Screening of Protein Related to Wool Development and Fineness in Gansu Alpine Fine-Wool Sheep
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animals and Sample Collection
2.3. Total Protein Extraction and Mass Spectrometry Sample Preparation
2.4. DIA Mode Liquid Analysis
2.5. DIA Protein Data Analysis
2.6. Functional Enrichment Analysis
2.7. Correlation Analysis
2.8. Statistical Analyses
3. Results
3.1. Data Quality Control and Protein Identification
3.2. Screening for Differentially Expressed Proteins
3.3. Functional Enrichment Analysis of Differentially Expressed Proteins
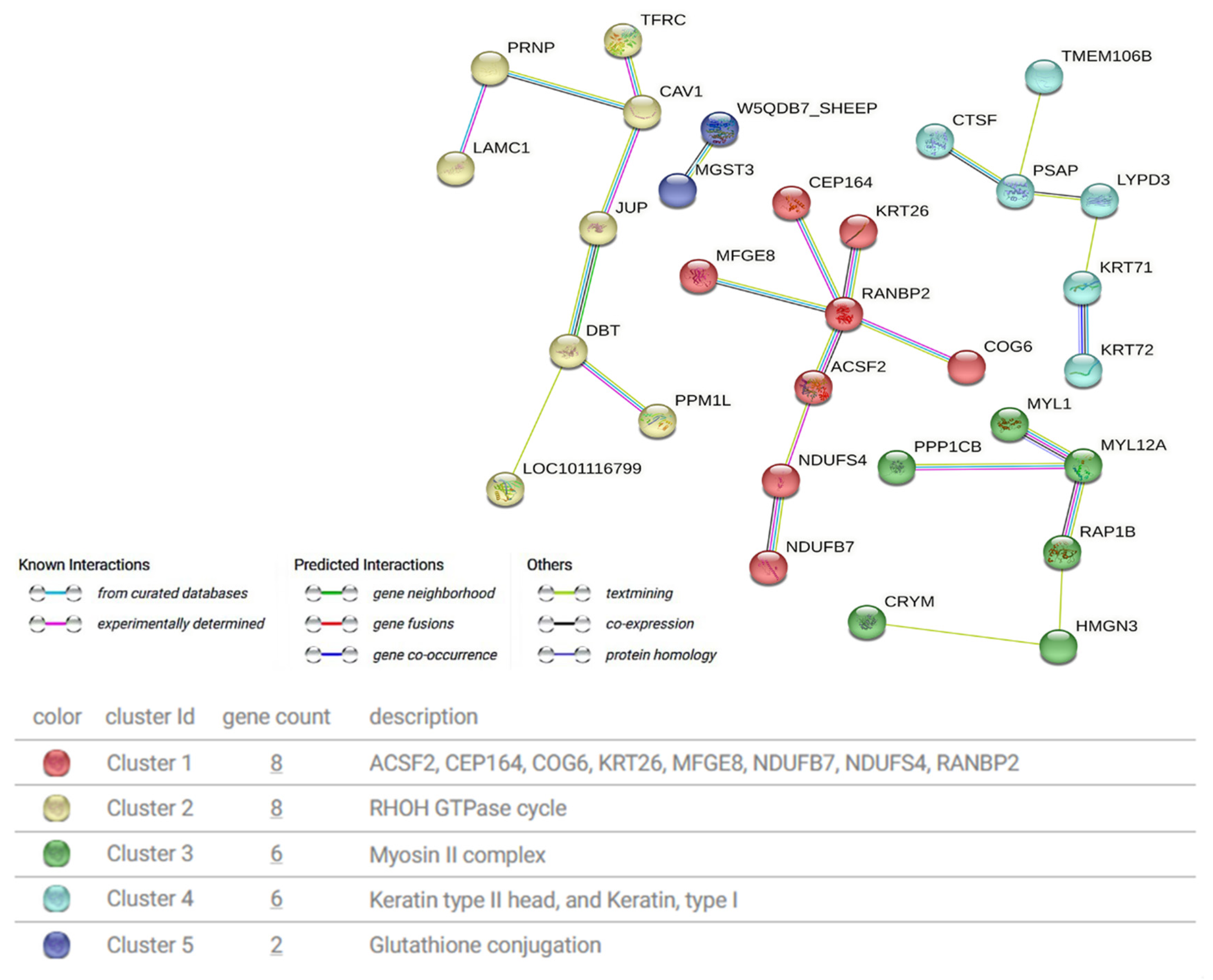
3.4. Protein Interaction Network Analysis
3.5. Identification of Keratin and Keratin-Associated Proteins Related to Wool Fineness
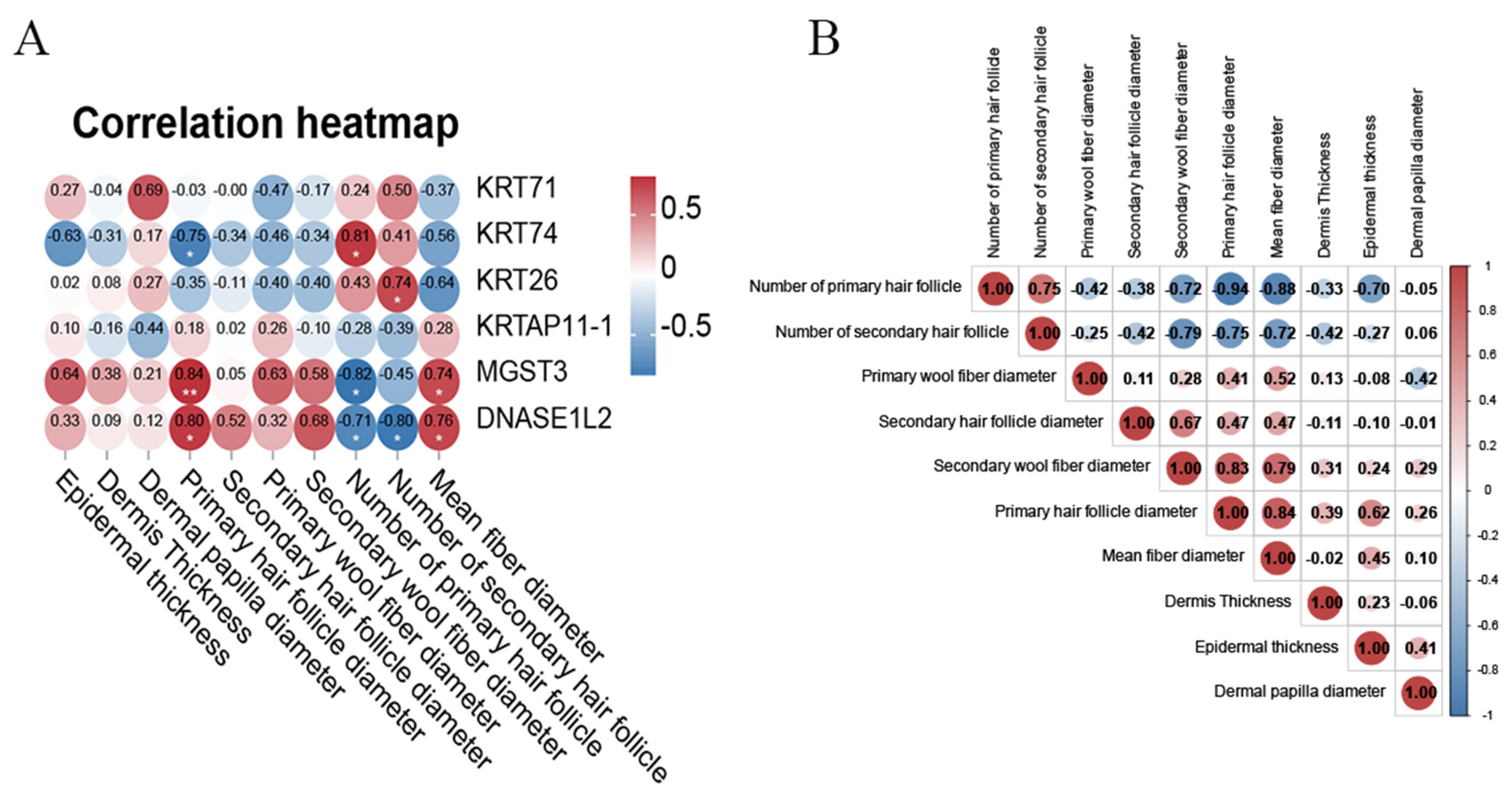
3.6. Correlation Analysis Between Key Differential Proteins and Wool Fiber and Hair Follicle Characteristics
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, W.H.; Purvis, I.W. Genetic parameter estimates for growth traits of Gansu Alpine Finewool sheep. Anim. Prod. Sci. 2012, 52, 321–328. [Google Scholar] [CrossRef]
- Patnaik, A.; Mvubu, M.; Muniyasamy, S.; Botha, A.; Anandjiwala, R.D. Thermal and sound insulation materials from waste wool and recycled polyester fibers and their biodegradation studies. Energy Build. 2015, 92, 161–169. [Google Scholar] [CrossRef]
- Kicinska-Jakubowska, A.; Morales Villavicencio, A.; Zimniewska, M.; Przybylska, P.; Kwiatkowska, E. Evaluation of Wool Quality Parameters of Polish Sheep Breeds. J. Nat. Fibers 2022, 19, 5880–5887. [Google Scholar] [CrossRef]
- Zhang, W.; Jin, M.; Li, T.; Lu, Z.; Wang, H.; Yuan, Z.; Wei, C. Whole-Genome Resequencing Reveals Selection Signal Related to Sheep Wool Fineness. Animals 2023, 13, 2944. [Google Scholar] [CrossRef]
- Dayon, L.; Cominetti, O.; Affolter, M. Proteomics of human biological fluids for biomarker discoveries: Technical advances and recent applications. Expert Rev. Proteom. 2022, 19, 131–151. [Google Scholar] [CrossRef]
