Rumen Fermentation, Digestive Enzyme Activity, and Bacteria Composition between Pre-Weaning and Post-Weaning Dairy Calves
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experiment Design and Animal Management
2.2. Sample Collection and Measurement
2.2.1. Rumen Fluid Collection and Measurement
2.2.2. 16S rRNA Sequencing
2.3. Statistics
3. Results
3.1. Fermentation Profile and Digestive Enzyme Activity
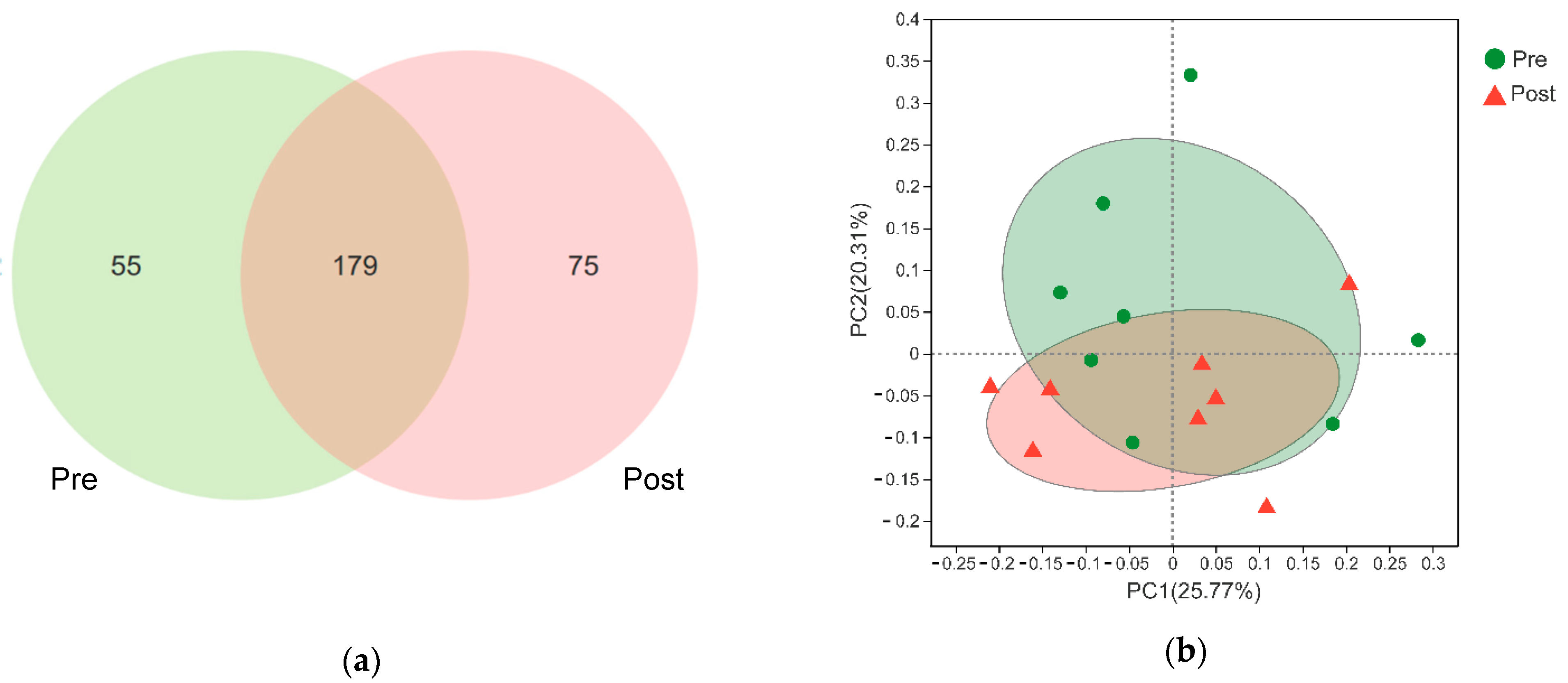
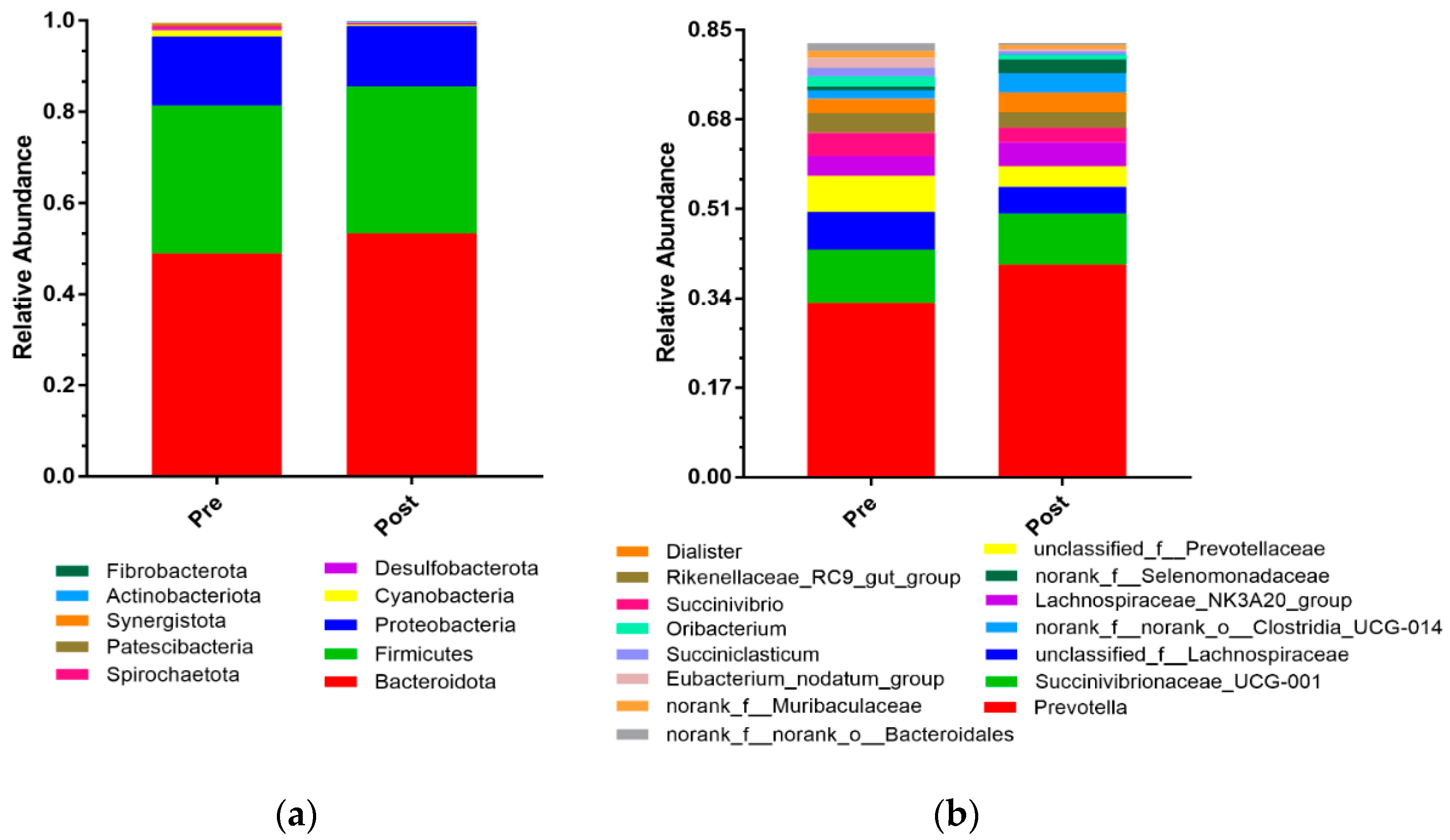
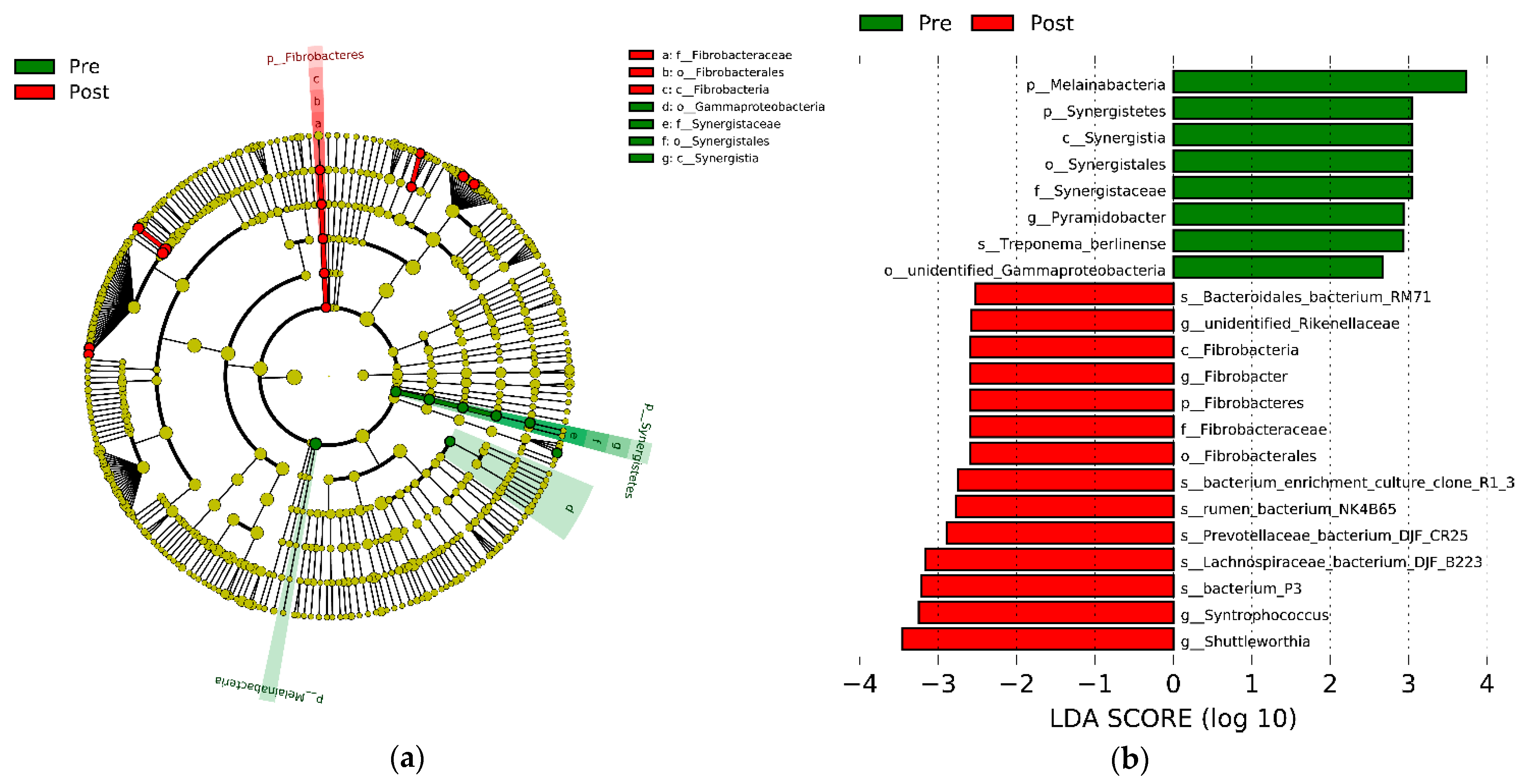
3.2. Rumen Bacteria Composition
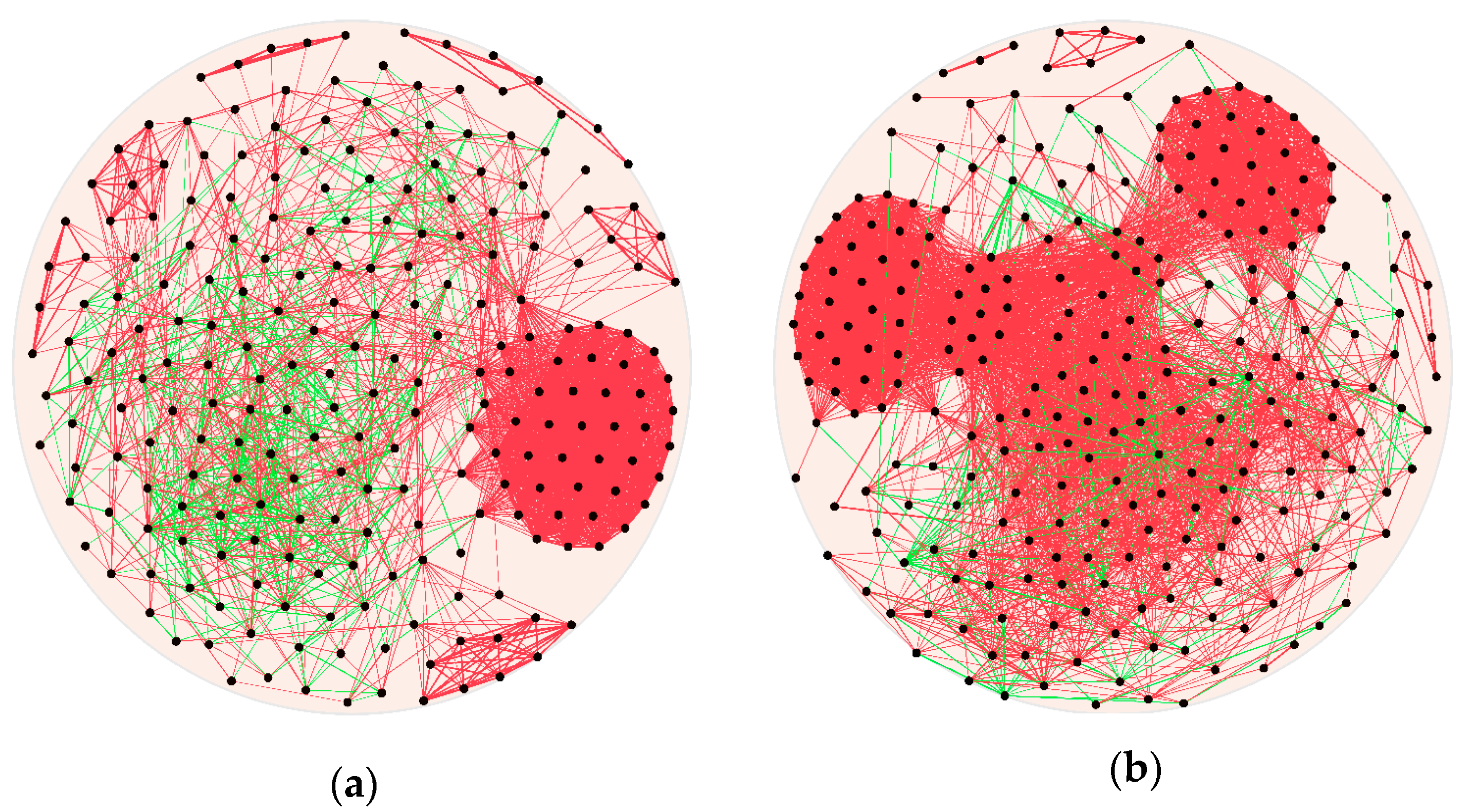
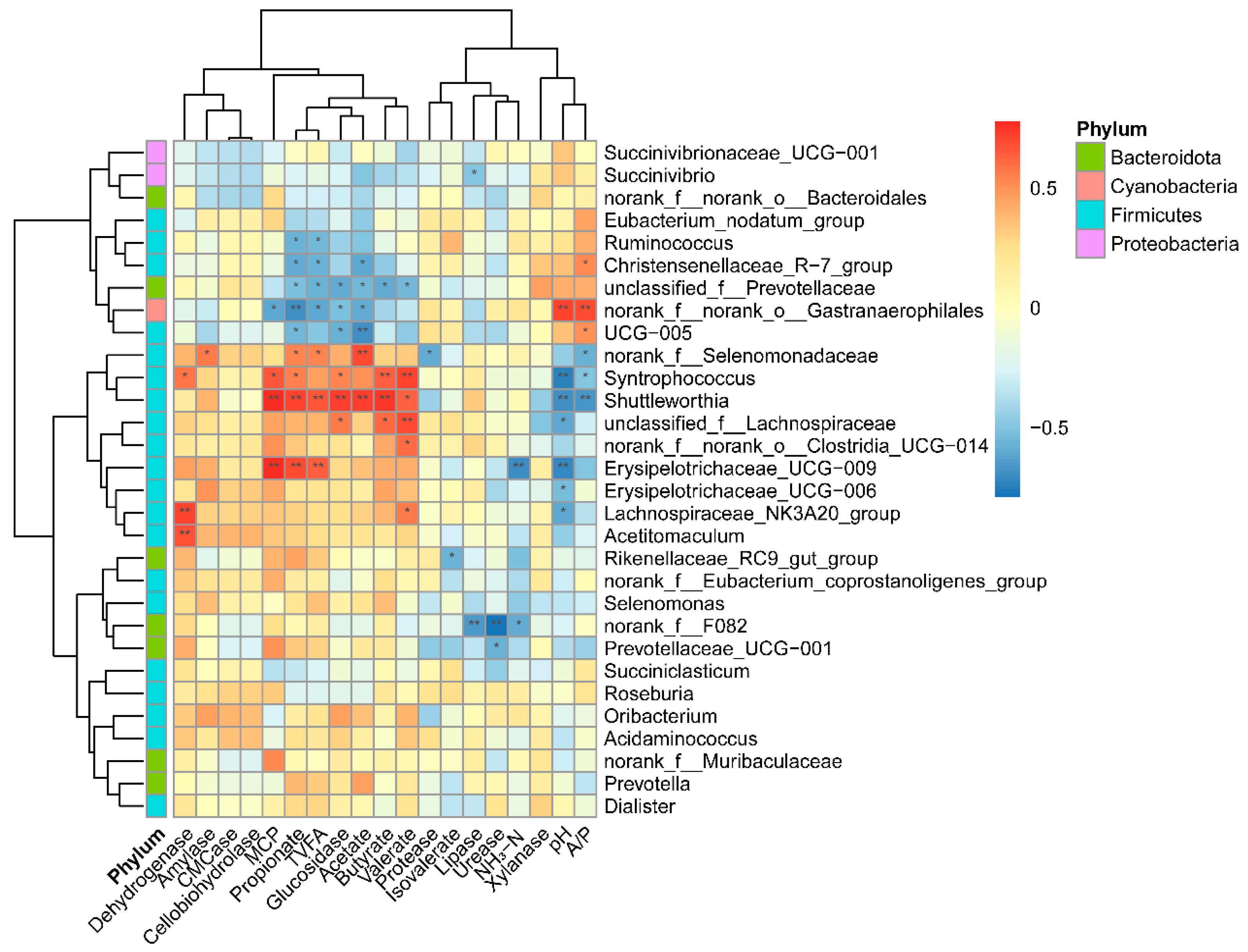
3.3. The Network and Correlation between Bacteria and Its Byproducts
4. Discussion
4.1. Rumen Fermentation Profile and Enzyme Activity
4.2. Rumen Bacteria Composition and Its Effects on Rumen Function
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCurdy, D.E.; Wilkins, K.R.; Hiltz, R.L.; Moreland, S.; Klanderman, K.; Laarman, A.H. Effects of supplemental butyrate and weaning on rumen fermentation in Holstein calves. J. Dairy Sci. 2019, 102, 8874–8882. [Google Scholar] [CrossRef]
- Meale, S.J.; Li, S.; Azevedo, P.; Derakhshani, H.; Plaizier, J.C.; Khafipour, E.; Steele, M.A. Development of ruminal and fecal microbiomes are affected by weaning but not weaning strategy in dairy calves. Front Microbiol. 2016, 7, 582. [Google Scholar] [CrossRef]
- Laarman, A.H.; Ruiz-Sanchez, A.L.; Sugino, T.; Guan, L.L.; Oba, M. Effects of feeding a calf starter on molecular adaptations in the ruminal epithelium and liver of Holstein dairy calves. J. Dairy Sci. 2012, 95, 2585–2594. [Google Scholar] [CrossRef]
- Connor, E.E.; Baldwin, R.L.t.; Li, C.J.; Li, R.W.; Chung, H. Gene expression in bovine rumen epithelium during weaning identifies molecular regulators of rumen development and growth. Funct. Integr. Genomics. 2013, 13, 133–142. [Google Scholar] [CrossRef]
