Three Novel KIT Polymorphisms Found in Horses with White Coat Color Phenotypes
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Horses
2.2. Genotyping and Variant Screening
3. Results and Discussion
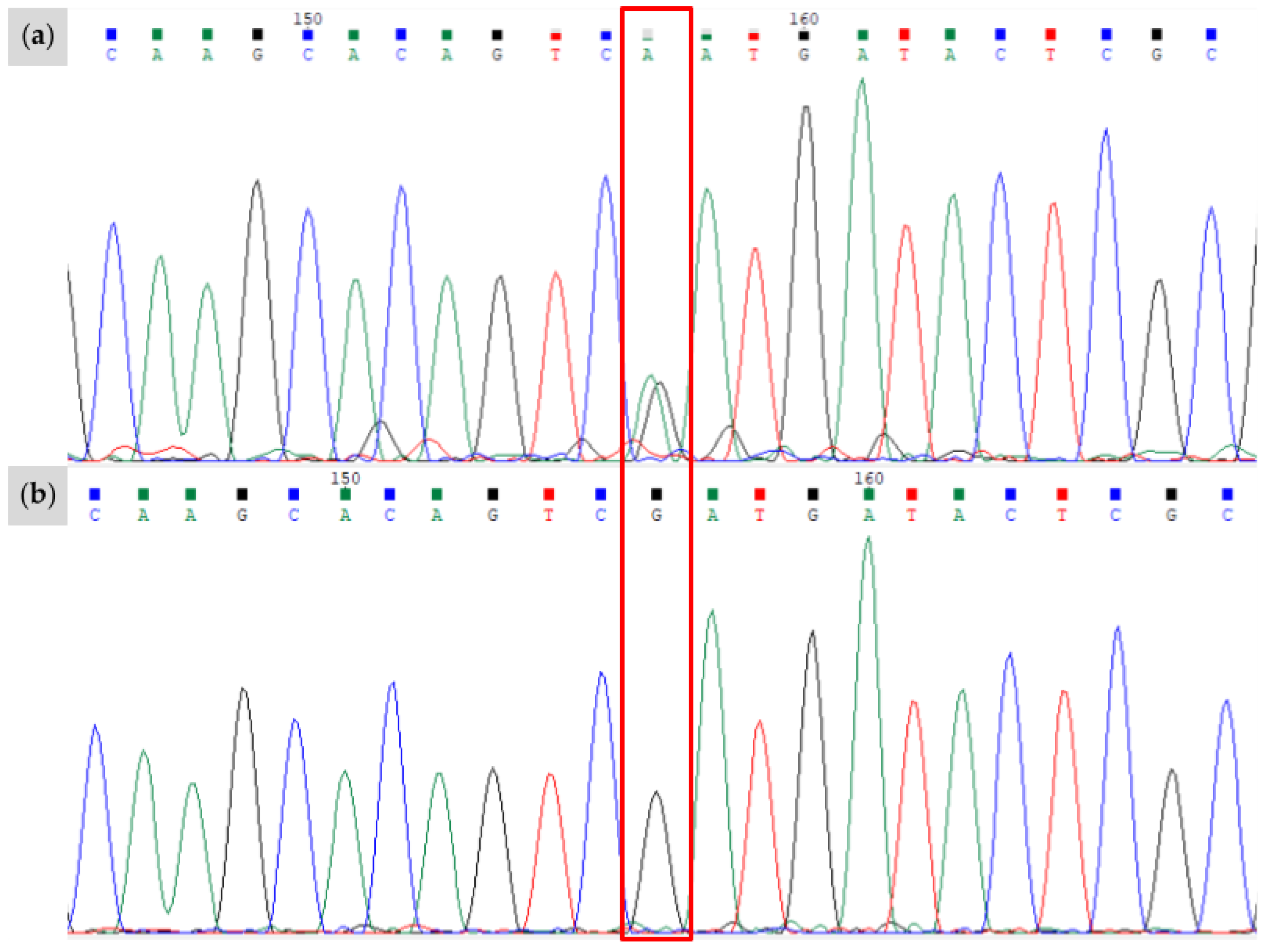
3.1. Novel KIT Insertion Identified in Family 1
3.2. Novel KIT Splice Site Identified in Family 2
3.3. Novel Stop-Gain Variant in a Stock-Type Mare
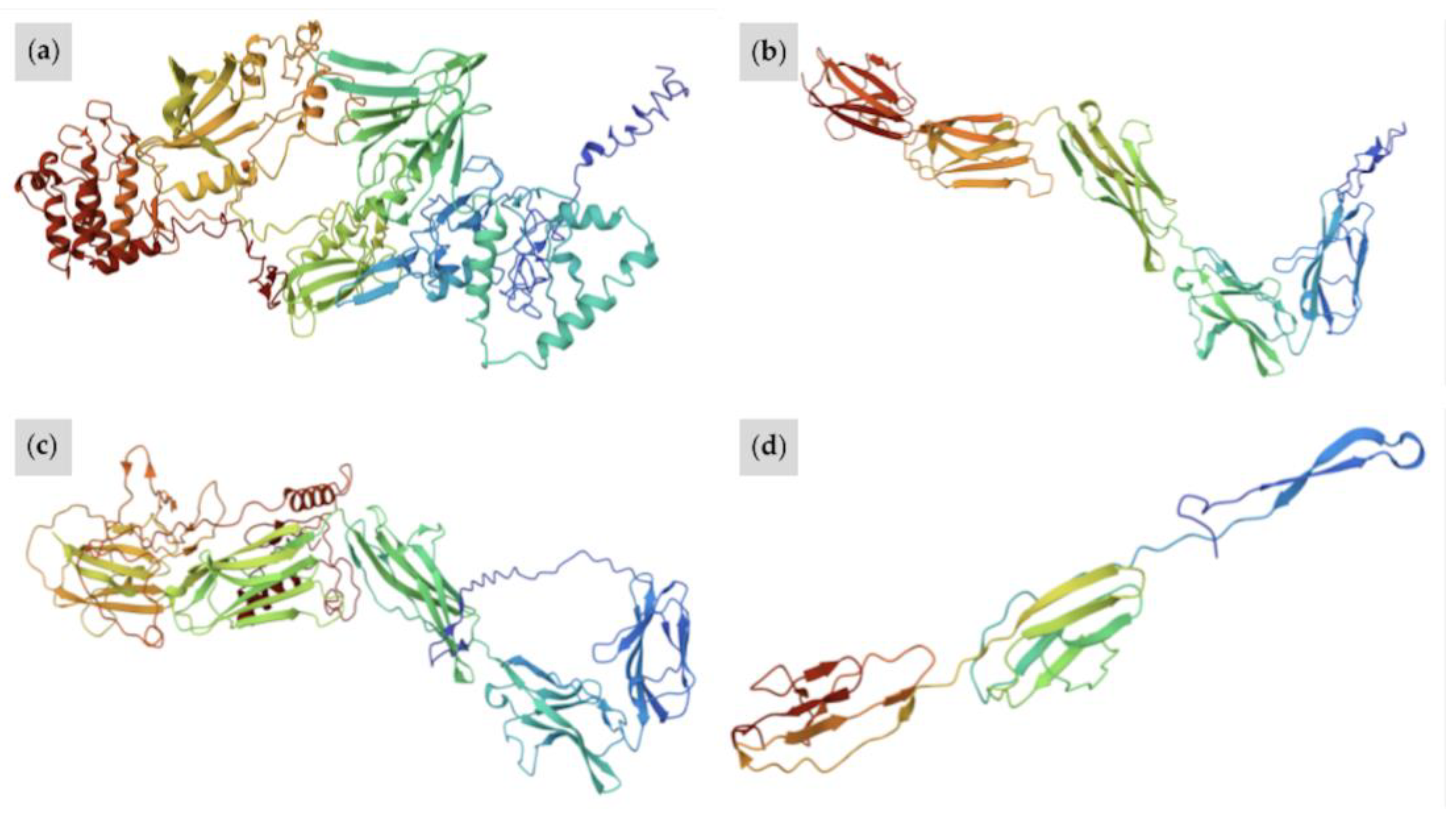
3.4. Predicted Functional Impacts of Novel Variants
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McFadden, A.; Vierra, M.; Martin, K.; Brooks, S.A.; Everts, R.E.; Lafayette, C. Spotting the pattern: A review on white coat color in the domestic horse. Animals 2024, 14, 451. [Google Scholar] [CrossRef]
- Brooks, S.A.; Palermo, K.M.; Khan, A.; Hein, J. Impact of white-spotting alleles, including W20, on phenotype in the American Paint Horse. Anim. Genet. 2020, 51, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Santschi, E.M.; Vrotsos, P.D.; Purdy, A.K.; Mikelson, J.R. Incidence of the endothelin receptor B mutation that causes lethal white foal syndrome in white-patterned horses. Am. J. Vet. Res. 2001, 62, 97–103. [Google Scholar] [CrossRef]
- Hauswirth, R.; Haase, B.; Blatter, M.; Brooks, S.A.; Burger, D.; Drögemüller, C.; Gerber, V.; Henke, D.; Janda, J.; Jude, R. Mutations in MITF and PAX3 cause “splashed white” and other white spotting phenotypes in horses. PLoS Genet. 2012, 8, e1002653. [Google Scholar] [CrossRef] [PubMed]