- Karpov, O.A.; Stotland, A.; Raedschelders, K.; Chazarin, B.; Ai, L.; Murray, C.I.; Van Eyk, J.E. Proteomics of the heart. Physiol. Rev. 2024, 104, 931–982. [Google Scholar] [CrossRef] [PubMed]
- Thongboonkerd, V. Proteomics. In Proceedings of the Nutrigenomics—Opportunities in Asia: 1st ILSI International Conference on Nutrigenomics, Singapore, 7–9 December 2005; Tai, E.S., Gillies, P.J., Eds.; S.Karger AG: Basel, Switzerland, 2007; Volume 60, pp. 80–90. [Google Scholar]
- Li, Z.B.; Flint, P.W.; Boluyt, M.O. Evaluation of several two-dimensional gel electrophoresis techniques in cardiac proteomics. Electrophoresis 2005, 26, 3572–3585. [Google Scholar] [CrossRef]
- Macklin, A.; Khan, S.; Kislinger, T. Recent advances in mass spectrometry based clinical proteomics: Applications to cancer research. Clin. Proteom. 2020, 17, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, P.K.; Li, L. A Tutorial Review of Labeling Methods in Mass Spectrometry-Based Quantitative Proteomics. ACS Meas. Sci. Au 2024, 4, 315–337. [Google Scholar] [CrossRef]
- Frankenfield, A.M.; Ni, J.; Ahmed, M.; Hao, L. Protein Contaminants Matter: Building Universal Protein Contaminant Libraries for DDA and DIA Proteomics. J. Proteome Res. 2022, 21, 2104–2113. [Google Scholar] [CrossRef]
- Sun, F.; Tan, H.; Li, Y.; De Boevre, M.; Zhang, H.; Zhou, J.; Li, Y.; Yang, S. An integrated data-dependent and data-independent acquisition method for hazardous compounds screening in foods using a single UHPLC-Q-Orbitrap run. J. Hazard. Mater. 2021, 401, 123266. [Google Scholar] [CrossRef]
- Kuster, B.; Tüshaus, J.; Bayer, F.P. A new mass analyzer shakes up the proteomics field. Nat. Biotechnol. 2024, 42, 1796–1797. [Google Scholar] [CrossRef] [PubMed]
- Viitaharju, J.; Polari, L.; Kauko, O.; Merilahti, J.; Rokka, A.; Toivola, D.M.; Laitinen, K. Improved breast milk proteome coverage by DIA based LC-MS/MS method. Proteomics 2024, 24, 2300340. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Y.; Wang, J.; Yang, Y.; Miao, C.; Guo, Y.; Zhang, Z.; Cao, Q.; Shui, W. Combining Untargeted and Targeted Proteomic Strategies for Discrimination and Quantification of Cashmere Fibers. PLoS ONE 2016, 11, e0147044. [Google Scholar] [CrossRef]
