G.; Yoon, I.; Ponder, M.; Escobar, J. Use of fermentation product on growth performance and microbiota of weaned pigs during infection. J. Anim. Sci. 2010, 88, 3896–3908. [Google Scholar] [CrossRef]
- Harrison, G.; Hemken, R.; Dawson, K.; Harmon, R.; Barker, K. Influence of addition of yeast culture supplement to diets of lactating cows on ruminal fermentation and microbial populations. J. Dairy Sci. 1988, 71, 2967–2975. [Google Scholar] [CrossRef]
- Mullins, C.; Mamedova, L.; Carpenter, A.; Ying, Y.; Allen, M.; Yoon, I.; Bradford, B. Analysis of rumen microbial populations in lactating dairy cattle fed diets varying in carbohydrate profiles and Saccharomyces cerevisiae fermentation product. J. Dairy Sci. 2013, 96, 5872–5881. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhou, Z.; Liu, Y.; Shi, P.; Yao, B.; Ringø, E.; Yoon, I. Effects of dietary Saccharomyces cerevisiae fermentation product (DVAQUA®) on growth performance, intestinal autochthonous bacterial community and non-specific immunity of hybrid tilapia (Oreochromis niloticus♀× O. aureus♂) cultured in cages. Aquaculture 2009, 294, 99–107. [Google Scholar] [CrossRef]
- Thoetkiattikul, H.; Mhuantong, W.; Laothanachareon, T.; Tangphatsornruang, S.; Pattarajinda, V.; Eurwilaichitr, L.; Champreda, V. Comparative analysis of microbial profiles in cow rumen fed with different dietary fiber by tagged 16S rRNA gene pyrosequencing. Curr. Microbiol. 2013, 67, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Golder, H.; Denman, S.; McSweeney, C.; Celi, P.; Lean, I. Ruminal bacterial community shifts in grain-, sugar-, and histidine-challenged dairy heifers. J. Dairy Sci. 2014, 97, 5131–5150. [Google Scholar] [CrossRef] [PubMed]
- Clavel, T.; Lepage, P.; Charrier, C. The Family Coriobacteriaceae. In The Prokaryotes; Springer: New York, NY, USA, 2014; pp. 201–238. [Google Scholar]
