Histone Methyltransferase SETD2 Is Required for Porcine Early Embryonic Development
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vitro Maturation of Oocytes
2.2. Parthenogenetic Activation (PA)
2.3. In Vitro Fertilization (IVF)
2.4. Real-Time Quantitative Polymerase Chain Reaction (qPCR)
2.5. Immunofluorescence
2.6. Microinjection
2.7. TdT (Terminal Deoxynucleotidyl Transferase)-Mediated dUDP Nick-End (TUNEL) Staining
2.8. Statistical Analysis
3. Results
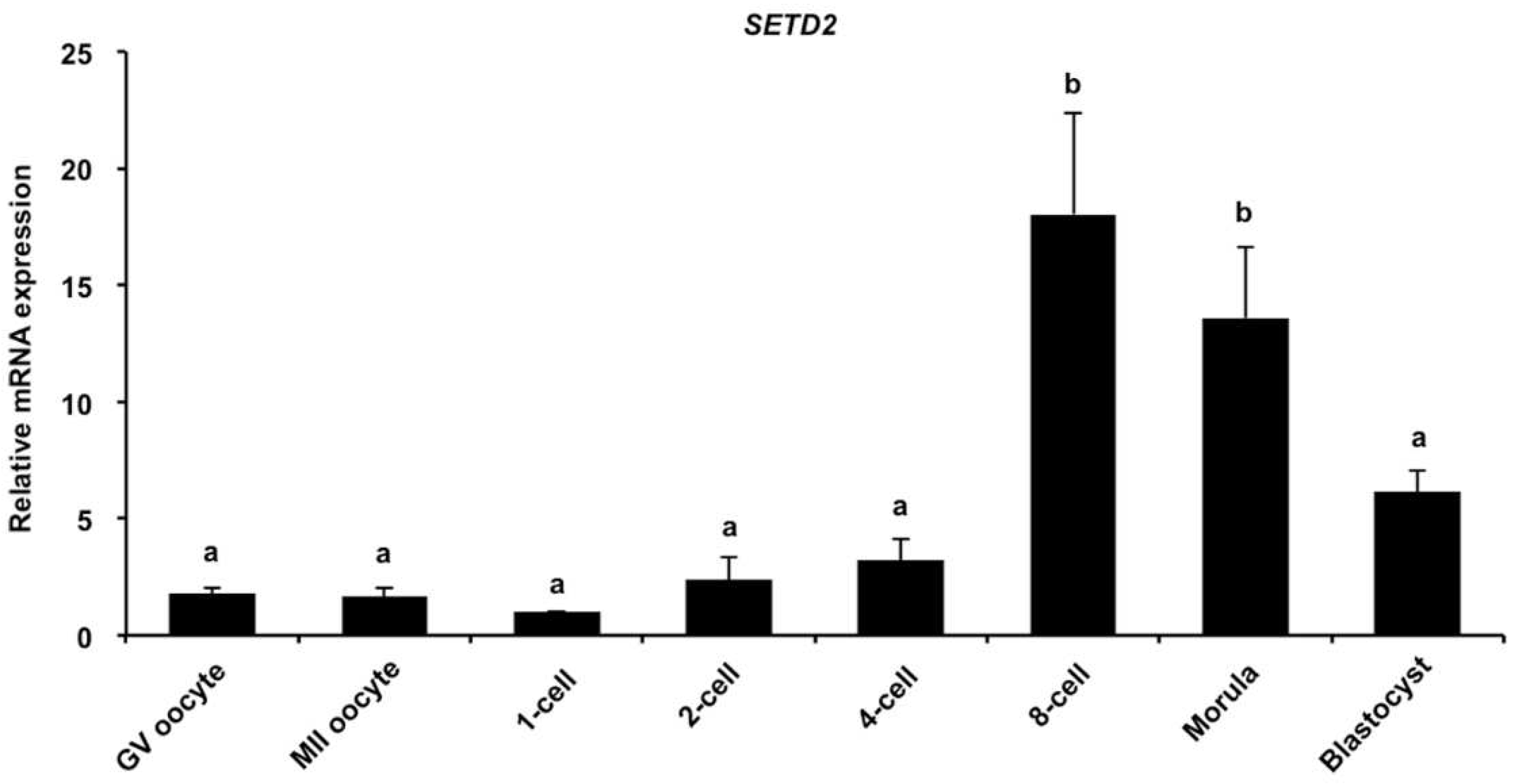
3.1. Dynamic Expression of SETD2 mRNA during Early Embryonic Development
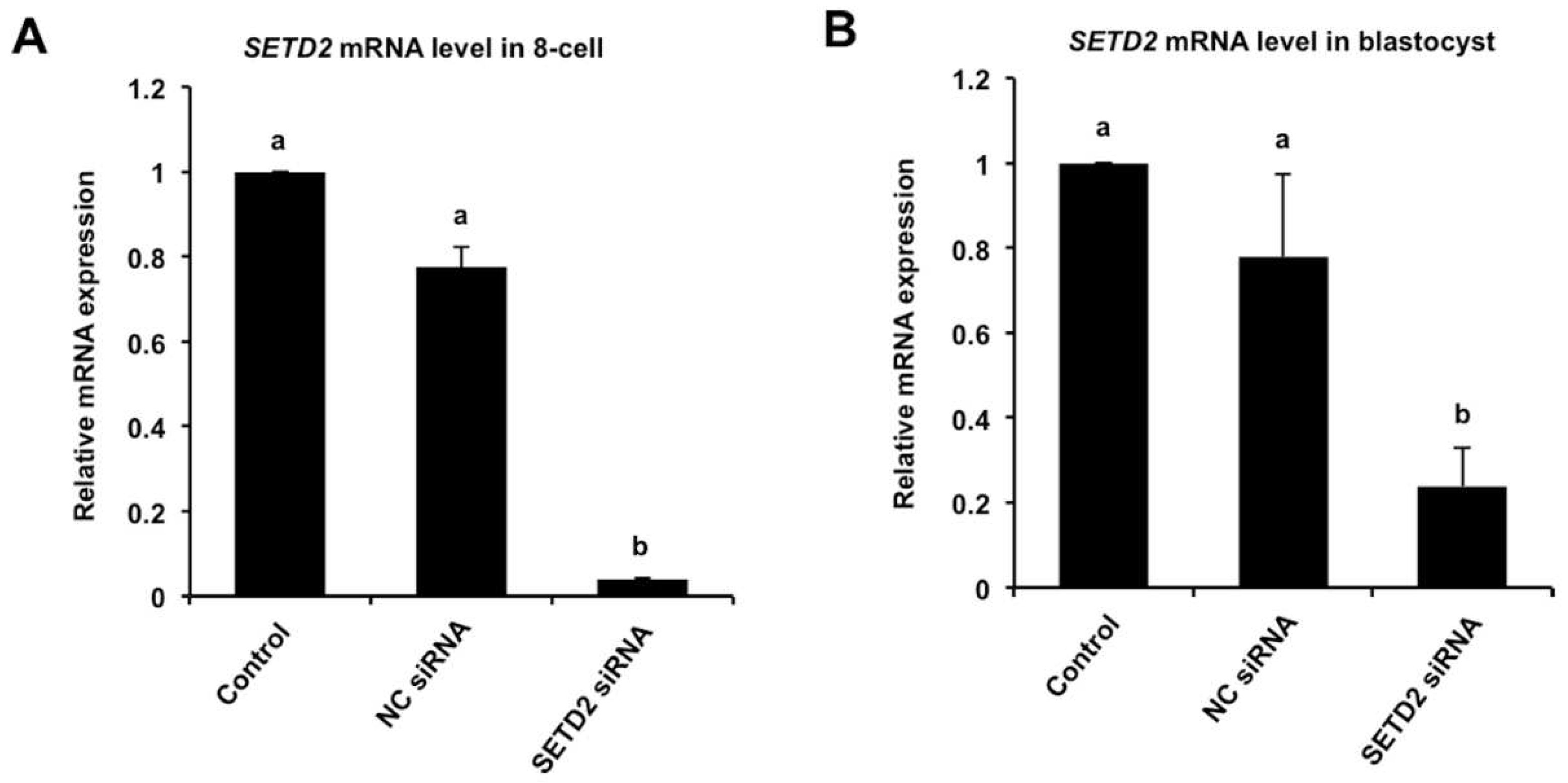
3.2. RNAi-Mediated Efficient Knockdown of SETD2 mRNA in Early Embryos
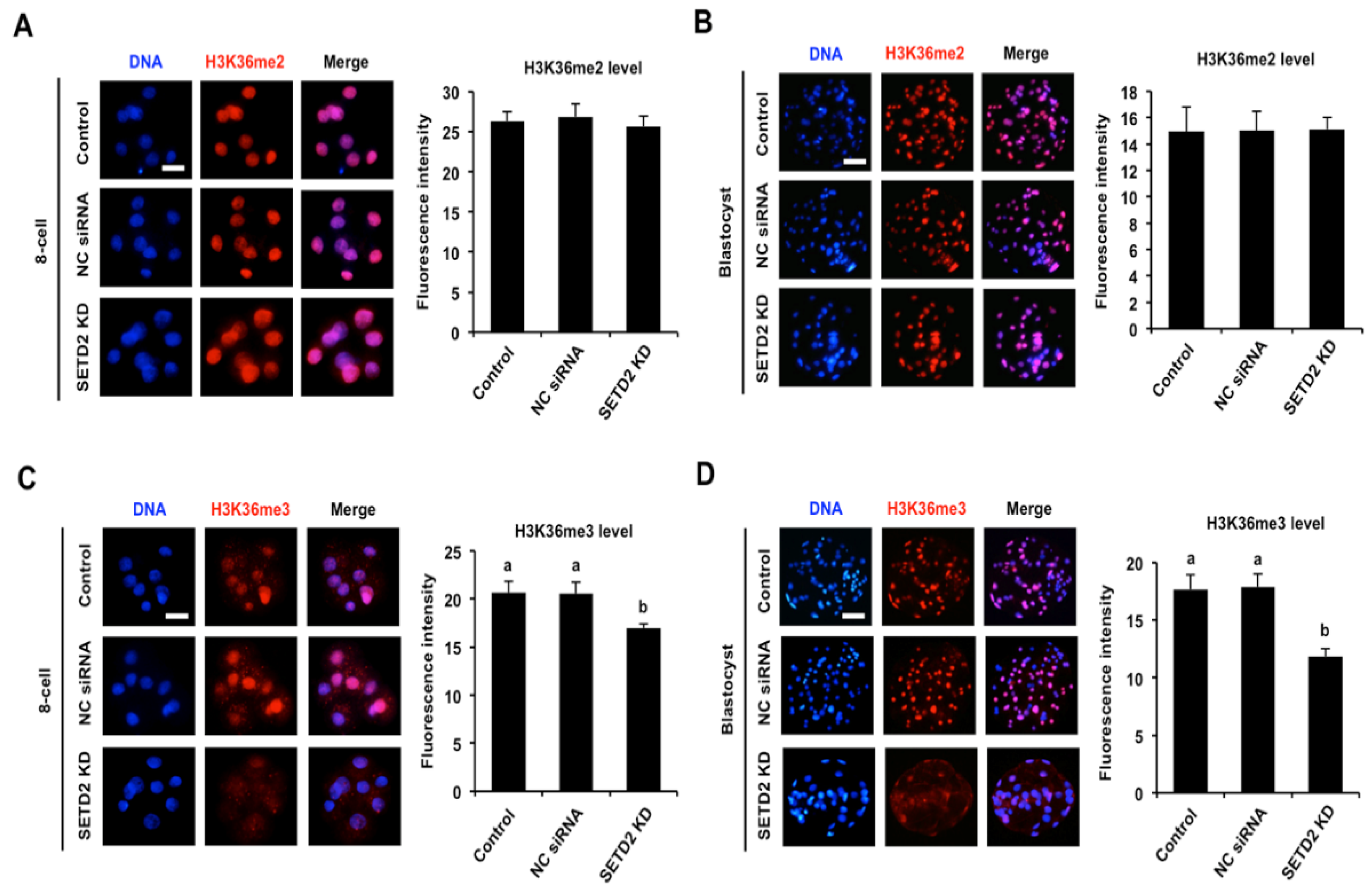
3.3. SETD2 Knockdown Efficiently Reduces H3K36me3 Levels in Early Embryos
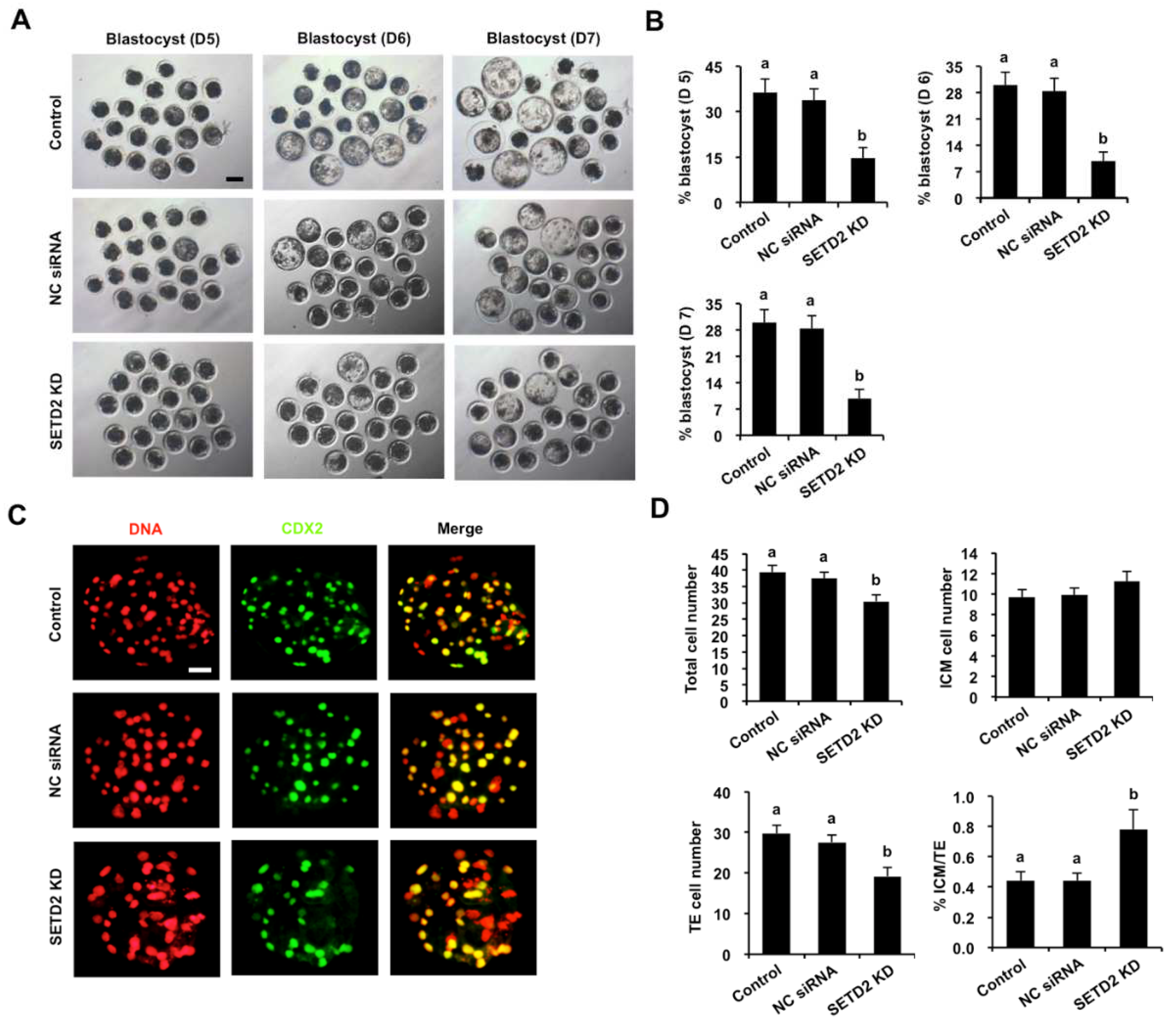
3.4. SETD2 Knockdown Blocks Blastocyst Development and Perturbs Lineage Allocation
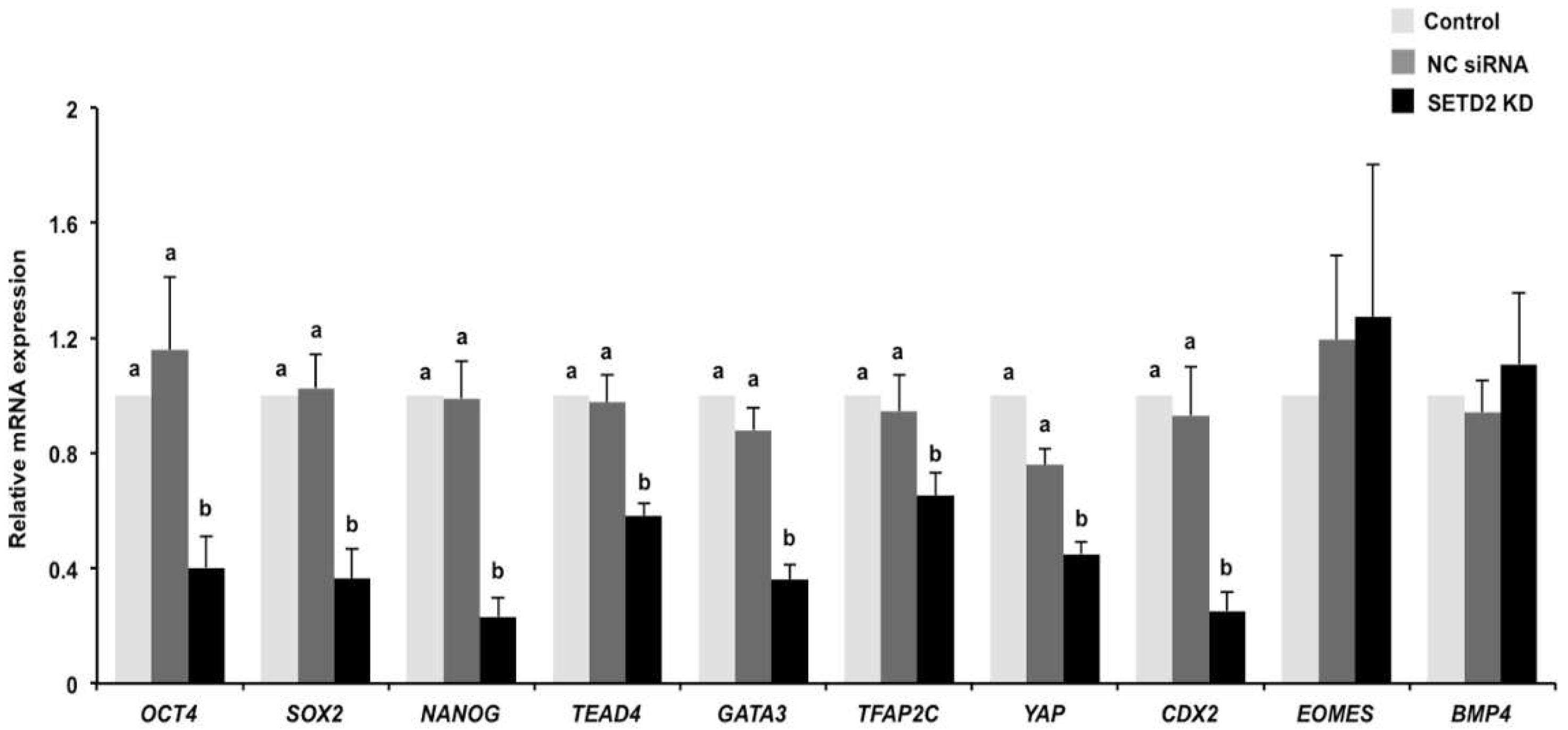
3.5. SETD2 Knockdown Perturbs the Expression of Genes Important for the First Cell Differentiation
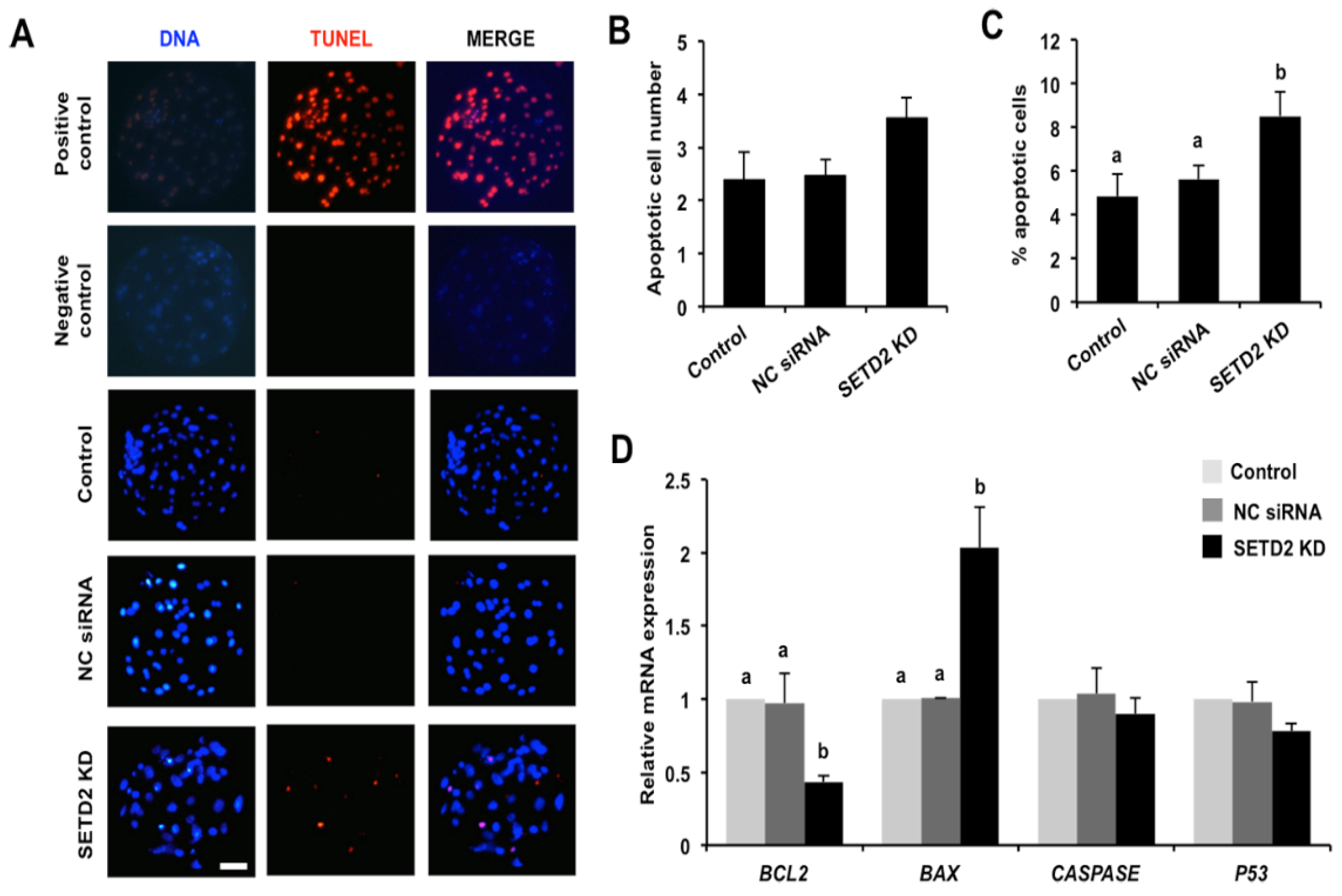
3.6. SETD2 Knockdown Elevates Apoptosis and Alters the Expression of Pro-Apoptotic and Anti-Apoptotic Genes in Blastocysts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marcho, C.; Cui, W.; Mager, J. Epigenetic dynamics during preimplantation development. Reproduction 2015, 150, R109–R120. [Google Scholar] [CrossRef]