- Laarman, A.H.; Sugino, T.; Oba, M. Effects of starch content of calf starter on growth and rumen pH in Holstein calves during the weaning transition. J. Dairy Sci. 2012, 95, 4478–4487. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Q.; Wang, G.; Niu, X.; Wang, W.; Li, F.; Li, F.; Zhang, Z. The functional development of the rumen is influenced by weaning and associated with ruminal microbiota in lambs. Anim. Biotechnol. 2020, 2, 1–17. [Google Scholar] [CrossRef]
- Bu, D.; Zhang, X.; Ma, L.; Park, T.; Wang, L.; Wang, M.; Xu, J.; Yu, Z. Repeated inoculation of young calves with rumen microbiota does not significantly modulate the rumen prokaryotic microbiota consistently but decreases diarrhea. Front. Microbiol. 2020, 11, 1403. [Google Scholar] [CrossRef]
- Choudhury, P.K.; Salem, A.Z.M.; Jena, R.; Kumar, S.; Singh, R.; Puniya, A.K. Rumen microbiology: An overview. In Rumen Microbiology: From Evolution to Revolution; Puniya, A.K., Singh, R., Kamra, D.N., Eds.; Springer: New Delhi, India, 2015; pp. 3–16. [Google Scholar] [CrossRef]
- Flint, H.J. The rumen microbial ecosystem—Some recent developments. Trends Microbiol. 1997, 5, 483–488. [Google Scholar] [CrossRef]
- Stewart, R.D.; Auffret, M.D.; Warr, A.; Walker, A.W.; Roehe, R.; Watson, M. Compendium of 4941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 2019, 37, 953–961. [Google Scholar] [CrossRef]
- Hess, M.; Sczyrba, A.; Egan, R.; Kim, T.-W.; Chokhawala, H.; Schroth, G.; Luo, S.; Clark, D.S.; Chen, F.; Zhang, T.; et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 2011, 331, 463–467. [Google Scholar] [CrossRef]
- Morais, S.; Mizrahi, I. Islands in the stream: From individual to communal fiber degradation in the rumen ecosystem. FEMS Microbiol. Rev. 2019, 43, 362–379. [Google Scholar] [CrossRef]