- Henkel, J.; Lafayette, C.; Brooks, S.A.; Martin, K.; Patterson Rosa, L.; Cook, D.; Jagannathan, V.; Leeb, T. Whole-genome sequencing reveals a large deletion in the MITF gene in horses with white spotted coat color and increased risk of deafness. Anim. Genet. 2019, 50, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Bailey, E.; Brooks, S.A. Horse Genetics, 3rd ed.; CABI: Wallingford, UK, 2020; ISBN 978-178-639-259-6. [Google Scholar]
- Bellone, R.R.; Holl, H.; Setaluri, V.; Devi, S.; Maddodi, N.; Archer, S.; Sandmeyer, L.; Ludwig, A.; Foerster, D.; Pruvost, M.; et al. Evidence for a retroviral insertion in TRPM1 as the cause of congenital stationary night blindness and leopard complex spotting in the horse. PLoS ONE 2013, 8, e78280. [Google Scholar] [CrossRef]
- Rockwell, H.; Mack, M.; Famula, T.; Sandmeyer, L.; Bauer, B.S.; Dwyer, A.; Lassline, M.; Beeson, S.; Archer, S.; McCue, M.; et al. Genetic investigation of equine recurrent uveitis in Appaloosa horses. Anim. Genet. 2020, 51, 111–116. [Google Scholar] [CrossRef]
- Sandmeyer, L.S.; Kingsley, N.B.; Walder, C.; Archer, S.; Leis, M.L.; Bellone, R.R.; Bauer, B.S. Risk factors for equine recurrent uveitis in a population of Appaloosa horses in western Canada. Vet. Ophthalmol. 2020, 23, 515–525. [Google Scholar] [CrossRef]
- Online Mendelian Inheritance in Animals (OMIA). Available online: https://omia.org/ (accessed on 17 January 2025).