- Burton, A.; Torres-Padilla, M.E. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 723–734. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, C.; Zhang, Y. Epigenetic regulation of mouse preimplantation embryo development. Curr. Opin. Genet. Dev. 2020, 64, 13–20. [Google Scholar] [CrossRef]
- Simmet, K.; Wolf, E.; Zakhartchenko, V. Manipulating the Epigenome in Nuclear Transfer Cloning: Where, When and How. Int. J. Mol. Sci. 2020, 22, 236. [Google Scholar] [CrossRef]
- Santos, F.; Dean, W. Epigenetic reprogramming during early development in mammals. Reproduction 2004, 127, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Ng, R.K.; Dean, W.; Dawson, C.; Lucifero, D.; Madeja, Z.; Reik, W.; Hemberger, M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 2008, 10, 1280–1290. [Google Scholar] [CrossRef] [PubMed]
- Torres-Padilla, M.E.; Parfitt, D.E.; Kouzarides, T.; Zernicka-Goetz, M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 2007, 445, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Sun, X.J.; Zhang, Y.L.; Kuang, Y.; Hu, C.Q.; Wu, W.L.; Shen, S.H.; Du, T.T.; Li, H.; He, F.; et al. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc. Natl. Acad. Sci. USA 2010, 107, 2956–2961. [Google Scholar] [CrossRef]