- Prive, F.; Newbold, C.J.; Kaderbhai, N.N.; Girdwood, S.G.; Golyshina, O.V.; Golyshin, P.N.; Scollan, N.D.; Huws, S.A. Isolation and characterization of novel lipases/esterases from a bovine rumen metagenome. Appl. Microbiol. Biotechnol. 2015, 99, 5475–5485. [Google Scholar] [CrossRef]
- Sun, Y.; Cheng, J. Hydrolysis of lignocellulosic materials for ethanol production: A review. Bioresour. Technol. 2002, 83, 1–11. [Google Scholar] [CrossRef]
- Pengpeng, W.; Tan, Z. Ammonia assimilation in rumen bacteria: A review. Anim. Biotechnol. 2013, 24, 107–128. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, S.; He, Y.; Zheng, N.; Yan, X.; Wang, J. Pipeline for targeted meta-proteomic analyses to assess the diversity of cattle rumen microbial urease. Front. Microbiol. 2020, 11, 573414. [Google Scholar] [CrossRef]
- Patra, A.K.; Aschenbach, J.R. Ureases in the gastrointestinal tracts of ruminant and monogastric animals and their implication in urea-N/ammonia metabolism: A review. J. Adv. Res. 2018, 13, 39–50. [Google Scholar] [CrossRef]
- Yanez-Ruiz, D.R.; Abecia, L.; Newbold, C.J. Manipulating rumen microbiome and fermentation through interventions during early life: A review. Front. Microbiol. 2015, 6, 1133. [Google Scholar] [CrossRef]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef]
- Kertz, A.F.; Hill, T.M.; Quigley, J.D., 3rd; Heinrichs, A.J.; Linn, J.G.; Drackley, J.K. A 100-Year Review: Calf nutrition and management. J. Dairy Sci. 2017, 100, 10151–10172. [Google Scholar] [CrossRef]
- Kim, Y.H.; Nagata, R.; Ohtani, N.; Ichijo, T.; Ikuta, K.; Sato, S. Effects of dietary forage and calf starter diet on ruminal pH and bacteria in Holstein calves during weaning transition. Front. Microbiol. 2016, 7, 1575. [Google Scholar] [CrossRef]
- Amin, N.; Schwarzkopf, S.; Kinoshita, A.; Troscher-Mussotter, J.; Danicke, S.; Camarinha-Silva, A.; Huber, K.; Frahm, J.; Seifert, J. Evolution of rumen and oral microbiota in calves is influenced by age and time of weaning. Anim. Microbiome. 2021, 3, 31. [Google Scholar] [CrossRef]
