Effects of Grazing in a Low Deciduous Forest on Rumen Microbiota and Volatile Fatty Acid Production in Lambs
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Estimating Consumption During Grazing
2.3. Collection of Rumen Fluid
2.4. Measurement of VFAs in the Ruminal Fluid
2.5. DNA Isolation and 16S rRNA Gene Sequencing
2.6. Statistical Analysis
2.7. Data Analysis
3. Results
3.1. Voluntary Intake and Daily Weight Gain
3.2. Concentrations of VFAs in Lambs Housed and Grazing LDF over Three Periods
3.3. Rumen Microbial Diversity in Lambs Grazing LDF
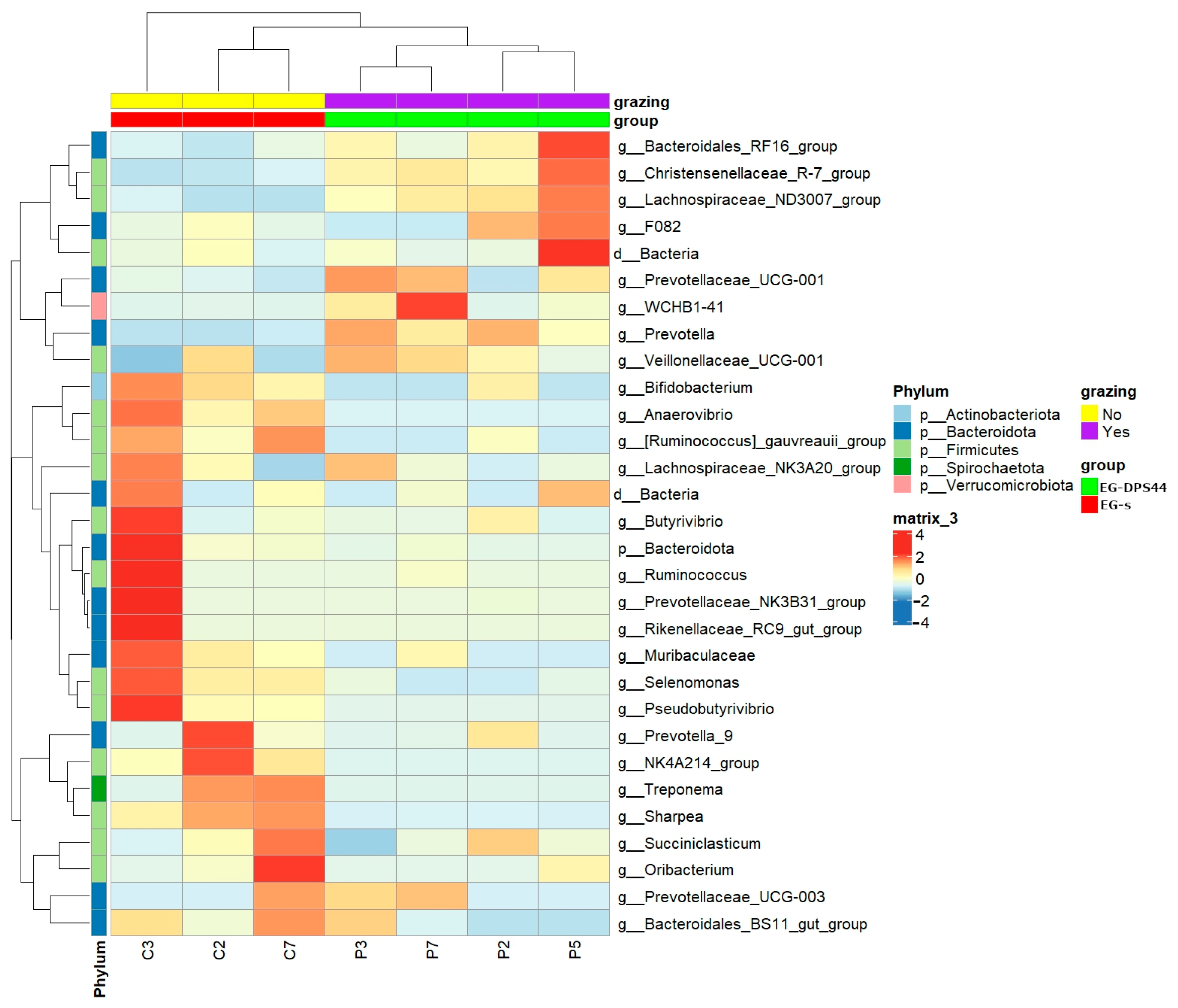
3.4. Rumen Microbial Community Composition and Functional Profiles in Lambs Grazing the LDF Compared to Housed Lambs
3.5. Correlations Between Microbial Biomarkers and VFAs in Lambs Grazing LDF
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BW | Body Weight |
| VFAs | Volatile Fatty Acids |
| LDF | Low Deciduous Forest |
| EG | Experimental Group |
| CG | Control Group |
| EGs | Experimental Group Stabilization |
| CGs | Control Group Stabilization |
| EGg14 | Experimental Group grazing 14 days |
| CGg14 | Control Group grazing 14 days |
| EG-DPS44 | Experimental Group grazing 44 days |
| CGg44 | Control Group grazing 44 days |
| VI | Voluntary Intake |
| DWG | Daily Weight Gain |
| TCA | Tricarboxylic Acid Cycle |
References
- Sanjorjo, R.A.; Tseten, T.; Kang, M.-K.; Kwon, M.; Kim, S.-W. In Pursuit of Understanding the Rumen Microbiome. Fermentation 2023, 9, 114. [Google Scholar] [CrossRef]
- McDonald, P.; Edwards, R.A.; Greenhalgh, J.F.D.; Morgan, C.A.; Sinclair, L.-A.; Wilkinson, R.G. Animal Nutrition, 7th ed.; Prentice Hall: Ciudad de Mexico, Mexico, 2010; ISBN 978-0-582-41906-3. [Google Scholar]
- Keum, G.B.; Pandey, S.; Kim, E.S.; Doo, H.; Kwak, J.; Ryu, S.; Choi, Y.; Kang, J.; Kim, S.; Kim, H.B. Understanding the Diversity and Roles of the Ruminal Microbiome. J. Microbiol. 2024, 62, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Janssen, P.H. Rumen Microbial Community Composition Varies with Diet and Host, but a Core Microbiome Is Found across a Wide Geographical Range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Li, K.; Li, Y.; Gu, M.; Omoor, I.N.A.; Liu, H.; Qiu, S.; Jiang, X.; Lu, J.; Ma, Z.; et al. Deciphering the Impact of Nutrient Composition and Tissue Structure on Rumen Microbiome Dynamics in Roughage Degradation. Res. Artic. 2024. [Google Scholar] [CrossRef]
- Zhou, G.; Li, J.; Liang, X.; Yang, B.; He, X.; Tang, H.; Guo, H.; Liu, G.; Cui, W.; Chen, Y.; et al. Multi-Omics Revealed the Mechanism of Feed Efficiency in Sheep by the Combined Action of the Host and Rumen Microbiota. Anim. Nutr. 2024, 18, 367–379. [Google Scholar] [CrossRef]
- Perez, H.G.; Stevenson, C.K.; Lourenco, J.M.; Callaway, T.R. Understanding Rumen Microbiology: An Overview. Encyclopedia 2024, 4, 148–157. [Google Scholar] [CrossRef]
- He, S.; Yuan, Z.; Dai, S.; Wang, Z.; Zhao, S.; Wang, R.; Li, Q.; Mao, H.; Wu, D. Intensive Feeding Alters the Rumen Microbiota and Its Fermentation Parameters in Natural Grazing Yaks. Front. Vet. Sci. 2024, 11, 1365300. [Google Scholar] [CrossRef]
- Torres-Fajardo, R.A.; González-Pech, P.G.; Torres-Acosta, J.F.D.J.; Sandoval-Castro, C.A. Nutraceutical Potential of the Low Deciduous Forest to Improve Small Ruminant Nutrition and Health: A Systematic Review. Agronomy 2021, 11, 1403. [Google Scholar] [CrossRef]
- Torres-Fajardo, R.A.; Ortíz-Domínguez, G.; González-Pech, P.G.; Sandoval-Castro, C.A.; Torres-Acosta, J.F.d.J. The Complexity of Goats’ Feeding Behaviour: An Overview of the Research in the Tropical Low Deciduous Forest. Small Rumin. Res. 2024, 231, 107199. [Google Scholar] [CrossRef]
- Ventura-Cordero, J.; González-Pech, P.G.; Torres-Acosta, J.F.J.; Sandoval-Castro, C.A.; Tun-Garrido, J. Sheep and Goat Browsing a Tropical Deciduous Forest during the Rainy Season: Why Does Similar Plant Species Consumption Result in Different Nutrient Intake? Anim. Prod. Sci. 2017, 59, 66–72. [Google Scholar] [CrossRef]