- Blume-Jensen, P.; Claesson-Welsh, L.; Siegbahn, A.; Zsebo, K.M.; Westermark, B.; Heldin, C.H. Activation of the human c-kit product by ligand-induced dimerization mediates circular actin reorganization and chemotaxis. EMBO J. 1991, 10, 4121–4128. [Google Scholar] [CrossRef]
- Phung, B.; Sun, J.; Schepsky, A.; Steingrimsson, E.; Rönnstrand, L. C-KIT signaling depends on microphthalmia-associated transcription factor for effects on cell proliferation. PLoS ONE 2011, 6, e24064. [Google Scholar] [CrossRef]
- Steel, K.P.; Davidson, D.R.; Jackson, I.J. TRP-2/DT, a new early melanoblast marker, shows that steel growth factor (c-kit ligand) is a survival factor. Development 1992, 115, 1111–1119. [Google Scholar] [CrossRef]
- Pulos, W.L.; Hutt, F.B. Lethal dominant white in horses. J. Hered. 1969, 60, 59–63. [Google Scholar] [CrossRef] [PubMed]
- McFadden, A.; Martin, K.; Vierra, M.; Robilliard, H.; Lundquist, E.; Everts, R.E.; Brooks, S.A.; Lafayette, C. Three Hps5 mutations associated with depigmentation in diverse horse breeds. Livest. Sci. 2023, 282, 105454. [Google Scholar] [CrossRef]
- Kalbfleisch, T.S.; Rice, E.S.; DePriest, M.S.; Walenz, B.P.; Hestand, M.S.; Vermeesch, J.R.; O′Connell, B.L.; Fiddes, I.T.; Vershinina, A.O.; Saremi, N.F.; et al. Improved reference genome for the domestic horse increases assembly contiguity and composition. Commun. Biol. 2018, 1, 197. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Brown, G.R.; Hiatt, S.M.; Thibaud-Nissen, F.; Astashyn, A.; Ermolaeva, O.; Farrell, C.M.; Hart, J.; Landrum, M.J.; McGarvey, K.M.; et al. RefSeq: An update on mammalian reference sequences. Nucleic Acids Res. 2014, 42, D756–D763. [Google Scholar] [CrossRef]