- Edmunds, J.W.; Mahadevan, L.C.; Clayton, A.L. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008, 27, 406–420. [Google Scholar] [CrossRef]
- Shirane, K.; Miura, F.; Ito, T.; Lorincz, M.C. NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat. Genet. 2020, 52, 1088–1098. [Google Scholar] [CrossRef]
- DiFiore, J.V.; Ptacek, T.S.; Wang, Y.; Li, B.; Simon, J.M.; Strahl, B.D. Unique and Shared Roles for Histone H3K36 Methylation States in Transcription Regulation Functions. Cell Rep. 2020, 31, 107751. [Google Scholar] [CrossRef]
- Krogan, N.J.; Kim, M.; Tong, A.; Golshani, A.; Cagney, G.; Canadien, V.; Richards, D.P.; Beattie, B.K.; Emili, A.; Boone, C.; et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell Biol. 2003, 23, 4207–4218. [Google Scholar] [CrossRef]
- Li, F.; Mao, G.; Tong, D.; Huang, J.; Gu, L.; Yang, W.; Li, G.M. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013, 153, 590–600. [Google Scholar] [CrossRef]
- Morselli, M.; Pastor, W.A.; Montanini, B.; Nee, K.; Ferrari, R.; Fu, K.; Bonora, G.; Rubbi, L.; Clark, A.T.; Ottonello, S.; et al. In Vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. eLife 2015, 4, e06205. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Zhou, K.; Wu, T.; Zhao, B.S.; Sun, M.; Chen, Z.; Deng, X.; Xiao, G.; Auer, F.; et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. [Google Scholar] [CrossRef]