- Belanche, A.; Kingston-Smith, A.H.; Griffith, G.W.; Newbold, C.J. A Multi-Kingdom Study Reveals the Plasticity of the Rumen Microbiota in Response to a Shift From Non-Grazing to Grazing Diets in Sheep. Front. Microbiol. 2019, 10, 122. [Google Scholar] [CrossRef]
- Guo, J.; Li, P.; Liu, S.; Miao, B.; Zeng, B.; Jiang, Y.; Li, L.; Wang, L.; Chen, Y.; Zhang, H. Characterization of the Rumen Microbiota and Volatile Fatty Acid Profiles of Weaned Goat Kids under Shrub-Grassland Grazing and Indoor Feeding. Animals 2020, 10, 176. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.; Wang, X.; Pang, X.; Liu, G. Effects of Supplementary Feeding on the Rumen Morphology and Bacterial Diversity in Lambs. PeerJ 2020, 8, e9353. [Google Scholar] [CrossRef] [PubMed]
- Van Gylswyk, N.O. The Effect of Supplementing a Low–Protein Hay on the Cellulolytic Bacteria in the Rumen of Sheep and on the Digestibility of Cellulose and Hemicellulose. J. Agric. Sci. 1970, 74, 169–180. [Google Scholar] [CrossRef]
- Hespell, R.B.; Wolf, R.; Bothast, R.J. Fermentation of Xylans by Butyrivibrio Fibrisolvens and Other Ruminal Bacteria. Appl. Environ. Microbiol. 1987, 53, 2849–2853. [Google Scholar] [CrossRef] [PubMed]
- Barraza, A.; Montes-Sánchez, J.J.; Caamal-Chan, M.G.; Loera-Muro, A. Characterization of Microbial Communities from Rumen and Large Intestine of Lactating Creole Goats Grazing in Arid Plant Communities. Microbiology 2021, 167, 001092. [Google Scholar] [CrossRef] [PubMed]
- González-Pech, P.G.; Marín-Tun, C.G.; Valladares-González, D.A.; Ventura-Cordero, J.; Ortiz-Ocampo, G.I.; Cámara-Sarmiento, R.; Sandoval-Castro, C.A.; Torres-Acosta, J.F.J. A Protocol of Human Animal Interaction to Habituate Young Sheep and Goats for Behavioural Studies. Behav. Process. 2018, 157, 632–637. [Google Scholar] [CrossRef]
- Agreil, C.; Meuret, M. An Improved Method for Quantifying Intake Rate and Ingestive Behaviour of Ruminants in Diverse and Variable Habitats Using Direct Observation. Small Rumin. Res. 2004, 54, 99–113. [Google Scholar] [CrossRef]
- Gonzalez-Pech, P.G.; Torres-Acosta, J.F.J.; Castro, C.A.S. Adapting a bite coding grid for small ruminants browsing a deciduous tropical forest. Trop. Subtrop. Agroecosyst. 2014, 17, 1. [Google Scholar] [CrossRef]
- Martín, E.; Pérez, E.; Cañón, S.; Rodríguez, J.; Rodriguez, F.d.S. Sonda oro-ruminal experimental como alternativa para la obtención de microorganismos anaeróbicos del rumen. Cienc. Tecnol. Agropecu. 2005, 6, 39–42. [Google Scholar] [CrossRef]
- Barros-Rodríguez, M.A.; Solorio-Sánchez, F.J.; Sandoval-Castro, C.A.; Klieve, A.; Rojas-Herrera, R.A.; Briceño-Poot, E.G.; Ku-Vera, J.C. Rumen Function In Vivo and In Vitro in Sheep Fed Leucaena leucocephala. Trop. Anim. Health Prod. 2015, 47, 757–764. [Google Scholar] [CrossRef]
- Rodríguez, G.F.; Llamas, L.G. Manual de Técnicas de Investigación En Rumiología. In Digestibilidad, Balance de Nutrimentos y Patrones de Fermentación Ruminal; Castellanos, R.A., Llamas, L.G., Shimada, A.S., Sistema de educación continua en producción animal en México, Eds.; PAIEPEME: Mexico City, Mexico, 1990. [Google Scholar]
- Ryan, J.P. Determination of Volatile Fatty Acids and Some Related Compounds in Ovine Rumen Fluid, Urine, and Blood Plasma, by Gas-Liquid Chromatography. Anal. Biochem. 1980, 108, 374–384. [Google Scholar] [CrossRef]
- Winer, B.J. Statistical Principles in Experimental Design, 2nd ed.; Mcgraw-Hill: New York, NY, USA, 1971; pp. 261–285. [Google Scholar]
- Kuehl, R.O. Diseño de Experimentos: Principios Estadísticos de Diseño Y Análisis De Investigación, 2nd ed.; México y América Central: Mexico City, Mexico, 2001; pp. 492–519. [Google Scholar]