- Broderick, G.A.; Kang, J.H. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Makkar, H.P.; Sharma, O.P.; Dawra, R.K.; Negi, S.S. Simple determination of microbial protein in rumen. J. Dairy Sci. 1982, 65, 2170–2173. [Google Scholar] [CrossRef]
- Cao, Z.J.; Li, S.L.; Xing, J.J.; Ma, M.; Wang, L.L. Effects of maize grain and lucerne particle size on ruminal fermentation, digestibility and performance of cows in midlactation. J. Anim. Physiol. Anim. Nutr. 2008, 92, 157–167. [Google Scholar] [CrossRef]
- Bailey, M.J.; Biely, P.; Poutanen, K. Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 1992, 23, 257–270. [Google Scholar] [CrossRef]
- Prins, R.A.; van Rheenen, D.L.; van’t Klooster, A.T. Characterization of microbial proteolytic enzymes in the rumen. Antonie Van Leeuwenhoek 1983, 49, 585–595. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, P.; Li, Z.; Yang, P.; Luo, H.; Bai, Y.; Wang, Y.; Yao, B. Two neutral thermostable cellulases from Phialophora sp. G5 act synergistically in the hydrolysis of filter paper. Bioresour. Technol. 2012, 121, 404–410. [Google Scholar] [CrossRef]
- Bernfield, P. Amylase, α and β Methods. Biochim. Biophys. Acta (BBA)-Enzymol. 1955, 65, 149–158. [Google Scholar]
- Henrissat, B.; Bairoch, A. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 1996, 316, 695–696. [Google Scholar] [CrossRef]
- Chang, L.; Ding, M.; Bao, L.; Chen, Y.; Zhou, J.; Lu, H. Characterization of a bifunctional xylanase/endoglucanase from yak rumen microorganisms. Appl. Microbiol. Biotechnol. 2011, 90, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Lenártová, V.; Holovská KFau-Javorský, P.; Javorský PFau-Rybosová, E.; Rybosová EFau-Havassy, I.; Havassy, I. Glutamate dehydrogenase and glutamine synthetase activity of the bacteria of the sheep’s rumen after different nitrogen intake. Physiol. Plant. 1987, 5, 471–476. [Google Scholar]