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by histone modifications. Science 2010, 327, 996–1000. [Google Scholar] [CrossRef]
- Liu, D.J.; Zhang, F.; Chen, Y.; Jin, Y.; Zhang, Y.L.; Chen, S.B.; Xie, Y.Y.; Huang, Q.H.; Zhao, W.L.; Wang, L.; et al. Setd2 knockout zebrafish is viable and fertile: Differential and developmental stress-related requirements for Setd2 and histone H3K36 trimethylation in different vertebrate animals. Cell Discov. 2020, 6, 72. [Google Scholar] [CrossRef]
- Mukai, M.; Hira, S.; Nakamura, K.; Nakamura, S.; Kimura, H.; Sato, M.; Kobayashi, S. H3K36 Trimethylation-Mediated Epigenetic Regulation is Activated by Bam and Promotes Germ Cell Differentiation During Early Oogenesis in Drosophila. Biol. Open 2015, 4, 119–124. [Google Scholar] [CrossRef]
- Xu, Q.; Xiang, Y.; Wang, Q.; Wang, L.; Brind’Amour, J.; Bogutz, A.B.; Zhang, Y.; Zhang, B.; Yu, G.; Xia, W.; et al. SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat. Genet. 2019, 51, 844–856. [Google Scholar] [CrossRef]