- Yue, L.; Lu, Z.; Guo, T.; Liu, J.; Yang, B.; Yuan, C. Proteome Analysis of Alpine Merino Sheep Skin Reveals New Insights into the Mechanisms Involved in Regulating Wool Fiber Diameter. Int. J. Mol. Sci. 2023, 24, 15227. [Google Scholar] [CrossRef]
- Zhao, B.; Wu, C.; Sammad, A.; Ma, Z.; Suo, L.; Wu, Y.; Fu, X. The fiber diameter traits of Tibetan cashmere goats are governed by the inherent differences in stress, hypoxic, and metabolic adaptations: An integrative study of proteome and transcriptome. BMC Genom. 2022, 23, 191. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, G.; Zhang, R.; Guo, J.; Li, C.; Martin, G.; Chen, Y.; Wang, X. Comparative proteomic analyses using iTRAQ-labeling provides insights into fiber diversity in sheep and goats. J. Proteom. 2018, 172, 82–88. [Google Scholar] [CrossRef]
- Buczak, K.; Kirkpatrick, J.M.; Truckenmueller, F.; Santinha, D.; Ferreira, L.; Roessler, S.; Singer, S.; Beck, M.; Ori, A. Spatially resolved analysis of FFPE tissue proteomes by quantitative mass spectrometry. Nat. Protoc. 2020, 15, 2956–2979. [Google Scholar] [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Sun, H.; Zhao, F.; Ma, L.; Wang, J.; Liu, X.; Li, M.; Hao, Z.; Li, S. MicroRNA expression profiles reveal wool development and fineness regulation in Gansu alpine fine-wool sheep. Genomics 2024, 116, 110922. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Han, J.; Yuan, C.; Liu, J.; Niu, C.; Lu, Z.; Yue, Y.; Yang, B. Comparative proteomics reveals genetic mechanisms underlying secondary hair follicle development in fine wool sheep during the fetal stage. J. Proteom. 2020, 223, 103827. [Google Scholar] [CrossRef]
- Plowman, J.; Thomas, A.; Perloiro, T.; Clerens, S.; de Almeida, A.M. Characterisation of white and black merino wools: A proteomics study. Animal 2019, 13, 659–665. [Google Scholar] [CrossRef]