- Guo, M.; Wu, F.; Hao, G.; Qi, Q.; Li, R.; Li, N.; Wei, L.; Chai, T. Bacillus subtilis improves immunity and disease resistance in rabbits. Front. Immunol. 2017, 8, 354. [Google Scholar] [CrossRef] [PubMed]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2012, 10, 57–59. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R Stat. Soc. Ser. A Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Shen, H.; Lu, Z.; Xu, Z.; Chen, Z.; Shen, Z.J.M. Associations among dietary non-fiber carbohydrate, ruminal microbiota and epithelium G-protein-coupled receptor, and histone deacetylase regulations in goats. Microbiome 2017, 5, 123. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.L.G.; Freitas, J.A.; Micai, B.; Azevedo, R.A.; Greco, L.F.; Santos, J.E.P. Effect of supplemental yeast culture and dietary starch content on rumen fermentation and digestion in dairy cows. J. Dairy Sci. 2018, 101, 201–221. [Google Scholar] [CrossRef]
- Gasiorek, M.; Stefanska, B.; Pruszynska-Oszmalek, E.; Taciak, M.; Komisarek, J.; Nowak, W. Effect of oat hay provision method on growth performance, rumen fermentation and blood metabolites of dairy calves during preweaning and postweaning periods. Animal 2020, 10, 2054–2062. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Yang, C.; Dong, L.; Diao, Q.; Si, B.; Ma, J.; Tu, Y. Rumen fermentation characteristics in pre- and post-weaning calves upon feeding with mulberry leaf flavonoids and candida tropicalis individually or in combination as a supplement. Animals 2019, 9, 990. [Google Scholar] [CrossRef] [PubMed]
- Aikman, P.C.; Henning, P.H.; Humphries, D.J.; Horn, C.H. Rumen pH and fermentation characteristics in dairy cows supplemented with Megasphaera elsdenii NCIMB 41125 in early lactation. J. Dairy Sci. 2011, 94, 2840–2849. [Google Scholar] [CrossRef]