- Cezard, T.; Cunningham, F.; Hunt, S.E.; Koylass, B.; Kumar, N.; Saunders, G.; Shen, A.; Silva, A.F.; Tsukanov, K.; Venkataraman, S.; et al. The European Variation Archive: A FAIR resource of genomic variation for all species. Nucleic Acids Res. 2022, 50, D1216–D1220. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Bennett, D.C.; Lamoreux, M.L. The color loci of mice—A genetic century. Pigment Cell Res. 2003, 16, 333–344. [Google Scholar] [CrossRef]
- Magdesian, K.G.; Tanaka, J.; Bellone, R.R. A de novo MITF deletion explains a novel splashed white phenotype in an American Paint Horse. J. Hered. 2020, 111, 287–293. [Google Scholar] [CrossRef]
- Patterson Rosa, L.; Martin, K.; Vierra, M.; Foster, G.; Brooks, S.A.; Lafayette, C. Non-frameshift deletion on MITF is associated with a novel splashed white spotting pattern in horses (Equus caballus). Anim. Genet. 2022, 53, 538–540. [Google Scholar] [CrossRef]
- Ebanks, J.P.; Wickett, R.R.; Boissy, R.E. Mechanisms regulating skin pigmentation: The rise and fall of complexion coloration. Int. J. Mol. Sci. 2009, 10, 4066–4087. [Google Scholar] [CrossRef] [PubMed]
- Hauswirth, R.; Jude, R.; Haase, B.; Bellone, R.R.; Archer, S.; Holl, H.; Brooks, S.A.; Tozaki, T.; Penedo, M.C.T.; Rieder, S. Novel variants in the KIT and PAX3 genes in horses with white-spotted coat colour phenotypes. Anim. Genet. 2014, 44, 763–765. [Google Scholar] [CrossRef]
- Metallinos, D.L.; Bowling, A.T.; Rine, J. A missense mutation in the endothelin-B receptor gene is associated with Lethal white foal syndrome: An equine version of Hirschsprung disease. Mamm. Genome 1998, 9, 426–431. [Google Scholar] [CrossRef]
- Wang, H.H.; Chen, H.S.; Li, H.B.; Zhang, H.; Mei, L.Y.; He, C.F.; Wang, X.W.; Men, M.C.; Jiang, L.; Liao, X.B.; et al. Identification and functional analysis of a novel mutation in the SOX10 gene associated with Waardenburg syndrome type IV. Gene 2014, 538, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Chandra Mohan, S.L.N. Case of Waardenburg Shah syndrome in a family with review of literature. J. Otol. 2018, 13, 105–110. [Google Scholar] [CrossRef]