- Zuo, X.; Rong, B.; Li, L.; Lv, R.; Lan, F.; Tong, M.H. The histone methyltransferase SETD2 is required for expression of acrosin-binding protein 1 and protamines and essential for spermiogenesis in mice. J. Biol. Chem. 2018, 293, 9188–9197. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Huang, Z.; Gu, L. SETD2 reduction adversely affects the development of mouse early embryos. J. Cell Biochem. 2020, 121, 797–803. [Google Scholar] [CrossRef]
- Wu, H.; Gordon, J.A.; Whitfield, T.W.; Tai, P.W.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Chromatin dynamics regulate mesenchymal stem cell lineage specification and differentiation to osteogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 438–449. [Google Scholar] [CrossRef]
- Abed, J.A.; Jones, R.S. H3K36me3 key to Polycomb-mediated gene silencing in lineage specification. Nat. Struct. Mol. Biol. 2012, 19, 1214–1215. [Google Scholar] [CrossRef]
- Diao, Y.F.; Lin, T.; Li, X.; Oqani, R.K.; Lee, J.E.; Kim, S.Y.; Jin, D.I. Dynamic changes of SETD2, a histone H3K36 methyltransferase, in porcine oocytes, IVF and SCNT embryos. PLoS ONE. 2018, 13, e0191816. [Google Scholar] [CrossRef]
- Li, C.; Diao, F.; Qiu, D.; Jiang, M.; Li, X.; Han, L.; Li, L.; Hou, X.; Ge, J.; Ou, X.; et al. Histone methyltransferase SETD2 is required for meiotic maturation in mouse oocyte. J. Cell Physiol. 2018, 234, 661–668. [Google Scholar] [CrossRef]