- Milliken, G.A.; Johnson, D.E. Analysis of Messy Data, 2nd ed.; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2009; Volume 1, pp. 499–529. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2′s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Kaehler, B.D.; Bokulich, N.A.; McDonald, D.; Knight, R.; Caporaso, J.G.; Huttley, G.A. Species Abundance Information Improves Sequence Taxonomy Classification Accuracy. Nat. Commun. 2019, 10, 4643. [Google Scholar] [CrossRef] [PubMed]
- Jaccard, P. Nouvelles Recherches Sur La Distribution Florale. Bull. Soc. Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Sokal, R.R. The Principles and Practice of Numerical Taxonomy. Taxon 1963, 12, 190–199. [Google Scholar] [CrossRef]
- Fisher, R.A.; Corbet, A.S.; Williams, C.B. The Relation Between the Number of Species and the Number of Individuals in a Random Sample of an Animal Population. J. Anim. Ecol. 1943, 12, 42–58. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative Beta Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Sørensen, T. A Method of Establishing Groups of Equal Amplitude in Plant Sociology Based on Similarity of Species Content and Its Application to Analyses of the Vegetation on Danish Commons; Munksgaard: Copenhagen, Denmark, 1948. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Peddada, S.D. Analysis of Compositions of Microbiomes with Bias Correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Yang, C.; Mai, J.; Cao, X.; Burberry, A.; Cominelli, F.; Zhang, L. Ggpicrust2: An R Package for PICRUSt2 Predicted Functional Profile Analysis and Visualization. Bioinformatics 2023, 39, btad470. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc Database of Metabolic Pathways and Enzymes—A 2019 Update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable Association Discovery in Population-Scale Meta-Omics Studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Duncan, A.J.; Poppi, D.P. Nutritional Ecology of Grazing and Browsing Ruminants. In The Ecology of Browsing and Grazing; Gordon, I.J., Prins, H.H.T., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 89–116. ISBN 978-3-540-72422-3. [Google Scholar]
- Sanon, H.O.; Kaboré-Zoungrana, C.; Ledin, I. Behaviour of Goats, Sheep and Cattle and Their Selection of Browse Species on Natural Pasture in a Sahelian Area. Small Rumin. Res. 2007, 67, 64–74. [Google Scholar] [CrossRef]
- Janis, C. An Evolutionary History of Browsing and Grazing Ungulates. In The Ecology of Browsing and Grazing; Gordon, I.J., Prins, H.H.T., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 21–45. ISBN 978-3-540-72422-3. [Google Scholar]
- Jaimez-Rodríguez, P.R.; González-Pech, P.G.; Ventura-Cordero, J.; Brito, D.R.B.; Costa-Júnior, L.M.; Sandoval-Castro, C.A.; Torres-Acosta, J.F.J. The Worm Burden of Tracer Kids and Lambs Browsing Heterogeneous Vegetation Is Influenced by Strata Harvested and Not Total Dry Matter Intake or Plant Life Form. Trop. Anim. Health Prod. 2019, 51, 2243–2251. [Google Scholar] [CrossRef]
- González-Pech, P.G.; Torres-Acosta, J.F.D.J.; Sandoval-Castro, C.A.; Tun-Garrido, J. Feeding Behavior of Sheep and Goats in a Deciduous Tropical Forest during the Dry Season: The Same Menu Consumed Differently. Small Rumin. Res. 2015, 133, 128–134. [Google Scholar] [CrossRef]
- González-Pech, P.G.; Ventura-Cordero, J.; Ortiz-Ocampo, G.I.; Jaimez-Rodríguez, P.R.; Tun Garrido, J.D.L.C.; Sandoval-Castro, C.A.; Torres-Acosta, J.F.D.J. Plantas Consumidas Por Ovinos y Caprinos En La Selva Baja Caducifolia de Yucatán. In Guía Ilustrada; UADY; Colofón ediciones academicas: Mexico City, Mexico, 2017; pp. 1–64. [Google Scholar]
- Camacho-Escobar, M.A.; Galicia-Jiménez, M.M.; Sánchez-Bernal, E.I.; Ávila-Serrano, N.Y.; López-Garrido, S.J.; Camacho-Escobar, M.A.; Galicia-Jiménez, M.M.; Sánchez-Bernal, E.I.; Ávila-Serrano, N.Y.; López-Garrido, S.J. Producción de metano y bióxido de carbono in vitro de pastos tropicales de la costa de Oaxaca, México. Terra Latinoam. 2020, 38, 425–434. [Google Scholar] [CrossRef]
- López Herrera, M.; Arias Gamboa, M.; Alpizar Naranjo, A.; Castillo Umaña, M. Calidad de fibra y producción de metano en ensilados de leguminosas con fuentes de carbohidratos. Nutr. Anim. Trop. 2021, 15, 1–24. [Google Scholar] [CrossRef]