- Šuľák, M.; Sikorová, L.; Jankuvová, J.; Javorský, P.; Pristaš, P. Variability of Actinobacteria, a minor component of rumen microflora. Folia Microbiol. 2012, 57, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Morotomi, M.; Nagai, F.; Sakon, H.; Tanaka, R. Paraprevotella clara gen. nov., sp. nov. and Paraprevotella xylaniphila sp. nov., members of the family ‘Prevotellaceae’isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2009, 59, 1895–1900. [Google Scholar] [CrossRef]
- Hobson, P.N.; Stewart, C.S. The Rumen Microbial Ecosystem; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar]
- Antharam, V.C.; Li, E.C.; Ishmael, A.; Sharma, A.; Mai, V.; Rand, K.H.; Wang, G.P. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 2013, 51, 2884–2892. [Google Scholar] [CrossRef]
- Biddle, A.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
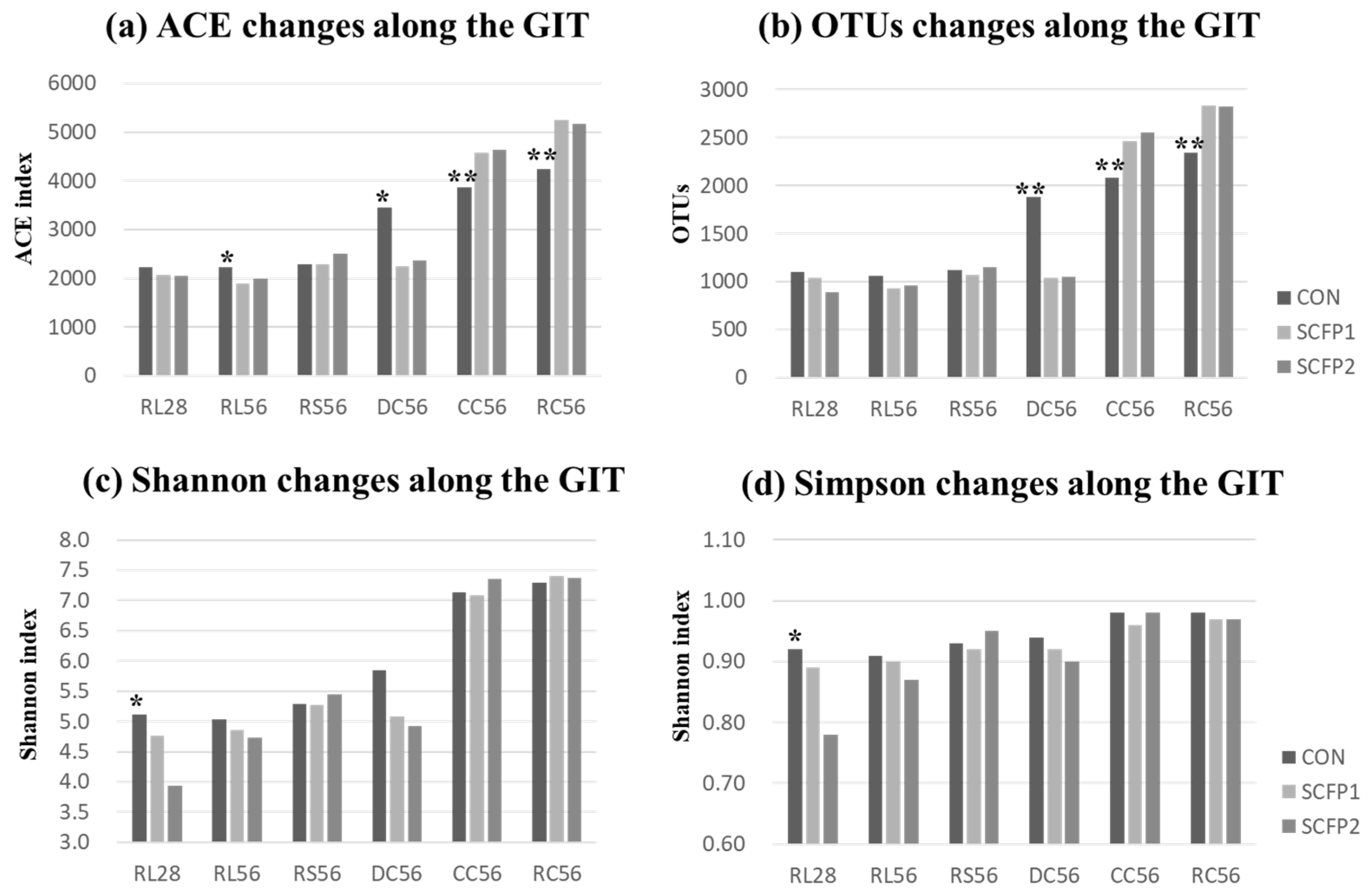
 ) CON, (
) CON, ( ) SCFP1 and (
) SCFP1 and ( ) SCFP2 through GITs. CON = No SmartCare and XPC; SCFP1 = 1 g/head/d SmartCare in milk + 0.5% XPC in the starter grains; SCFP2 = 1 g/head/d SmartCare in milk + 1% XPC in the starter grains (Diamond V, Cedar Rapids, Iowa). * p < 0.1, ** p < 0.05 between CON and SCFP (SCFP1 & SCFP2). RL28 = Rumen liquid portions at day 28 (n = 13), RL56 = Rumen liquid portions at day 56 (n = 15), RS56 = Rumen solid contents at day 56 (n = 15), DC56 = Duodenal contents at day 56 (n = 15), CC = Cecal contents at day 56 (n = 15), RC = Rectal contents at day 56 (n = 15).
) SCFP2 through GITs. CON = No SmartCare and XPC; SCFP1 = 1 g/head/d SmartCare in milk + 0.5% XPC in the starter grains; SCFP2 = 1 g/head/d SmartCare in milk + 1% XPC in the starter grains (Diamond V, Cedar Rapids, Iowa). * p < 0.1, ** p < 0.05 between CON and SCFP (SCFP1 & SCFP2). RL28 = Rumen liquid portions at day 28 (n = 13), RL56 = Rumen liquid portions at day 56 (n = 15), RS56 = Rumen solid contents at day 56 (n = 15), DC56 = Duodenal contents at day 56 (n = 15), CC = Cecal contents at day 56 (n = 15), RC = Rectal contents at day 56 (n = 15).