- Dahl, J.A.; Jung, I.; Aanes, H.; Greggains, G.D.; Manaf, A.; Lerdrup, M.; Li, G.; Kuan, S.; Li, B.; Lee, A.Y.; et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016, 537, 548–552. [Google Scholar] [CrossRef]
- Morey, L.; Santanach, A.; Di Croce, L. Pluripotency and Epigenetic Factors in Mouse Embryonic Stem Cell Fate Regulation. Mol. Cell Biol. 2015, 35, 2716–2728. [Google Scholar] [CrossRef]
- Boskovic, A.; Bender, A.; Gall, L.; Ziegler-Birling, C.; Beaujean, N.; Torres-Padilla, M.E. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of H2A.Z acetylation and H3K36 trimethylation from embryonic genome activation. Epigenetics 2012, 7, 747–757. [Google Scholar] [CrossRef]
- Alberio, R. Regulation of Cell Fate Decisions in Early Mammalian Embryos. Annu. Rev. Anim. Biosci. 2020, 8, 377–393. [Google Scholar] [CrossRef]
- Paul, S.; Knott, J.G. Epigenetic control of cell fate in mouse blastocysts: The role of covalent histone modifications and chromatin remodeling. Mol. Reprod. Dev. 2014, 81, 171–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Xie, S.; Zhou, Y.; Xie, Y.; Liu, P.; Sun, M.; Xiao, H.; Jin, Y.; Sun, X.; Chen, Z.; et al. H3K36 histone methyltransferase Setd2 is required for murine embryonic stem cell differentiation toward endoderm. Cell Rep. 2014, 8, 1989–2002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Haversat, J.M.; Mager, J. CTR9/PAF1c