- Nozière, P.; Ortigues-Marty, I.; Loncke, C.; Sauvant, D. Carbohydrate Quantitative Digestion and Absorption in Ruminants: From Feed Starch and Fibre to Nutrients Available for Tissues. Animal 2010, 4, 1057–1074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lian, Z.; Xu, B.; Shen, Q.; Bao, M.; Huang, Z.; Jiang, H.; Li, W. Gut Microbiome Variation Along A Lifestyle Gradient Reveals Threats Faced by Asian Elephants. Genom. Proteom. Bioinform. 2023, 21, 150–163. [Google Scholar] [CrossRef]
- Guo, N.; Wu, Q.; Shi, F.; Niu, J.; Zhang, T.; Degen, A.A.; Fang, Q.; Ding, L.; Shang, Z.; Zhang, Z.; et al. Seasonal Dynamics of Diet–Gut Microbiota Interaction in Adaptation of Yaks to Life at High Altitude. Npj Biofilms Microbiomes 2021, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353.e21. [Google Scholar] [CrossRef]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia Muciniphila Gen. Nov., Sp. Nov., a Human Intestinal Mucin-Degrading Bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Wei, H.; Ding, L.; Wang, X.; Yan, Q.; Jiang, C.; Hu, C.; Wang, G.; Zhou, Y.; Henkin, Z.; Degen, A.A. Astragalus Root Extract Improved Average Daily Gain, Immunity, Antioxidant Status and Ruminal Microbiota of Early Weaned Yak Calves. J. Sci. Food Agric. 2021, 101, 82–90. [Google Scholar] [CrossRef]
- Ghedini, C.P.; Silva, L.H.P.; Moura, D.C.; Brito, A.F. Supplementing Flaxseed Meal with Sucrose, Flaxseed Oil, or Both: Effects on Milk Enterolactone, Ruminal Microbiota Profile, Production Performance, and Nutrient Utilization in Dairy Cows. J. Dairy Sci. 2024, 107, 6834–6851. [Google Scholar] [CrossRef]
- Qiu, Q.; Zhang, J.; Qu, M.; Li, Y.; Zhao, X.; Ouyang, K. Effect of Energy Provision Strategy on Rumen Fermentation Characteristics, Bacterial Diversity and Community Composition. Bioengineering 2023, 10, 107. [Google Scholar] [CrossRef]
- Wang, Y.; Nan, X.; Zhao, Y.; Jiang, L.; Wang, H.; Zhang, F.; Hua, D.; Liu, J.; Yao, J.; Yang, L.; et al. Dietary Supplementation of Inulin Ameliorates Subclinical Mastitis via Regulation of Rumen Microbial Community and Metabolites in Dairy Cows. Microbiol. Spectr. 2021, 9, e0010521. [Google Scholar] [CrossRef]
- El Otmani, S.; Chebli, Y.; Taminiau, B.; Chentouf, M.; Hornick, J.-L.; Cabaraux, J.-F. Effect of Olive Cake and Cactus Cladodes Incorporation in Goat Kids’ Diet on the Rumen Microbial Community Profile and Meat Fatty Acid Composition. Biology 2021, 10, 1237. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Zhang, Y.; Li, Z.; Guo, M.; Liu, B.; Wang, Z.; Cui, Y.; Wang, C.; Li, D.; Shi, Y. Roughage Quality Determines the Production Performance of Post-Weaned Hu Sheep via Altering Ruminal Fermentation, Morphology, Microbiota, and the Global Methylome Landscape of the Rumen Wall. Front. Microbiomes 2024, 2, 1272625. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Y.; Zhang, M.; Pang, X.; Xu, J.; Kang, C.; Li, M.; Zhang, C.; Zhang, Z.; Zhang, Y.; et al. Structural Changes of Gut Microbiota during Berberine-Mediated Prevention of Obesity and Insulin Resistance in High-Fat Diet-Fed Rats. PLoS ONE 2012, 7, e42529. [Google Scholar] [CrossRef]
- McLoughlin, S.; Spillane, C.; Campion, F.P.; Claffey, N.; Sosa, C.C.; McNicholas, Y.; Smith, P.E.; Diskin, M.G.; Waters, S.M. Breed and Ruminal Fraction Effects on Bacterial and Archaeal Community Composition in Sheep. Sci. Rep. 2023, 13, 3336. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Ji, S.; Duan, C.; Tian, P.; Ju, S.; Yan, H.; Zhang, Y.; Liu, Y. Age-Related Changes in the Ruminal Microbiota and Their Relationship With Rumen Fermentation in Lambs. Front. Microbiol. 2021, 12, 679135. [Google Scholar] [CrossRef]
- Schwab, C.G.; Broderick, G.A. A 100-Year Review: Protein and Amino Acid Nutrition in Dairy Cows. J. Dairy Sci. 2017, 100, 10094–10112. [Google Scholar] [CrossRef]