- Steffes, G.; Lorente-Cánovas, B.; Pearson, S.; Brooker, R.H.; Spiden, S.; Kiernan, A.E.; Guénet, J.L.; Steel, K.P. Mutanlallemand (mtl) and belly spot and deadness (bsd) are two new mutations of Lmx1a causing severe cochlear and vestibular defects. PLoS ONE 2012, 7, e51065. [Google Scholar] [CrossRef]
- Baynash, A.G.; Hosoda, K.; Giaid, A.; Richardson, J.A.; Emoto, N.; Hammer, R.E.; Yanagisawa, M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell 1994, 79, 1277–1285. [Google Scholar] [CrossRef]
- Di Palma, F.; Belyantseva, I.A.; Kim, H.J.; Vogt, T.F.; Kachar, B.; Noben-Trauth, K. Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc. Natl. Acad. Sci. USA 2002, 99, 14994–14999. [Google Scholar] [CrossRef]
- Shinohara, M.; Terada, Y.; Iwamatsu, A.; Shinohara, A.; Mochizuki, N.; Higuchi, M.; Gotoh, Y.; Ihara, S.; Nagata, S.; Itoh, H.; et al. SWAP-70 is a guanine-nucleotide-exchange factor that mediates signalling of membrane ruffling. Nature 2002, 416, 759–763. [Google Scholar] [CrossRef]
- Blasius, A.L.; Brandl, K.; Crozat, K.; Xia, Y.; Khovananth, K.; Krebs, P.; Smart, N.G.; Zampolli, A.; Ruggeri, Z.M.; Beutler, B.A. Mice with mutations of Dock7 have generalized hypopigmentation and white-spotting but show normal neurological function. Proc. Natl. Acad. Sci. USA 2009, 106, 2706–2711. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Bell, E.W.; Zhang, Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep. Methods 2021, 1, 100014. [Google Scholar] [CrossRef]
- Burge, C.; Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Dürig, N.; Jude, R.; Holl, H.; Brooks, S.A.; Lafayette, C.; Jagannathan, V.; Leeb, T. Whole genome sequencing reveals a novel deletion variant in the KIT gene in horses with white spotted coat colour phenotypes. Anim. Genet. 2017, 48, 483–485. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT—The BLAST-Like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]
