Identification of Ovarian Circular RNAs and Differential Expression Analysis between MeiShan and Large White Pigs
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. RNA Sequencing
2.3. Sequence Mapping and Identification of Differentially Expressed Genes
2.4. Identification and Verification of circRNAs
2.5. Identification of Potential Target miRNAs
2.6. Gene Ontology (GO) Enrichment Analysis
2.7. Transfections and Luciferase Reporter Assays
3. Results
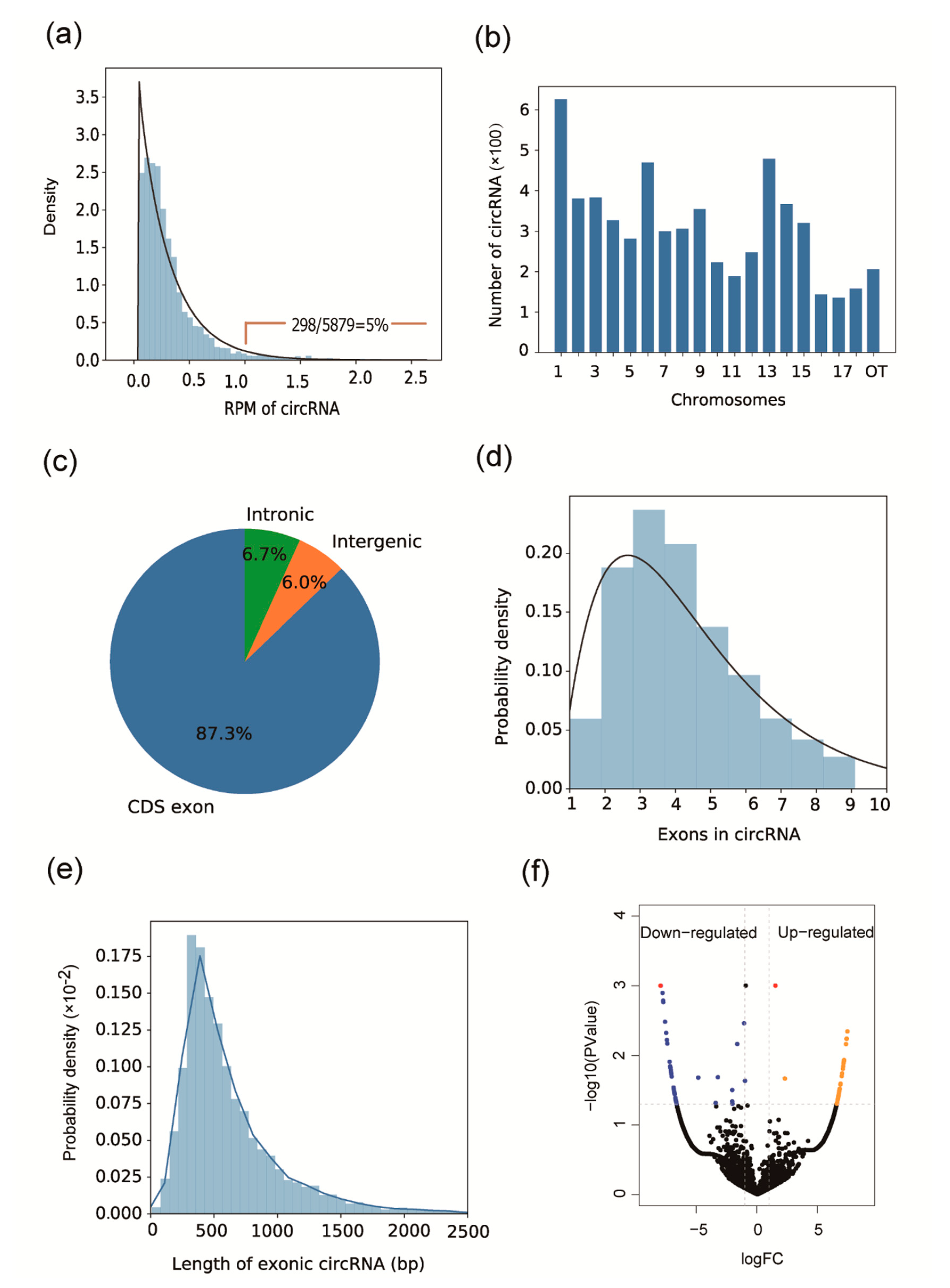
3.1. Identifying circRNA and Differential Expression Analysis
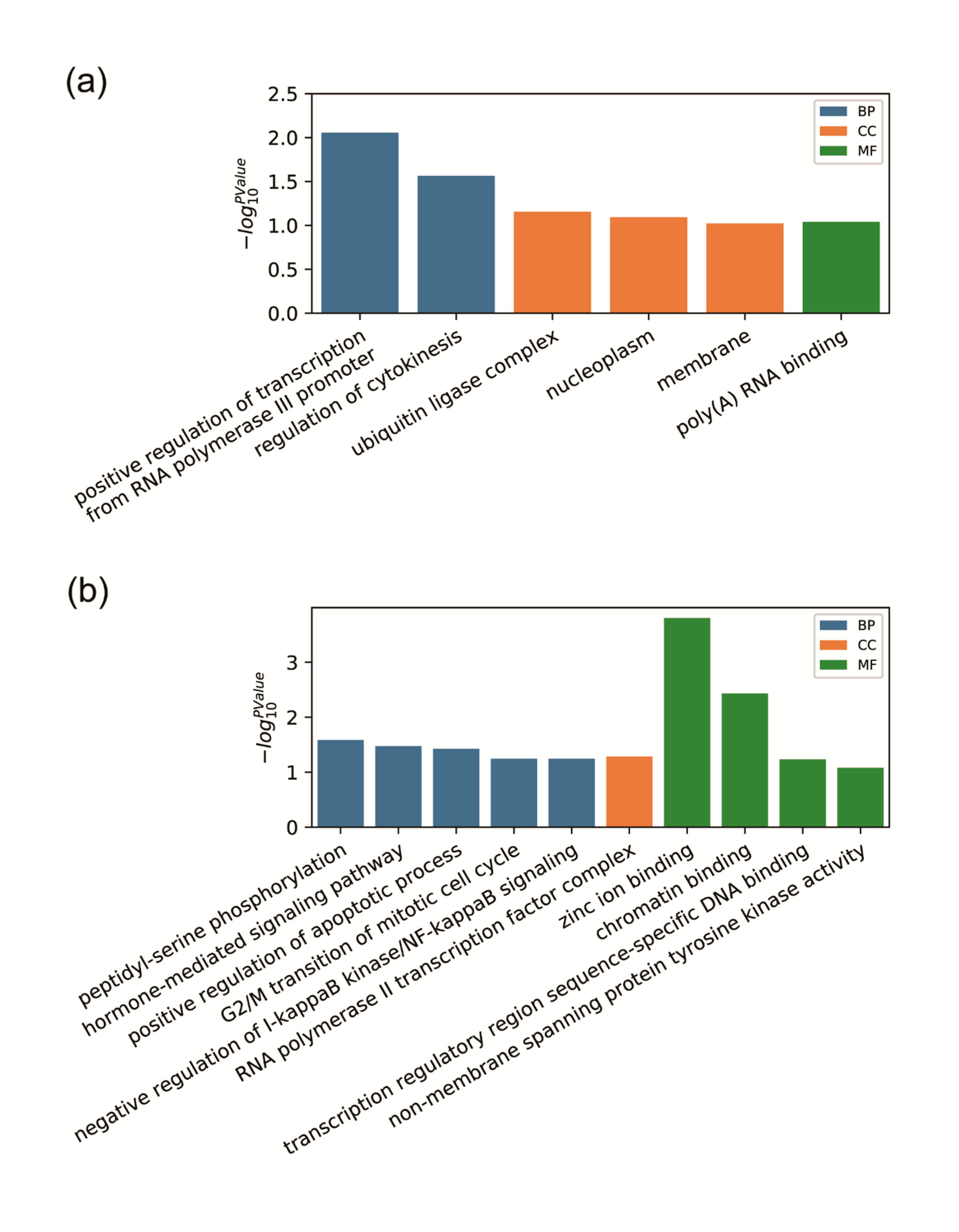