- Wang, D.; Tang, G.; Wang, Y.; Yu, J.; Chen, L.; Chen, J.; Wu, Y.; Zhang, Y.; Cao, Y.; Yao, J. Rumen Bacterial Cluster Identification and Its Influence on Rumen Metabolites and Growth Performance of Young Goats. Anim. Nutr. 2023, 15, 34–44. [Google Scholar] [CrossRef]
- Li, L.; Qu, J.; Zhu, H.; Liu, Y.; Wu, J.; Shao, G.; Guan, X.; Qu, Y. Effects of Feeding Different Levels of Dietary Corn Silage on Growth Performance, Rumen Fermentation and Bacterial Community of Post-Weaning Dairy Calves. Anim. Biosci. 2024, 37, 261–273. [Google Scholar] [CrossRef]
- Schären, M.; Frahm, J.; Kersten, S.; Meyer, U.; Hummel, J.; Breves, G.; Dänicke, S. Interrelations between the Rumen Microbiota and Production, Behavioral, Rumen Fermentation, Metabolic, and Immunological Attributes of Dairy Cows. J. Dairy Sci. 2018, 101, 4615–4637. [Google Scholar] [CrossRef]
- Khiaosa-ard, R.; Mahmood, M.; Mickdam, E.; Pacífico, C.; Meixner, J.; Traintinger, L.-S. Winery By-Products as a Feed Source with Functional Properties: Dose–Response Effect of Grape Pomace, Grape Seed Meal, and Grape Seed Extract on Rumen Microbial Community and Their Fermentation Activity in RUSITEC. J. Anim. Sci. Biotechnol. 2023, 14, 92. [Google Scholar] [CrossRef]
- Wang, S.; Chai, J.; Zhao, G.; Zhang, N.; Cui, K.; Bi, Y.; Ma, T.; Tu, Y.; Diao, Q. The Temporal Dynamics of Rumen Microbiota in Early Weaned Lambs. Microorganisms 2022, 10, 144. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Liu, L.; Liu, X.; Wang, Y.; Chen, B.; Wu, S.; Yao, J.; Xu, X. A Novel Identified Pseudomonas Aeruginosa, Which Exhibited Nitrate- and Nitrite-Dependent Methane Oxidation Abilities, Could Alleviate the Disadvantages Caused by Nitrate Supplementation in Rumen Fluid Fermentation. Microb. Biotechnol. 2021, 14, 1397–1408. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.S.; Hippie, H. Ruminobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley Online Library: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Zhou, M.; Yan, B.; Wong, J.W.C.; Zhang, Y. Enhanced Volatile Fatty Acids Production from Anaerobic Fermentation of Food Waste: A Mini-Review Focusing on Acidogenic Metabolic Pathways. Bioresour. Technol. 2018, 248, 68–78. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Paster, B.J.; Fontaine, S.L.; Rood, J.I. Transfer of Kingella Indologenes (Snell and Lapage 1976) to the Genus Suttonella Gen. Nov. as Suttonella Indologenes Comb. Nov.; Transfer of Bacteroides Nodosus (Beveridge 1941) to the Genus Dichelobacter Gen. Nov. as Dichelobacter Nodosus Comb. Nov.; and Assignment of the Genera Cardiobacterium, Dichelobacter, and Suttonella to Cardiobacteriaceae Fam. Nov. in the Gamma Division of Proteobacteria on the Basis of 16S rRNA Sequence Comparisons. Int. J. Syst. Evol. Microbiol. 1990, 40, 426–433. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Li, C.; Tian, H.; Weng, X.; Lin, C.; Zhang, D.; Zhao, Y.; Li, X.; Cheng, J.; et al. Rumen Microbiome and Fat Deposition in Sheep: Insights from a Bidirectional Mendelian Randomization Study. Npj Biofilms Microbiomes 2024, 10, 129. [Google Scholar] [CrossRef]
- Ma, T.; Wu, W.; Tu, Y.; Zhang, N.; Diao, Q. Resveratrol Affects in Vitro Rumen Fermentation, Methane Production and Prokaryotic Community Composition in a Time- and Diet-Specific Manner. Microb. Biotechnol. 2020, 13, 1118–1131. [Google Scholar] [CrossRef]
- Dai, H.; Huang, Q.; Li, S.; Du, D.; Yu, W.; Guo, J.; Zhao, Z.; Yu, X.; Ma, F.; Sun, P. Effect of Dietary Benzoic Acid Supplementation on Growth Performance, Rumen Fermentation, and Rumen Microbiota in Weaned Holstein Dairy Calves. Animals 2024, 14, 2823. [Google Scholar] [CrossRef]
- Li, C.; Chen, N.; Zhang, X.; Shahzad, K.; Qi, R.; Zhang, Z.; Lu, Z.; Lu, Y.; Yu, X.; Zafar, M.H.; et al. Mixed Silage with Chinese Cabbage Waste Enhances Antioxidant Ability by Increasing Ascorbate and Aldarate Metabolism through Rumen Prevotellaceae UCG-004 in Hu Sheep. Front. Microbiol. 2022, 13, 978940. [Google Scholar] [CrossRef]