- Liang, Y.-F.; Long, Z.-X.; Zhang, Y.-J.; Luo, C.-Y.; Yan, L.-T.; Gao, W.-Y.; Li, H. The chemical mechanisms of the enzymes in the branched-chain amino acids biosynthetic pathway and their applications. Biochimie 2021, 184, 72–87. [Google Scholar] [CrossRef]
- Neinast, M.; Murashige, D.; Arany, Z. Branched Chain Amino Acids. Annu. Rev. Physiol. 2019, 81, 139–164. [Google Scholar] [CrossRef]
- Ahmad, I.; Ahmed, I.; Fatma, S.; Peres, H. Role of branched-chain amino acids on growth, physiology and metabolism of different fish species: A review. Aquacult. Nutr. 2021, 27, 1270–1289. [Google Scholar] [CrossRef]
- Van Midden, K.P.; Mantz, M.; Fonovič, M.; Gazvoda, M.; Svete, J.; Huesgen, P.F.; van der Hoorn, R.A.L.; Klemenčič, M. Mechanistic insights into CrCEP1: A dual-function cysteine protease with endo- and transpeptidase activity. Int. J. Biol. Macromol. 2024, 271, 132505. [Google Scholar] [CrossRef]
- Saborowski, R.; Schatte, J.; Gimenez, L. Catalytic properties and polymorphism of serine endopeptidases from the midgut gland of the brown shrimp Crangon crangon (Decapoda, Caridea). Mar. Biol. 2012, 159, 1107–1118. [Google Scholar] [CrossRef]
- Buono, R.A.; Hudecek, R.; Nowack, M.K. Plant proteases during developmental programmed cell death. J. Exp. Bot. 2019, 70, 2097–2112. [Google Scholar] [CrossRef]
- Westrip, C.A.E.; Zhuang, Q.; Hall, C.; Eaton, C.D.; Coleman, M.L. Developmentally regulated GTPases: Structure, function and roles in disease. Cell. Mol. Life Sci. 2021, 78, 7219–7235. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Aguilera, A.; Rattmann, I.; Drew, D.Z.; Müller, L.U.W.; Summey, V.; Lucas, D.M.; Byrd, J.C.; Croce, C.M.; Gu, Y.; Cancelas, J.A.; et al. Involvement of RhoH GTPase in the development of B-cell chronic lymphocytic leukemia. Leukemia 2010, 24, 97–104. [Google Scholar] [CrossRef]
- Fleger-Weckmann, A.; Üstün, Y.; Kloepper, J.; Paus, R.; Bloch, W.; Chen, Z.-L.; Wegner, J.; Sorokin, L.; Langbein, L.; Eckes, B.; et al. Deletion of the epidermis derived laminin γ1 chain leads to defects in the regulation of late hair morphogenesis. Matrix Biol. 2016, 56, 42–56. [Google Scholar] [CrossRef]