- Jin, D.; Zhao, S.; Zheng, N.; Beckers, Y.; Wang, J. Urea metabolism and regulation by rumen bacterial urease in ruminants—A Review. Ann. Anim. Sci. 2018, 18, 303–318. [Google Scholar] [CrossRef]
- Arriola, K.G.; Oliveira, A.S.; Ma, Z.X.; Lean, I.J.; Giurcanu, M.C.; Adesogan, A.T. A meta-analysis on the effect of dietary application of exogenous fibrolytic enzymes on the performance of dairy cows. J. Dairy Sci. 2017, 100, 4513–4527. [Google Scholar] [CrossRef]
- Adesogan, A.T.; Arriola, K.G.; Jiang, Y.; Oyebade, A.; Paula, E.M.; Pech-Cervantes, A.A.; Romero, J.J.; Ferraretto, L.F.; Vyas, D. Symposium review: Technologies for improving fiber utilization. J. Dairy Sci. 2019, 102, 5726–5755. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; Colombatto, D.; Morgavi, D.P.; Yang, W.Z. Use of exogenous fibrolytic enzymes to improve feed utilization by ruminants. J. Anim. Sci. 2003, 87, 37–47. [Google Scholar] [CrossRef][Green Version]
- Li, F.; Guan, L.L. Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Appl. Environ. Microbiol. 2017, 83, e00061-17. [Google Scholar] [CrossRef]
- Carberry, C.A.; Kenny, D.A.; Han, S.; McCabe, M.S.; Waters, S.M. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 2012, 78, 4949–4958. [Google Scholar] [CrossRef]
- Neumann Anthony, P.; Suen, G.; Rodrigues Jorge, M. The phylogenomic diversity of herbivore-associated Fibrobacter spp. is correlated to lignocellulose-degrading potential. Msphere 2018, 3, e00593-518. [Google Scholar] [CrossRef]
- Jiang, F.G.; Lin, X.Y.; Yan, Z.G.; Hu, Z.Y.; Liu, G.M.; Sun, Y.D.; Liu, X.W.; Wang, Z.H. Effect of dietary roughage level on chewing activity, ruminal pH, and saliva secretion in lactating Holstein cows. J. Dairy Sci. 2017, 100, 2660–2671. [Google Scholar] [CrossRef]