regulates molecular lineage identity, histone H3K36 trimethylation and genomic imprinting during preimplantation development. Dev. Biol. 2013, 383, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Kohn, M.J.; Karavanova, I.; Kaneko, K.J.; Vullhorst, D.; DePamphilis, M.L.; Buonanno, A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 2007, 134, 3827–3836. [Google Scholar] [CrossRef] [PubMed]
- Home, P.; Ray, S.; Dutta, D.; Bronshteyn, I.; Larson, M.; Paul, S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J. Biol. Chem. 2009, 284, 28729–28737. [Google Scholar] [CrossRef]
- Cao, Z.; Carey, T.S.; Ganguly, A.; Wilson, C.A.; Paul, S.; Knott, J.G. Transcription factor AP-2gamma induces early Cdx2 expression and represses HIPPO signaling to specify the trophectoderm lineage. Development 2015, 142, 1606–1615. [Google Scholar]
- Nishioka, N.; Inoue, K.; Adachi, K.; Kiyonari, H.; Ota, M.; Ralston, A.; Yabuta, N.; Hirahara, S.; Stephenson, R.O.; Ogonuki, N.; et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 2009, 16, 398–410. [Google Scholar] [CrossRef]
- Niwa, H.; Toyooka, Y.; Shimosato, D.; Strumpf, D.; Takahashi, K.; Yagi, R.; Rossant, J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 2005, 123, 917–929. [Google Scholar] [CrossRef]