- Takaya, K.; Asou, T.; Kishi, K. Cathepsin F is a potential marker for senescent human skin fibroblasts and keratinocytes associated with skin aging. GeroScience 2023, 45, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Pečar Fonović, U.; Kos, J.; Mitrović, A. Compensational role between cathepsins. Biochimie 2024, 226, 62–76. [Google Scholar] [CrossRef]
- Yan, J.; Kahyo, T.; Zhang, H.; Ping, Y.; Zhang, C.; Jiang, S.; Ji, Q.; Ferdous, R.; Islam, M.S.; Oyama, S.; et al. Alpha-Synuclein Interaction with UBL3 Is Upregulated by Microsomal Glutathione S-Transferase 3, Leading to Increased Extracellular Transport of the Alpha-Synuclein under Oxidative Stress. Int. J. Mol. Sci. 2024, 25, 7353. [Google Scholar] [CrossRef]
- Jeon, G.; Kim, C.; Cho, U.M.; Hwang, E.T.; Hwang, H.S.; Min, J. Melanin-Decolorizing Activity of Antioxidant Enzymes, Glutathione Peroxidase, Thiol Peroxidase, and Catalase. Mol. Biotechnol. 2021, 63, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Zernov, N.V.; Skoblov, M.Y.; Marakhonov, A.V.; Shimomura, Y.; Vasilyeva, T.A.; Konovalov, F.A.; Abrukova, A.V.; Zinchenko, R.A. Autosomal Recessive Hypotrichosis with Woolly Hair Caused by a Mutation in the Keratin 25 Gene Expressed in Hair Follicles. J. Investig. Dermatol. 2016, 136, 1097–1105. [Google Scholar] [CrossRef]
- Grilz-Seger, G.; Neuditschko, M.; Ricard, A.; Velie, B.; Lindgren, G.; Mesarič, M.; Cotman, M.; Horna, M.; Dobretsberger, M.; Brem, G.; et al. Genome-Wide Homozygosity Patterns and Evidence for Selection in a Set of European and Near Eastern Horse Breeds. Genes 2019, 10, 491. [Google Scholar] [CrossRef]
- Tanaka, S.; Miura, I.; Yoshiki, A.; Kato, Y.; Yokoyama, H.; Shinogi, A.; Masuya, H.; Wakana, S.; Tamura, M.; Shiroishi, T. Mutations in the helix termination motif of mouse type I IRS keratin genes impair the assembly of keratin intermediate filament. Genomics 2007, 90, 703–711. [Google Scholar] [CrossRef]