- Zhang, R.; Ye, H.; Liu, J.; Mao, S. High-grain diets altered rumen fermentation and epithelial bacterial community and resulted in rumen epithelial injuries of goats. Appl. Microbiol. Biotechnol. 2017, 101, 6981–6992. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Oba, M. Characteristics of dairy cows with a greater or lower risk of subacute ruminal acidosis: Volatile fatty acid absorption, rumen digestion, and expression of genes in rumen epithelial cells. J. Dairy Sci. 2016, 99, 8733–8745. [Google Scholar] [CrossRef]
- Won, M.Y.; Oyama, L.B.; Courtney, S.J.; Creevey, C.J.; Huws, S.A. Can rumen bacteria communicate to each other? Microbiome 2020, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Morais, S.; Mizrahi, I. The Road Not Taken: The Rumen Microbiome, Functional Groups, and Community States. Trends Microbiol. 2019, 27, 538–549. [Google Scholar] [CrossRef]
- Shabat, S.K.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef]
- Weimer, P.J. Redundancy, resilience, and host specificity of the ruminal microbiota: Implications for engineering improved ruminal fermentations. Front. Microbiol. 2015, 6, 296. [Google Scholar] [CrossRef]
- Wallace, R.J.; Sasson, G.; Garnsworthy, P.C.; Tapio, I.; Gregson, E.; Bani, P.; Huhtanen, P.; Bayat, A.R.; Strozzi, F.; Biscarini, F.; et al. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 2019, 5, eaav8391. [Google Scholar] [CrossRef] [PubMed]
| Items 1 | Pre | Post | SEM | p |
|---|---|---|---|---|
| pH | 6.63 | 6.18 | 0.08 | 0.003 |
| NH3-N (mg/dL) | 7.68 | 8.78 | 1.94 | 0.711 |
| MCP (μg/mL) | 109.43 | 157.72 | 6.41 | <0.001 |
| Acetate (mmol/L) | 44.73 | 59.62 | 2.95 | 0.012 |
| Propionate (mmol/L) | 24.26 | 50.94 | 4.26 | 0.001 |
| Butyrate (mmol/L) | 4.21 | 9.19 | 0.87 | 0.001 |
| Isovalerate (mmol/L) | 0.73 | 0.58 | 0.07 | 0.650 |
| Valerate (mmol/L) | 1.06 | 2.25 | 0.26 | 0.383 |
| TVFA (mmol/L) | 80.61 | 124.22 | 6.96 | 0.001 |
| A/P | 2.11 | 1.26 | 0.13 | <0.001 |
| Items 1 | Pre | Post | SEM | p |
|---|---|---|---|---|
| Dehydrogenase (μg/min/mL) | 0.76 | 0.85 | 0.03 | 0.139 |