 ) CON, (
) CON, ( ) SCFP1 and (
) SCFP1 and ( ) SCFP2 through GITs. CON = No SmartCare and XPC; SCFP1 = 1 g/head/d SmartCare in milk + 0.5% XPC in the starter grains; SCFP2 = 1 g/head/d SmartCare in milk + 1% XPC in the starter grains (Diamond V, Cedar Rapids, Iowa). * p < 0.1, ** p < 0.05 between CON and SCFP (SCFP1 & SCFP2). RL28 = Rumen liquid portions at day 28 (n = 13), RL56 = Rumen liquid portions at day 56 (n = 15), RS56 = Rumen solid contents at day 56 (n = 15), DC56 = Duodenal contents at day 56 (n = 15), CC = Cecal contents at day 56 (n = 15), RC = Rectal contents at day 56 (n = 15).
) SCFP2 through GITs. CON = No SmartCare and XPC; SCFP1 = 1 g/head/d SmartCare in milk + 0.5% XPC in the starter grains; SCFP2 = 1 g/head/d SmartCare in milk + 1% XPC in the starter grains (Diamond V, Cedar Rapids, Iowa). * p < 0.1, ** p < 0.05 between CON and SCFP (SCFP1 & SCFP2). RL28 = Rumen liquid portions at day 28 (n = 13), RL56 = Rumen liquid portions at day 56 (n = 15), RS56 = Rumen solid contents at day 56 (n = 15), DC56 = Duodenal contents at day 56 (n = 15), CC = Cecal contents at day 56 (n = 15), RC = Rectal contents at day 56 (n = 15).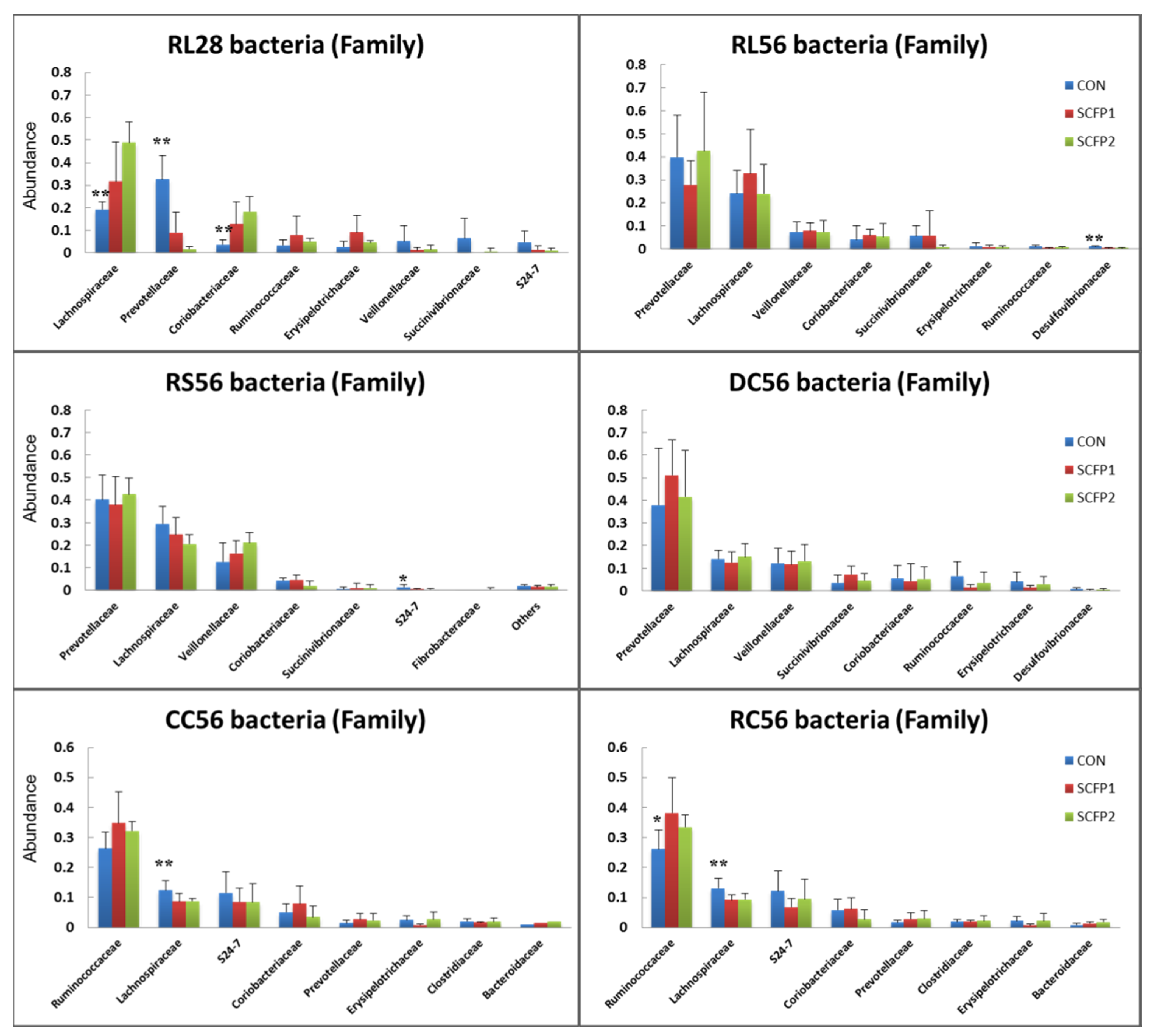
| Sites/Groups a | N b | Average Reads | Average OTUs c |
|---|---|---|---|
| RL28 | |||
| CON | 4 | 160,855 | 1101 |
| SCFP1 | 5 | 155,655 | 1042 |
| SCFP2 | 4 | 133,215 | 891 |
| RL56 | |||
| CON | 5 | 158,254 | 1059 |
| SCFP1 | 5 | 177,990 | 928 |
| SCFP2 | 5 | 161,029 | 962 |
| RS56 | |||
| CON | 5 | 144,101 | 1120 |
| SCFP1 | 5 | 119,776 | 1075 |
| SCFP2 | 5 | 141,623 | 1153 |
| DC56 | |||
| CON | 5 | 156,828 | 1877 |
| SCFP1 | 5 | 118,575 | 1045 |
| SCFP2 | 5 | 161,956 | 1053 |
| CC56 | |||
| CON | 5 | 183,365 | 2084 |
| SCFP1 | 5 | 185,415 | 2460 |
| SCFP2 | 5 | 178,122 | 2561 |
| RC56 | |||