- Yu, Z.; Gordon, S.W.; Nixon, A.J.; Bawden, C.S.; Rogers, M.A.; Wildermoth, J.E.; Maqbool, N.J.; Pearson, A.J. Expression patterns of keratin intermediate filament and keratin associated protein genes in wool follicles. Differentiation 2009, 77, 307–316. [Google Scholar] [CrossRef]
- Wasif, N.; Naqvi, S.K.u.-H.; Basit, S.; Ali, N.; Ansar, M.; Ahmad, W. Novel mutations in the keratin-74 (KRT74) gene underlie autosomal dominant woolly hair/hypotrichosis in Pakistani families. Hum. Genet. 2011, 129, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, A.; Farooq, M.; Fujikawa, H.; Inoue, A.; Ohyama, M.; Ehama, R.; Nakanishi, J.; Hagihara, M.; Iwabuchi, T.; Aoki, J.; et al. A Missense Mutation within the Helix Initiation Motif of the Keratin K71 Gene Underlies Autosomal Dominant Woolly Hair/Hypotrichosis. J. Investig. Dermatol. 2012, 132, 2342–2349. [Google Scholar] [CrossRef]
- Kuramoto, T.; Hirano, R.; Kuwamura, M.; Serikawa, T. Identification of the Rat Rex Mutation as a 7-bp Deletion at Splicing Acceptor Site of the Krt71 Gene. J. Vet. Med. Sci. 2010, 72, 909–912. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Sheng, S.D.; Hui, T.Y.; Yue, C.; Sun, J.M.; Guo, D.; Guo, S.L.; Li, B.J.; Xue, H.L.; Wang, Z.Y.; et al. An Integrated Analysis of Cashmere Fineness lncRNAs in Cashmere Goats. Genes 2019, 10, 266. [Google Scholar] [CrossRef]
- Erjavec, S.O.; Gelfman, S.; Abdelaziz, A.R.; Lee, E.Y.; Monga, I.; Alkelai, A.; Ionita-Laza, I.; Petukhova, L.; Christiano, A.M. Whole exome sequencing in Alopecia Areata identifies rare variants in KRT82. Nat. Commun. 2022, 13, 800. [Google Scholar] [CrossRef]
- Huang, D.; Ding, H.; Wang, Y.; Wang, X.; Zhao, H. Integration Analysis of Hair Follicle Transcriptome and Proteome Reveals the Mechanisms Regulating Wool Fiber Diameter in Angora Rabbits. Int. J. Mol. Sci. 2024, 25, 3260. [Google Scholar] [CrossRef]
- Gong, H.; Zhou, H.; Dyer, J.M.; Hickford, J.G.H. Identification of the ovine KAP11-1 gene (KRTAP11-1) and genetic variation in its coding sequence. Mol. Biol. Rep. 2011, 38, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yao, H.; Wang, H.; Sui, T. Development of Woolly Hair and Hairlessness in a CRISPR−Engineered Mutant Mouse Model with KRT71 Mutations. Cells 2023, 12, 1781. [Google Scholar] [CrossRef] [PubMed]