| Urease (μg/min/mL) | 3.37 | 2.86 | 0.31 | 0.449 |
| Protease (μg/min/mL) | 17.93 | 13.11 | 1.27 | 0.063 |
| CMCase (μg/min/mL) | 83.61 | 55.93 | 7.78 | 0.085 |
| Cellobiohydrolase (μg/min/mL) | 56.82 | 37.68 | 5.43 | 0.088 |
| Glucosidase (nmol/min/mL) | 120.84 | 200.21 | 21.00 | 0.065 |
| Amylase (mg/min/mL) | 1.04 | 1.11 | 0.13 | 0.796 |
| Lipase (nmol/min/mL) | 160.28 | 160.10 | 12.30 | 0.994 |
| Xylanase (nmol/min/mL) | 216.51 | 200.73 | 19.24 | 0.706 |
| Items 1 | Pre | Post | SEM | p |
|---|---|---|---|---|
| Sobs | 124.38 | 131.13 | 6.89 | 0.75 |
| Ace | 127.42 | 134.08 | 6.84 | 0.79 |
| Chao1 | 126.53 | 133.70 | 6.83 | 0.87 |
| Shannon | 2.52 | 2.52 | 0.10 | 0.96 |
| Simpson | 0.18 | 0.21 | 0.02 | 0.79 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Y.; Guo, C.; Gong, Y.; Sun, X.; Wang, W.; Wang, Y.; Yang, H.; Cao, Z.; Li, S. Rumen Fermentation, Digestive Enzyme Activity, and Bacteria Composition between Pre-Weaning and Post-Weaning Dairy Calves. Animals 2021, 11, 2527. https://doi.org/10.3390/ani11092527
Hao Y, Guo C, Gong Y, Sun X, Wang W, Wang Y, Yang H, Cao Z, Li S. Rumen Fermentation, Digestive Enzyme Activity, and Bacteria Composition between Pre-Weaning and Post-Weaning Dairy Calves. Animals. 2021; 11(9):2527. https://doi.org/10.3390/ani11092527
Chicago/Turabian StyleHao, Yangyi, Chunyan Guo, Yue Gong, Xiaoge Sun, Wei Wang, Yajing Wang, Hongjian Yang, Zhijun Cao, and Shengli Li. 2021. "Rumen Fermentation, Digestive Enzyme Activity, and Bacteria Composition between Pre-Weaning and Post-Weaning Dairy Calves" Animals 11, no. 9: 2527. https://doi.org/10.3390/ani11092527
APA StyleHao, Y., Guo, C., Gong, Y., Sun, X., Wang, W., Wang, Y., Yang, H., Cao, Z., & Li, S. (2021). Rumen Fermentation, Digestive Enzyme Activity, and Bacteria Composition between Pre-Weaning and Post-Weaning Dairy Calves. Animals, 11(9), 2527. https://doi.org/10.3390/ani11092527