- Shen, J.; Li, Z.; Yu, Z.; Zhu, W. Effects of Dietary Replacement of Soybean Meal with Dried Distillers Grains with Solubles on the Microbiota Occupying Different Ecological Niches in the Rumen of Growing Hu Lambs. J. Anim. Sci. Biotechnol. 2020, 11, 93. [Google Scholar] [CrossRef]
- Zhang, H.; Elolimy, A.A.; Akbar, H.; Thanh, L.P.; Yang, Z.; Loor, J.J. Association of Residual Feed Intake with Peripartal Ruminal Microbiome and Milk Fatty Acid Composition during Early Lactation in Holstein Dairy Cows. J. Dairy Sci. 2022, 105, 4971–4986. [Google Scholar] [CrossRef]
- Hao, Y.; Ouyang, T.; Wang, W.; Wang, Y.; Cao, Z.; Yang, H.; Guan, L.L.; Li, S. Competitive Analysis of Rumen and Hindgut Microbiota Composition and Fermentation Function in Diarrheic and Non-Diarrheic Postpartum Dairy Cows. Microorganisms 2024, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Cusa, E.; Obradors, N.; Baldomà, L.; Badía, J.; Aguilar, J. Genetic Analysis of a Chromosomal Region Containing Genes Required for Assimilation of Allantoin Nitrogen and Linked Glyoxylate Metabolism in Escherichia coli. J. Bacteriol. 1999, 181, 7479–7484. [Google Scholar] [CrossRef] [PubMed]
- Rabin, R.; Reeves, H.C.; Wegener, W.S.; Megraw, R.E.; Ajl, S.J. Glyoxylate in Fatty-Acid Metabolism. Science 1965, 150, 1548–1558. [Google Scholar] [CrossRef]
- Li, Y.; Peng, R.; Kunz, C.; Wang, K.; Terranova, M.; Zhang, Y.; Macsai, M.; Frossard, E.; Niu, M. Hydroponic Fodders as Alternative Feeds for Ruminants to Reduce Ruminal Methane Emissions: An in Vitro Study. J. Dairy Sci. 2024, 107, 10932–10944. [Google Scholar] [CrossRef]
- Dessalegn, G.; Tesfay, G. The Role of Bacteria in Nitrogen Metabolism in the Rumen with Emphasis of Cattle. Res. J. Agric. Environ. Manag. 2015, 4, 282–290. [Google Scholar]
- Russell, J.B.; O’Connor, J.D.; Fox, D.G.; Van Soest, P.J.; Sniffen, C.J. A Net Carbohydrate and Protein System for Evaluating Cattle Diets: I. Ruminal Fermentation. J. Anim. Sci. 1992, 70, 3551–3561. [Google Scholar] [CrossRef]
- Baldwin, J.E.; Krebs, H. The Evolution of Metabolic Cycles. Nature 1981, 291, 381–382. [Google Scholar] [CrossRef]
- Kornberg, H.L.; Krebs, H.A. Synthesis of Cell Constituents from C2-Units by a Modified Tricarboxylic Acid Cycle. Nature 1957, 179, 988–991. [Google Scholar] [CrossRef]
- Liu, Z.; Jiang, A.; Lv, X.; Zhou, C.; Tan, Z. Metabolic Changes in Serum and Milk of Holstein Cows in Their First to Fourth Parity Revealed by Biochemical Analysis and Untargeted Metabolomics. Anim. Open Access J. 2024, 14, 407. [Google Scholar] [CrossRef]
- Eastmond, P.J.; Graham, I.A. Re-Examining the Role of the Glyoxylate Cycle in Oilseeds. Trends Plant Sci. 2001, 6, 72–78. [Google Scholar] [CrossRef]
- Asanuma, N.; Iwamoto, M.; Hino, T. The Production of Formate, a Substrate for Methanogenesis, from Compounds Related with the Glyoxylate Cycle by Mixed Ruminal Microbes. Nihon Chikusan Gakkaiho 1999, 70, 67–73. [Google Scholar] [CrossRef]
- Zrenner, R.; Stitt, M.; Sonnewald, U.; Boldt, R. Pyrimidine and Purine Biosynthesis and Degradation in Plants. Annu. Rev. Plant Biol. 2006, 57, 805–836. [Google Scholar] [CrossRef] [PubMed]
- Kheirandish, P.; Petri, R.M.; Sener-Aydemir, A.; Schwartz-Zimmermann, H.E.; Berthiller, F.; Zebeli, Q.; Pacífico, C. Characterization of Microbial Intolerances and Ruminal Dysbiosis towards Different Dietary Carbohydrate Sources Using an In Vitro Model. J. Appl. Microbiol. 2022, 133, 458–476. [Google Scholar] [CrossRef] [PubMed]



| % | DM [%] | CP [%] | EE [%] | aNDF [%] | ADF [%] | Ash [%] |
|---|---|---|---|---|---|---|
| Grass | 29.35 | 6.97 | 63.46 | 37.74 | 37.74 | 5.35 |
| Concentrate feed | 91.2 | 17.87 | 11.49 | 4.36 | 4.36 | 4.25 |
| Group | DM | p | CP | p | ADF | p | aNDF | p | |
|---|---|---|---|---|---|---|---|---|---|
| Stabilization | CG | 620.50 ± 38.86 A | 0.3123 | 68.12 ± 5.18 A | 0.5758 | 165.69 ± 12.37 A | 0.6650 | 292.26 ± 21.82 A | 0.6650 |
| EG | 616.47 ± 34.93 A | 67.75 ± 2.12 A | 164.32 ± 6.42 A | 289.85 ± 11.32 A | |||||