- Jacinto, J.G.P.; Markey, A.D.; Veiga, I.M.B.; Paris, J.M.; Welle, M.; Beever, J.E.; Drögemüller, C. A KRT71 Loss-of-Function Variant Results in Inner Root Sheath Dysplasia and Recessive Congenital Hypotrichosis of Hereford Cattle. Genes 2021, 12, 1038. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Zhang, Y.; Tang, X.M. A case of KRT74 variation induced hair loss. Zhonghua Er Ke Za Zhi 2021, 59, 1086–1087. [Google Scholar] [PubMed]
- Xu, Y.; Zhang, X.; Hui, T.; Sun, J.; Cai, W.; Lin, G.; Wang, L.; Dou, X.; Wang, Z.; Han, D.; et al. Association analysis for SNPs of KRT26 and TCHH genes with cashmere production performance, body measurement traits and milk production traits in Liaoning cashmere goats. Anim. Biotechnol. 2023, 34, 698–708. [Google Scholar] [CrossRef]
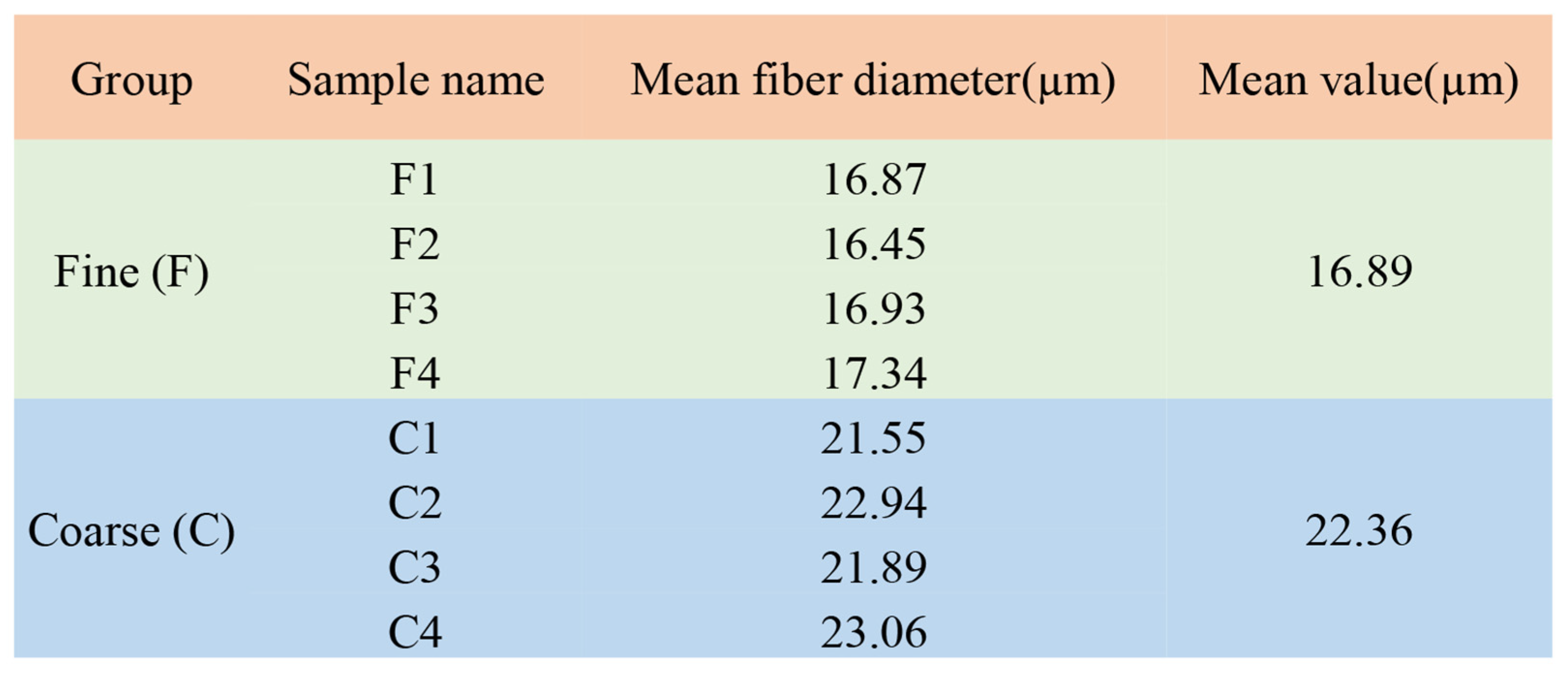
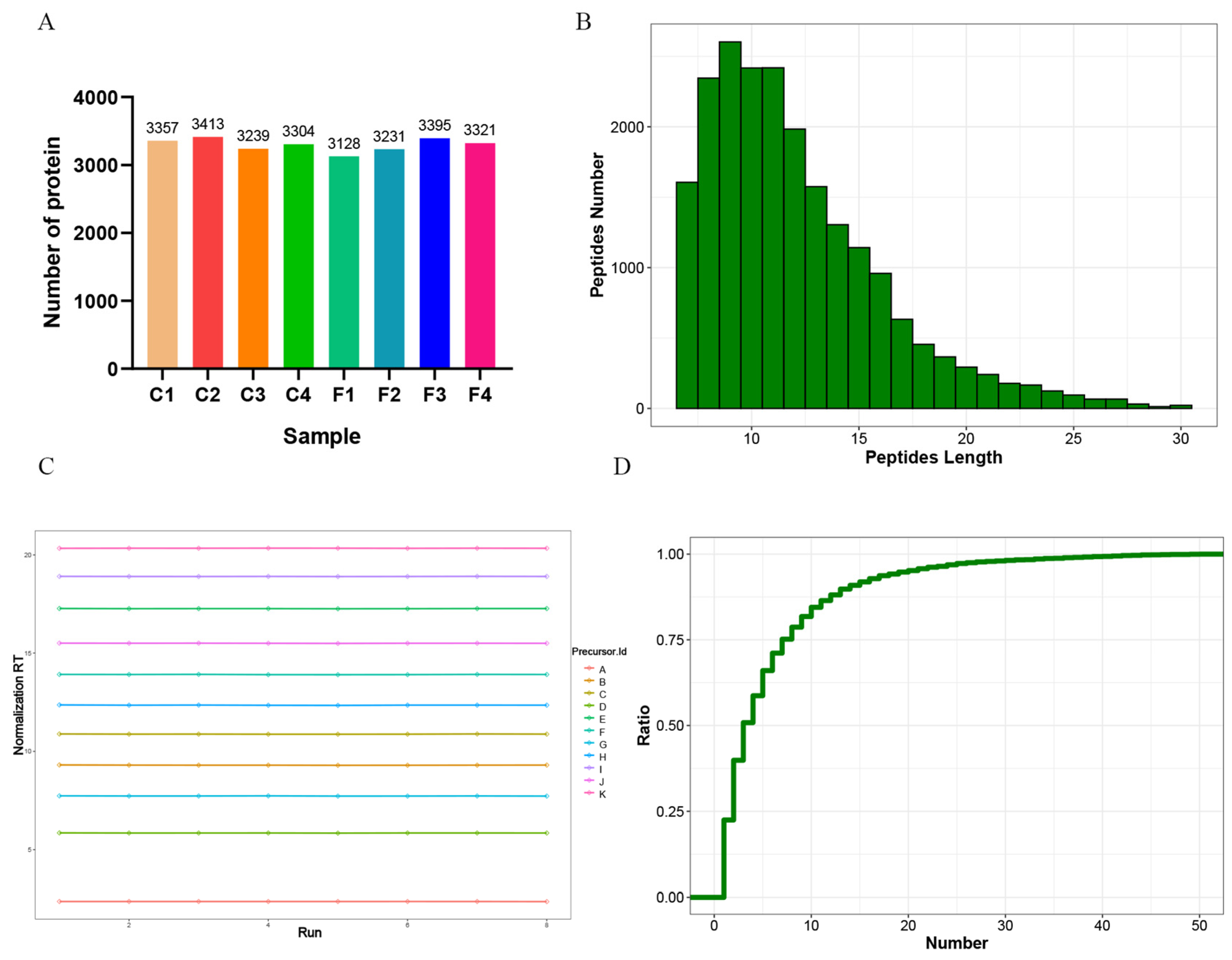
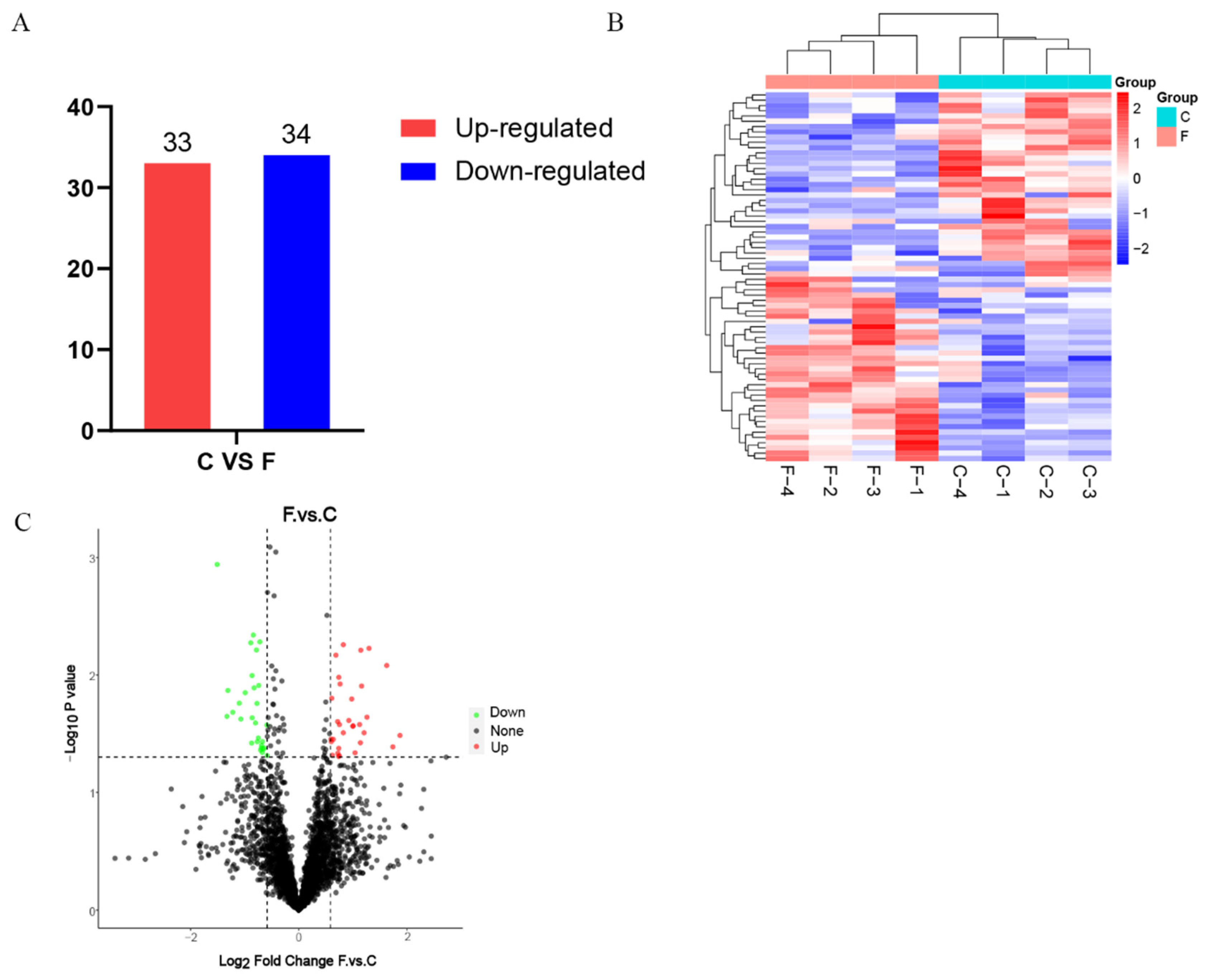
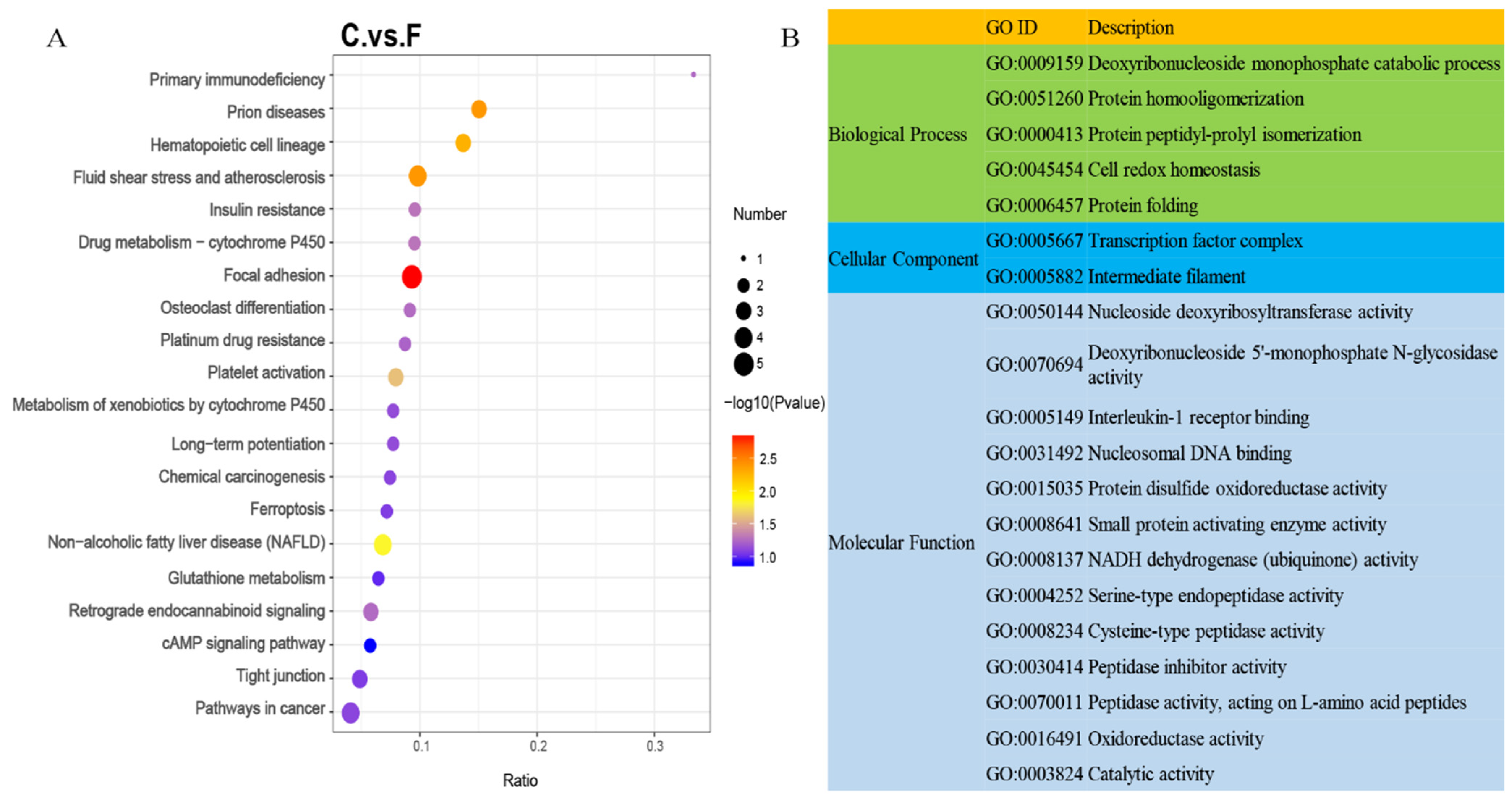

| Sample | Protein Concentration (μg/μL) | Total Protein (μg) | Protein Fluid Volume (μL) |
|---|---|---|---|
| F1 | 0.18 | 30.46 | 170.00 |
| F2 | 0.21 | 35.02 | 170.00 |
| F3 | 0.25 | 42.50 | 170.00 |
| F4 | 0.21 | 36.04 | 170.00 |
| C1 | 0.36 | 61.88 | 170.00 |
| C2 | 0.19 | 32.30 | 170.00 |
| C3 | 0.18 | 31.01 | 170.00 |
| C4 | 0.18 | 31.28 | 170.00 |
| Term/Pathway | Protein |
|---|---|
| GO:0005882 Intermediate filament | KRT26, KRT72, KRT74, KRT71, JUP |
| CL25737 Intermediate filament protein, and Nail development | KRT26, KRT72, KRT74, KRT71, DNASE1L2 |
| CL25742 Keratin, type I, and Keratin, type II | KRT26, KRT72, KRT74, KRT71 |
| ko00480 Glutathione metabolism | XP_012033428.1, MGST3 |
| ko00190 Oxidative phosphorylation | NDUFB7, NDUFS4 |
| ko04974 Protein digestion and absorption | KLK14 |
| ko00280 Valine, leucine and isoleucine degradation | DBT |
| ko04024 cAMP signaling pathway | RAP1B, PPP1CB |
| Classifications | Protein Name | |||