- Torella, A.; Zanobio, M.; Zeuli, R.; del Vecchio Blanco, F.; Savarese, M.; Giugliano, T.; Garofalo, A.; Piluso, G.; Politano, L.; Nigro, V. The position of nonsense mutations can predict the phenotype severity: A survey on the DMD gene. PLoS ONE 2020, 15, e0237803. [Google Scholar] [CrossRef]
- Attanasio, C.; David, A.; Neerman-Arbez, M. Outcome of donor splice site mutations accounting for congenital afibrinogenemia reflects order of intron removal in the fibrinogen alpha gene (FGA). Blood 2003, 101, 1851–1856. [Google Scholar] [CrossRef]
- Ma, S.L.; Vega-Warner, V.; Gillies, C.; Sampson, M.G.; Kher, V.; Sethi, S.K.; Otto, E.A. Whole exome sequencing reveals novel PHEX splice site mutations in patients with hypophosphatemic rickets. PLoS ONE 2015, 10, e0130729. [Google Scholar] [CrossRef]
- Hori, T.; Fukao, T.; Murase, K.; Sakaguchi, N.; Harding, C.O.; Kondo, N. Molecular basis of two-exon skipping (exons 12 and 13) by c.1248+5g>a in OXCT1 gene: Study on intermediates of OXCT1 transcripts in fibroblasts. Hum. Mutat. 2013, 34, 473–480. [Google Scholar] [CrossRef]
- Holl, H.; Brooks, S.A.; Bailey, E. De novo mutation of KIT discovered as a result of a non-hereditary white coat colour pattern. Anim. Genet. 2010, 41, 196–198. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
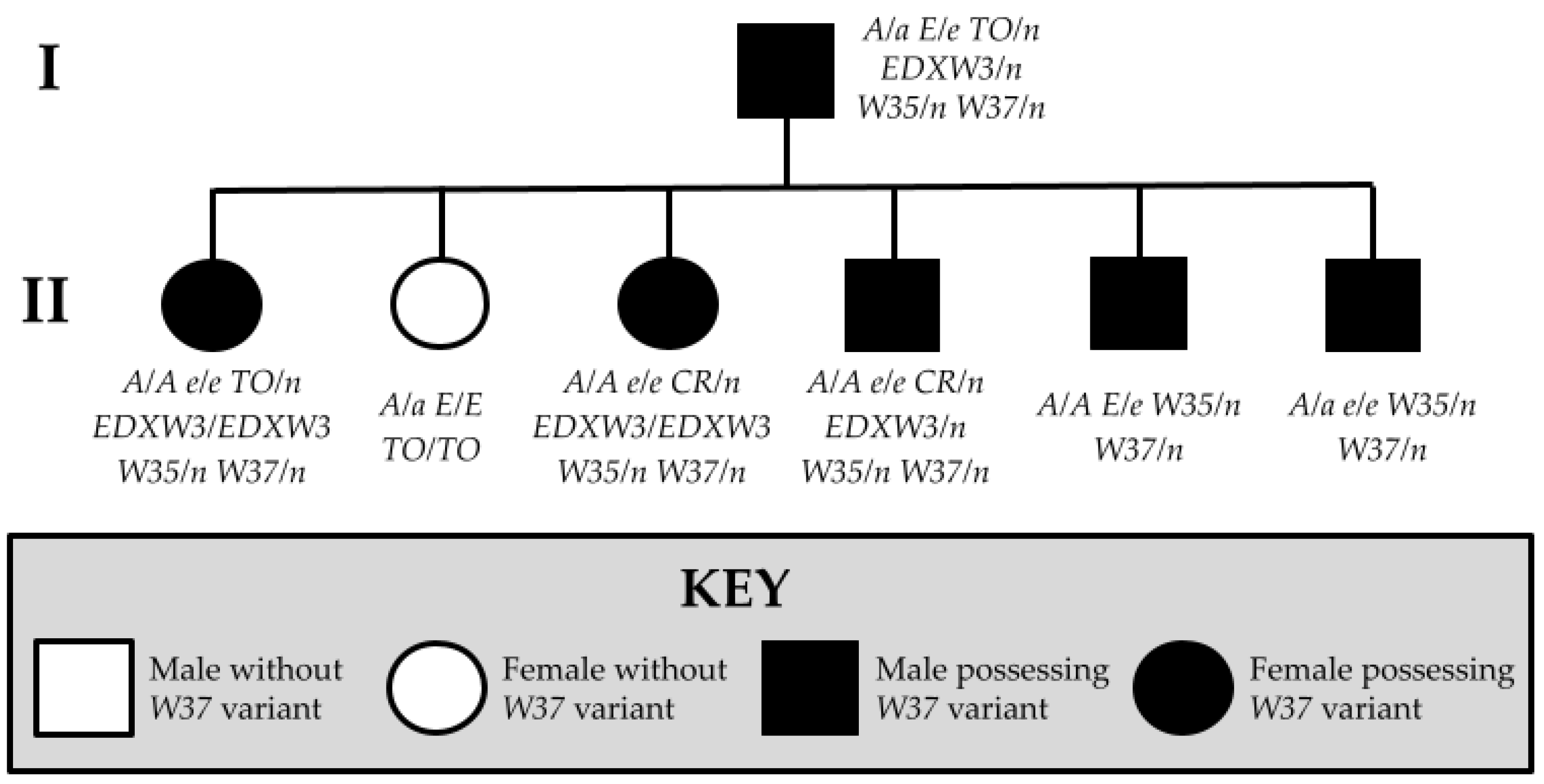
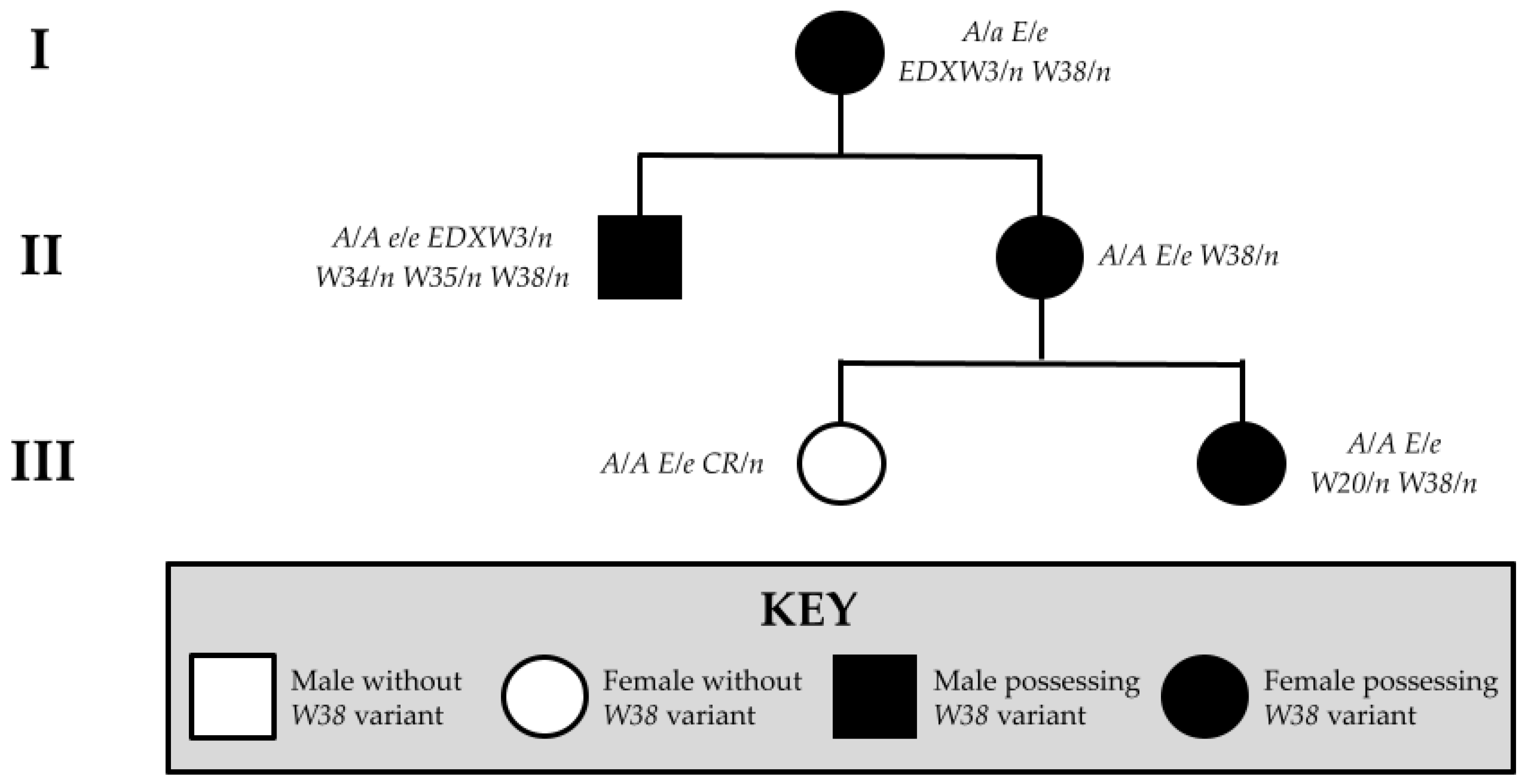
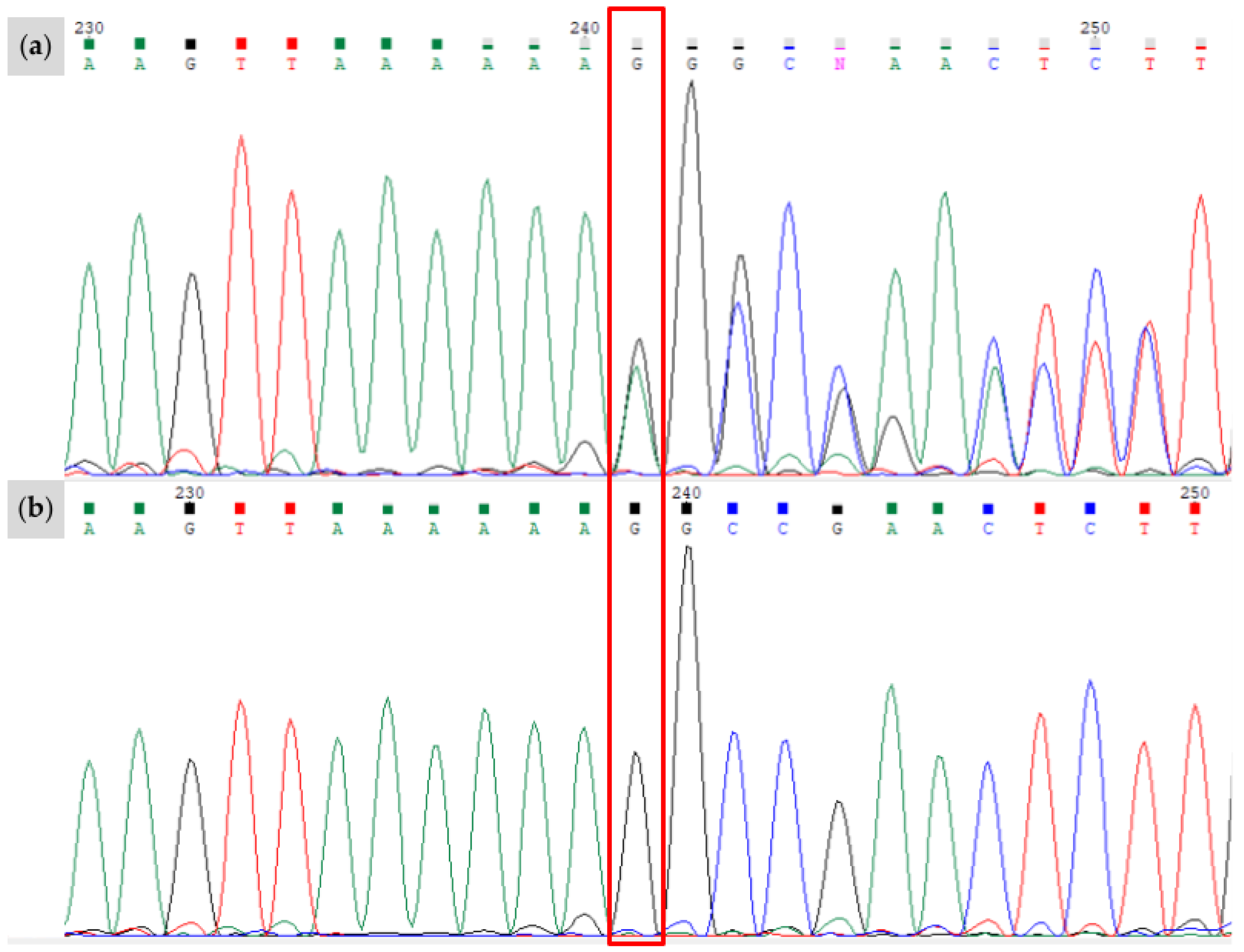

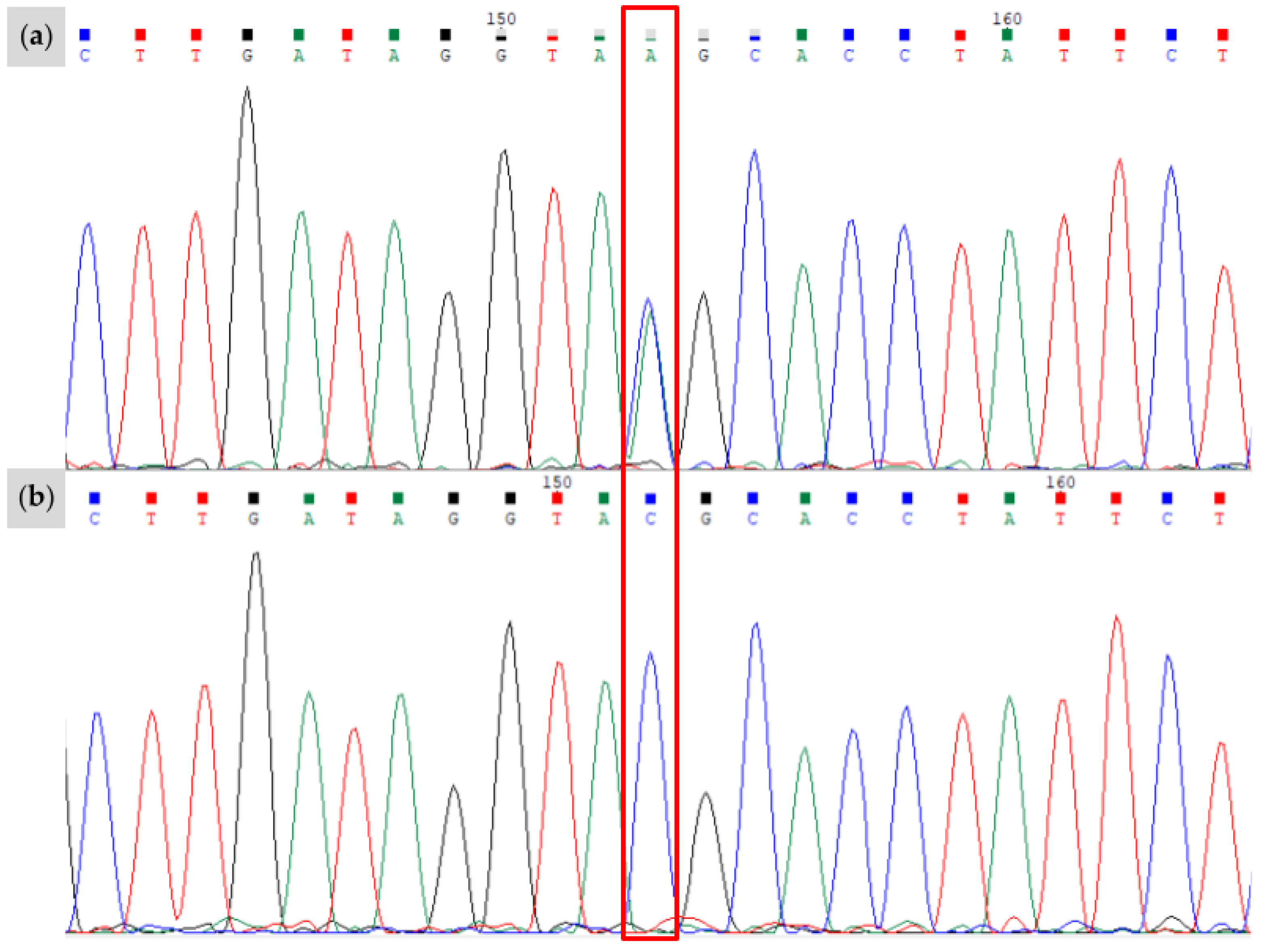


| Novel Variant | Relationship of Individual to Proband | Genotype | Associated Phenotype | Predicted Functional Impact |
|---|---|---|---|---|
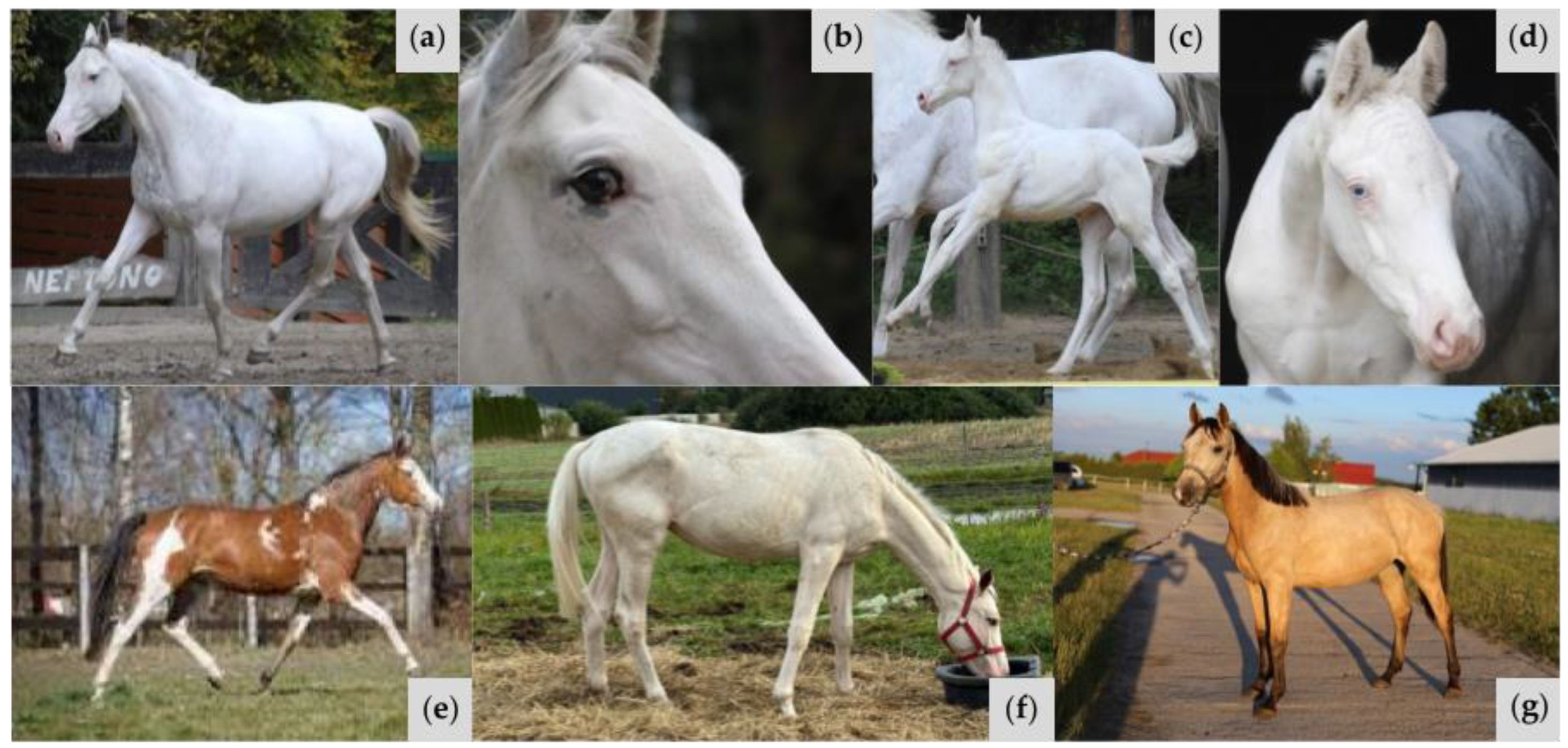