| DPS14 | CG | 639.52 ± 21.81 A | 0.11233 | 69.25 ± 2.28 A | 0.4974 | 168.12 ± 4.63 A | 0.0303 | 296.54 ± 8.17 A | 0.0303 |
| EG | 736.20 ± 31.96 A | 70.37 ± 3.29 A | 246.78 ± 1.61 B | 414.51 ± 2.72 B | |||||
| DPS44 | CG | 692.02 ± 22.66 A | 0.1123 | 135.66 ± 3.08 A | 0.0303 | 150.81 ± 7.62 A | 0.0303 | 312.98 ± 9.49 A | 0.0303 |
| EG | 746.20 ± 21.93 A | 135.16 ± 1.25 B | 245.55 ± 0.89 B | 412.44 ± 1.50 B |
| Productive Parameters | Group | Stabilization | DPS14 | DPS44 | EE | P Group | P Time | P Interaction | Orthogonal Contrast | |
|---|---|---|---|---|---|---|---|---|---|---|
| Linear | Quadratic | |||||||||
| Total VFA (mmol/100 mL) | CG | 55.56 ± 12.32 A–A | 56.57 ± 12.18 A–A | 45.61 ± 4.82 A–A | 3.3705 | 0.6472 | 0.3166 | 0.7782 | 0.4119 | 0.5653 |
| EG | 52.68 ± 2.83 A–A | 65.42 ± 9.75 A–A | 50.05 ± 1.71 A–A | 0.8256 | 1906 | |||||
| Rumen VFA (molar %) | ||||||||||
| Acetate | CG | 63.98 ± 2.58 A–A | 62.50 ± 1.67 A–A | 61.30 ± 1.98 A–A | 0.8212 | 0.1146 | 0.6442 | 0.5461 | 0.3644 | 0.9575 |
| EG | 65.84 ± 2.31 A–A | 63.47 ± 1.82 A–A | 66.56 ± 0.68 A–A | 0.8030 | 0.2894 | |||||
| Propionate | CG | 18.71 ± 2.27 A–A | 16.93 ± 2.08 A–A | 19.99 ± 1.50 A–A | 0.7722 | 0.6696 | 0.3924 | 0.1075 | 0.6400 | 0.3167 |
| EG | 14.74 ± 1.86 A–A | 21.35 ± 1.71A–A | 17.93 ± 0.60 A–A | 0.2561 | 0.0513 | |||||
| Butyrate | CG | 15.24 ± 1.41 A–A | 17.72 ± 1.27 A–A | 16.33 ± 1.38 A–A | 0.3623 | 0.14057 | 0.3294 | 0.0130 | 0.4045 | 0.1008 |
| EG | 16.50 ± 0.89 A–A | 13.05 ± 1.57 AB–B | 12.68 ± 0.27 B–A | 0.0102 | 0.1800 | |||||
| Iso-butyrate | CG | 0.11 ± 0.11 A–A | 0.05 ± 0.05 A–A | 0.34 ± 0.11 A–A | 0.0640 | 0.0013 | 0.1645 | 0.8713 | 0.3269 | 0.3955 |
| EG | 0.42 ± 0.18 A–A | 0.48 ± 0.02 A–A | 0.76 ± 0.22 A–A | 0.1423 | 0.5668 | |||||
| Valerate | CG | 2.69 ± 0.56 A–B | 1.83 ± 0.33 A–A | 1.37 ± 0.44 B–A | 0.1275 | 0.0557 | 0.1854 | 0.0907 | 0.3239 | 0.0149 |
| EG | 1.30 ± 0.38 A–B | 0.95 ± 0.10 A–B | 1.04 ± 0.03 A–A | 0.5707 | 0.5807 | |||||
| Iso-valerate | CG | 0.12 ± 0.12 A–A | 0.16 ± 0.05 A–A | 0.49 ± 0.17 A–B | 0.0497 | 0.0004 | 0.0580 | 0.0729 | 0.0577 | 0.3518 |
| EG | 1.20 ± 0.18 AB–B | 0.70 ± 0.07 AB–B | 1.01 ± 0.03 B–B | 0.3021 | 0.0189 | |||||
| Acetate: propionate | CG | 3.62 ± 0.58 A–A | 3.95 ± 0.71 A–A | 3.13 ± 0.31 A–A | 0.2295 | 0.5453 | 0.3433 | 0.2016 | 0.5525 | 0.4203 |
| EG | 4.79 ± 0.88 A–A | 3.03 ± 0.27 A–A | 3.73 ± 0.15 A–A | 0.2060 | 0.1010 | |||||
| Jaccard | Sokal-Sneath | Yule Y | UniFrac-W | Bray-Curtis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ps-F | p | Ps-F | p | Ps-F | p | Ps-F | p | Ps-F | p | ||
| PERMANOVA | 1.33 | 0.004 | 1.17 | 0.003 | 4.24 | 0.004 | 2.63 | 0.001 | 1.74 | 0.001 | |
| Pairwise comparison | |||||||||||
| CG-s | CG-DPS44 | 1.05 | 0.32 | 1.01 | 0.35 | 2.55 | 0.22 | 2.45 | 0.12 | 1.25 | 0.36 |
| CG-s | EG-s | 1.01 | 0.42 | 1.01 | 0.43 | 0.93 | 0.36 | 0.75 | 0.51 | 0.97 | 0.47 |
| CG-DPS44 | EG-DPS44 | 1.39 | 0.02 | 1.21 | 0.02 | 4.27 | 0.02 | 3.89 | 0.09 | 2.02 | 0.03 |
| EG-s | EG-DPS44 | 1.64 | 0.03 | 1.34 | 0.03 | 6.53 | 0.02 | 4.10 | 0.02 | 2.61 | 0.03 |
| Taxonomy | EG-s | EG-DPS44 | W | p | q | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 50 | 75 | 100 | 25 | 50 | 75 | 100 | |||||
| Phylum | Verrucomicrobiota | 9.5 | 16 | 29.75 | 53 | 569.25 | 1093.5 | 1482.5 | 1571 | 4 | 3.08 × 10−05 | 4.3 × 10−04 |
| Actinobacteriota | 52.5 | 121.5 | 216.25 | 307 | 13.75 | 18.5 | 22.5 | 24 | 3 | 7.10 × 10−04 | 4.9 × 10−03 | |
| Class | Kiritimatiellae | 1 | 3 | 16 | 49 | 562.25 | 1087 | 1476 | 1569 | 4 | 3.08 × 10−05 | 3.0 × 10−04 |
| Alphaproteobacteria | 1 | 1 | 1 | 1 | 6 | 9.5 | 35.25 | 102 | 4 | 4.24 × 10−04 | 2.1 × 10−04 | |
| Actinobacteria | 30 | 105 | 184.5 | 198 | 1 | 3 | 6 | 9 | 4 | 8.69 × 10−06 | 1.7 × 10−04 | |
| Negativicutes | 483 | 589.5 | 671 | 731 | 184.25 | 211.5 | 233.5 | 247 | 4 | 1.02 × 10−04 | 6.7 × 10−04 | |
| Bacilli | 232.5 | 1113 | 2097.75 | 2457 | 33 | 52.5 | 81.5 | 116 | 3 | 8.99 × 10−04 | 3.5 × 10−043 | |