|---|---|---|---|---|
| Keratin-associated protein | KRTAP6-1 | LOC101106296 | LOC114110489 | LOC101106558 |
| LOC101103772 | LOC101116371 | LOC114113348 | LOC114113904 | |
| LOC101104027 | LOC101116882 | LOC114113380 | LOC101104203 | |
| LOC101106046 | LOC114110486 | LOC114113396 | LOC114118004 | |
| LOC101107617 | ||||
| Keratin | KRT25 | KRT82 | KRT8 | KRT5 |
| KRT27 | LOC101112716 | KRT78 | LOC101111178 | |
| KRTAP4.3 | KRT24 | KRT6A | LOC114113976 | |
| KRT2.11 | KRT26 | LOC101111440 | LOC101112555 | |
| KRT33A | KRT28 | KRT12 | LOC101112805 | |
| LOC100526780 | KRT20 | KRT39 | LOC100526784 | |
| LOC100526782 | KRT23 | KRT13 | LOC101112469 | |
| KRT71 | KRT40 | LOC101111791 | KRT80 | |
| KRT17 | LOC101112657 | KRT1 | KRT35 | |
| KRTCAP2 | LOC100526781 | LOC100141295 | LOC101118712 | |
| KRT2 | KRT32 | KRT14 | LOC101118712 | |
| LOC101109951 | KRT36 | KRT18 | KRT19 | |
| LOC101110219 | KRT15 | KRT3 | LOC105610157 | |
| KRT74 | KRTDAP | KRT4 | LOC101115571 | |
| KRT84 | KRT9 | KRT79 | KRT77 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.; Tian, L.; Wang, G.; Zhao, F.; Zhao, P.; Zhang, S.; Li, S.; Yang, G. Screening of Protein Related to Wool Development and Fineness in Gansu Alpine Fine-Wool Sheep. Animals 2025, 15, 2578. https://doi.org/10.3390/ani15172578
He Z, Tian L, Wang G, Zhao F, Zhao P, Zhang S, Li S, Yang G. Screening of Protein Related to Wool Development and Fineness in Gansu Alpine Fine-Wool Sheep. Animals. 2025; 15(17):2578. https://doi.org/10.3390/ani15172578
Chicago/Turabian StyleHe, Zhaohua, Liming Tian, Guan Wang, Fangfang Zhao, Pengfei Zhao, Shuhong Zhang, Shaobin Li, and Guangli Yang. 2025. "Screening of Protein Related to Wool Development and Fineness in Gansu Alpine Fine-Wool Sheep" Animals 15, no. 17: 2578. https://doi.org/10.3390/ani15172578
APA StyleHe, Z., Tian, L., Wang, G., Zhao, F., Zhao, P., Zhang, S., Li, S., & Yang, G. (2025). Screening of Protein Related to Wool Development and Fineness in Gansu Alpine Fine-Wool Sheep. Animals, 15(17), 2578. https://doi.org/10.3390/ani15172578