- Huppertz, B.; Herrler, A. Regulation of proliferation and apoptosis during development of the preimplantation embryo and the placenta. Birth Defects Res. Part C Embryo Today 2005, 75, 249–261. [Google Scholar] [CrossRef]
- Straszewski-Chavez, S.L.; Abrahams, V.M.; Mor, G. The role of apoptosis in the regulation of trophoblast survival and differentiation during pregnancy. Endocr. Rev. 2005, 26, 877–897. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, W.; Ning, W.; Liu, C.; Zou, Y.; Yao, Y.; Kang, J.; Cao, Z. Histone Methyltransferase SETD2 Is Required for Porcine Early Embryonic Development. Animals 2022, 12, 2226. https://doi.org/10.3390/ani12172226
Shao W, Ning W, Liu C, Zou Y, Yao Y, Kang J, Cao Z. Histone Methyltransferase SETD2 Is Required for Porcine Early Embryonic Development. Animals. 2022; 12(17):2226. https://doi.org/10.3390/ani12172226
Chicago/Turabian StyleShao, Weini, Wei Ning, Chang Liu, Yuanyuan Zou, Yurui Yao, Jiaxin Kang, and Zubing Cao. 2022. "Histone Methyltransferase SETD2 Is Required for Porcine Early Embryonic Development" Animals 12, no. 17: 2226. https://doi.org/10.3390/ani12172226
APA StyleShao, W., Ning, W., Liu, C., Zou, Y., Yao, Y., Kang, J., & Cao, Z. (2022). Histone Methyltransferase SETD2 Is Required for Porcine Early Embryonic Development. Animals, 12(17), 2226. https://doi.org/10.3390/ani12172226