| Order | WCHB1-41 | 1 | 3 | 16 | 49 | 562.25 | 1087 | 1476 | 1569 | 4 | 3.10 × 10−05 | 6.35 × 10−04 |
| Christensenellales | 230.5 | 284 | 419.5 | 739 | 744.75 | 856.5 | 1155.2 | 1753 | 3 | 6.52 × 10−03 | 4.4 × 10−02 | |
| Rhodospirillales | 1 | 1 | 1 | 1 | 6 | 8 | 33 | 102 | 3 | 6.92 × 10−04 | 7.0 × 10−03 | |
| Erysipelotrichales | 220.75 | 1091 | 2069.5 | 2443 | 22.75 | 34 | 54.75 | 90 | 4 | 2.85 × 10−04 | 3.8 × 10−03 | |
| Bifidobacteriales | 30 | 105 | 184 | 196 | 1 | 3 | 6 | 9 | 4 | 8.70 × 10−06 | 3.57 × 10−04 | |
| Veillonellales-Selenomonadales | 258.75 | 353 | 475.5 | 588 | 135.25 | 149.5 | 164 | 194 | 3 | 3.28 × 10−03 | 2.6 × 10−02 | |
| Family | Bacteroidales RF16_group | 39.5 | 45 | 46.25 | 50 | 592 | 768 | 987 | 1191 | 10 | 8.79 × 10−22 | 5.71 × 10−20 |
| WCHB1-41 | 1 | 3 | 16 | 49 | 562.25 | 1087 | 1476 | 1569 | 4 | 3.10 × 10−05 | 6.71 × 10−04 | |
| Erysipelatoclostridiaceae | 219.5 | 1089.5 | 2061.7 | 2424 | 12.75 | 20.5 | 38.25 | 78 | 4 | 3.67 × 10−04 | 5.96 × 10−03 | |
| Bifidobacteriaceae | 30 | 105 | 184 | 196 | 1 | 3 | 6 | 9 | 4 | 8.70 × 10−06 | 2.83 × 10−04 | |
| Muribaculaceae | 236 | 565 | 875.75 | 899 | 66.25 | 73.5 | 93.75 | 144 | 3 | 1.25 × 10−03 | 1.35 × 10−02 | |
| Genus | Bacteroidales RF16_group | 39.5 | 45 | 46.25 | 50 | 592 | 768 | 987 | 1191 | 10 | 8.79 × 10−22 | 1.21 × 10−19 |
| Lachnospiraceae ND3007_group | 1 | 4 | 8.25 | 12 | 75 | 90 | 103 | 112 | 5 | 2.61 × 10−07 | 1.20 × 10−05 | |
| WCHB1-41 | 1 | 3 | 16 | 49 | 562.25 | 1087 | 1476 | 1569 | 4 | 3.10 × 10−05 | 6.64 × 10−04 | |
| Roseburia | 1 | 1 | 2 | 5 | 7.75 | 8.5 | 10.25 | 14 | 4 | 3.37 × 10−05 | 6.64 × 10−04 | |
| Rikenellaceae RC9_gut_group | 76.5 | 108 | 144 | 183 | 270.75 | 354.5 | 414.75 | 426 | 3 | 2.4 × 10−03 | 2.59 × 10−02 | |
| Sharpea | 198.25 | 217.5 | 724 | 2239 | 1 | 3 | 5.75 | 8 | 6 | 8.65 × 10−10 | 5.97 × 10−08 | |
| Prevotella_7 | 28 | 33.5 | 263.25 | 948 | 1 | 1.5 | 3.25 | 7 | 4 | 4.41 × 10−04 | 7.61 × 10−03 | |
| Bifidobacterium | 30 | 105 | 184 | 196 | 1 | 3 | 6 | 9 | 4 | 8.7 × 10−06 | 3.00 × 10−04 | |
| FD2005 | 12 | 18 | 37.75 | 79 | 1 | 1 | 2.25 | 6 | 4 | 3.02 × 10−05 | 6.64 × 10−04 | |
| Erysipelotrichaceae UCG-002 | 17 | 97 | 553.25 | 1691 | 1 | 1 | 1.5 | 3 | 3 | 7.97 × 10−04 | 1.09 × 10−02 | |
| Muribaculaceae | 236 | 565 | 875.75 | 899 | 66.25 | 73.5 | 93.75 | 144 | 3 | 1.2 × 10−03 | 1.43 × 10−02 | |
| Anaerovibrio | 22.75 | 36.5 | 60 | 96 | 1 | 4 | 8.5 | 13 | 3 | 1.2 × 10−03 | 1.43 × 10−02 | |
| [Eubacterium]ruminantium_group | 8.5 | 11 | 12.75 | 18 | 1 | 1 | 1 | 1 | 3 | 4.7 × 10−03 | 4.71 × 10−02 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ávila-Cervantes, R.; González-Pech, P.; Sandoval-Castro, C.; Torres-Acosta, F.; Ramos-Zapata, J.; Galicia-Jiménez, M.; Pacheco-Arjona, R. Effects of Grazing in a Low Deciduous Forest on Rumen Microbiota and Volatile Fatty Acid Production in Lambs. Animals 2025, 15, 1565. https://doi.org/10.3390/ani15111565
Ávila-Cervantes R, González-Pech P, Sandoval-Castro C, Torres-Acosta F, Ramos-Zapata J, Galicia-Jiménez M, Pacheco-Arjona R. Effects of Grazing in a Low Deciduous Forest on Rumen Microbiota and Volatile Fatty Acid Production in Lambs. Animals. 2025; 15(11):1565. https://doi.org/10.3390/ani15111565
Chicago/Turabian StyleÁvila-Cervantes, Raúl, Pedro González-Pech, Carlos Sandoval-Castro, Felipe Torres-Acosta, José Ramos-Zapata, Mónica Galicia-Jiménez, and Ramón Pacheco-Arjona. 2025. "Effects of Grazing in a Low Deciduous Forest on Rumen Microbiota and Volatile Fatty Acid Production in Lambs" Animals 15, no. 11: 1565. https://doi.org/10.3390/ani15111565
APA StyleÁvila-Cervantes, R., González-Pech, P., Sandoval-Castro, C., Torres-Acosta, F., Ramos-Zapata, J., Galicia-Jiménez, M., & Pacheco-Arjona, R. (2025). Effects of Grazing in a Low Deciduous Forest on Rumen Microbiota and Volatile Fatty Acid Production in Lambs. Animals, 15(11), 1565. https://doi.org/10.3390/ani15111565