| W37 | Proband (Sire) (Figure 4a,b) | A/a E/e TO/n EDXW3/n W35/n W37/n | Near all-white with small patches of black pigment on face; one blue eye | Truncation of KIT protein prior to active kinase domain inhibits signal transduction processes Typical protein folding is repressed; W37 KIT is more linear in structure than wild-type KIT, featuring no alpha helices and fewer pleated beta sheets |
| Offspring 1 (Figure 4c) | A/A e/e CR/n EDXW3/EDXW3 W35/n W37/n | All-white; pink skin | ||
| Offspring 2 (Figure 4d,e) | A/A e/e CR/n EDXW3/n W35/n W37/n | All-white; pink skin; two partial blue eyes | ||
| Offspring 3 (Figure 4f) | A/A E/e W35/n W37/n | Bay with partial bald face and sabino-like depigmentation observed on belly and hind legs | ||
| Offspring 4 (Figure 4g) | A/a e/e W35/n W37/n | Full bald face with extensive sabino-like depigmentation observed on body and legs with red pigment on upper body, mane, and tail | ||
| Offspring 5 (Figure 4h) | A/A e/e TO/n EDXW3/EDXW3 W35/n W37/n | Near all-white with small patches of red pigmentation on body and face | ||
| Offspring 6 (Figure 4i) | A/a E/E TO/TO | Bay with large white patches on body, typical of Tobiano | ||
| W38 | Proband (Figure 6a,b) | A/A E/e W38/n | All-white; pink skin with small pigmented spots on skin of body and face; at least one blue eye | Truncated KIT protein with conversion of active tyrosine kinase domain to a general kinase; decreased specificity for tyrosine disrupts pathways upregulating pigmentation W38 KIT lacks structural complexity of wild-type protein, with fewer alpha helices and only a number of pleated beta sheets |
| Offspring 1 (Figure 6c,d) | A/A E/e W20/n W38/n | All-white; pink skin; at least one blue eye | ||
| Dam (Figure 6e) | A/a E/e EDXW3/n W38/n | Sabino-like with patches of white on body, high white socks, and wide blaze | ||