| CON | 5 | 184,172 | 2338 |
| SCFP1 | 5 | 203,071 | 2829 |
| SCFP2 | 5 | 196,179 | 2827 |
| Item/Index 1 | N 2 | RL56 | RS56 | DC56 | CC56 | RC56 | SEM |
|---|---|---|---|---|---|---|---|
| Mean 3 | |||||||
| ACE | 15 | 2038.2 a | 2360.9 ab | 2685.3 b | 4358.6 c | 4893.5 c | 156.6 |
| Chao1 | 15 | 1998.9 a | 2293.6 a | 2537.7 a | 4271.7 b | 4810.1 b | 153.4 |
| Shannon | 15 | 4.88 a | 5.33 a | 5.28 a | 7.18 b | 7.35 b | 0.16 |
| Simpson | 15 | 0.90 a | 0.93 ab | 0.92 a | 0.97 b | 0.97 b | 0.01 |
| OTUs | 15 | 982.9 a | 1116.0 a | 1325.0 a | 2363.5 b | 2664.7 b | 94.6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, J.; Alugongo, G.M.; Ji, S.; Wu, Z.; Dong, S.; Li, S.; Yoon, I.; Chung, R.; Cao, Z. Effects of Saccharomyces Cerevisiae Fermentation Products on the Microbial Community throughout the Gastrointestinal Tract of Calves. Animals 2019, 9, 4. https://doi.org/10.3390/ani9010004
Xiao J, Alugongo GM, Ji S, Wu Z, Dong S, Li S, Yoon I, Chung R, Cao Z. Effects of Saccharomyces Cerevisiae Fermentation Products on the Microbial Community throughout the Gastrointestinal Tract of Calves. Animals. 2019; 9(1):4. https://doi.org/10.3390/ani9010004
Chicago/Turabian StyleXiao, Jianxin, Gibson M. Alugongo, Shoukun Ji, Zhaohai Wu, Shuangzhao Dong, Shengi Li, Ilkyu Yoon, Ruby Chung, and Zhijun Cao. 2019. "Effects of Saccharomyces Cerevisiae Fermentation Products on the Microbial Community throughout the Gastrointestinal Tract of Calves" Animals 9, no. 1: 4. https://doi.org/10.3390/ani9010004
APA StyleXiao, J., Alugongo, G. M., Ji, S., Wu, Z., Dong, S., Li, S., Yoon, I., Chung, R., & Cao, Z. (2019). Effects of Saccharomyces Cerevisiae Fermentation Products on the Microbial Community throughout the Gastrointestinal Tract of Calves. Animals, 9(1), 4. https://doi.org/10.3390/ani9010004