| Half-sibling (Figure 6f) | A/A e/e EDXW3/n W34/n W35/n W38/n | All-white; pink skin | ||
| Offspring 2 (Figure 6g) | A/A E/e CR/n | Diluted coat; minimal white marking on muzzle | ||
| W39 | Proband (Figure 8) | A/a e/e W39/n | Roan-like with red hair mixed in with the white hair on body, mane, and tail; Full bald face with small spots of red pigment; black pigment below knees and hocks | KIT protein is truncated after just one IG domain and active domain is lost, impairing biological function W39 KIT is the most linear of all superimposed proteins, with no alpha helices and few pleated beta sheets |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obradovic, N.A.; McFadden, A.; Martin, K.; Vierra, M.; McLoone, K.; Martin, E.; Thomas, A.; Everts, R.E.; Brooks, S.A.; Lafayette, C. Three Novel KIT Polymorphisms Found in Horses with White Coat Color Phenotypes. Animals 2025, 15, 915. https://doi.org/10.3390/ani15070915
Obradovic NA, McFadden A, Martin K, Vierra M, McLoone K, Martin E, Thomas A, Everts RE, Brooks SA, Lafayette C. Three Novel KIT Polymorphisms Found in Horses with White Coat Color Phenotypes. Animals. 2025; 15(7):915. https://doi.org/10.3390/ani15070915
Chicago/Turabian StyleObradovic, Nikol A., Aiden McFadden, Katie Martin, Micaela Vierra, Kaitlyn McLoone, Erik Martin, Adelaide Thomas, Robin E. Everts, Samantha A. Brooks, and Christa Lafayette. 2025. "Three Novel KIT Polymorphisms Found in Horses with White Coat Color Phenotypes" Animals 15, no. 7: 915. https://doi.org/10.3390/ani15070915
APA StyleObradovic, N. A., McFadden, A., Martin, K., Vierra, M., McLoone, K., Martin, E., Thomas, A., Everts, R. E., Brooks, S. A., & Lafayette, C. (2025). Three Novel KIT Polymorphisms Found in Horses with White Coat Color Phenotypes. Animals, 15(7), 915. https://doi.org/10.3390/ani15070915