3.2. Gene Ontology Enrichment Analysis of Differentially Expressed circRNAs
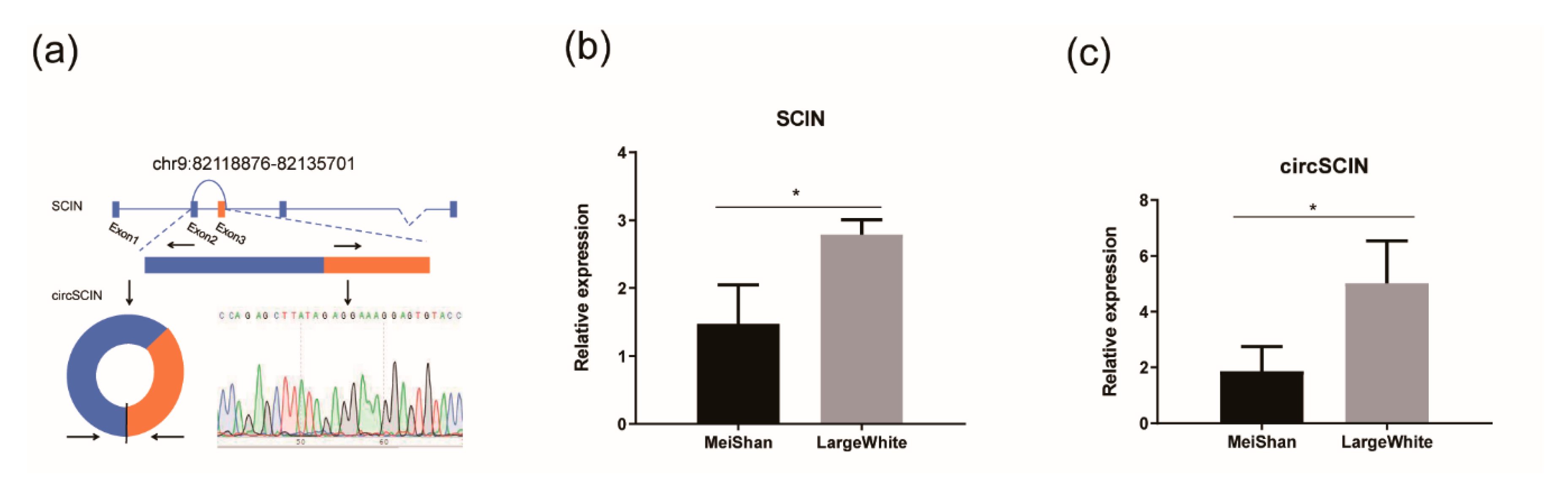
3.3. Verifying circSCIN and SCIN Differential Expression
3.4. CircSCIN binds to miR-133 and miR-148a/b
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Ethics Statement
References
- Christenson, R.K. Ovulation rate and embryonic survival in Chinese Meishan and white crossbred pigs. J. Anim. Sci. 1993, 71, 3060–3066. [Google Scholar] [CrossRef]
- Rothschild, M.; Jacobson, C.; Vaske, D.; Tuggle, C.; Wang, L.; Short, T.; Eckardt, G.; Sasaki, S.; Vincent, A.; McLaren, D.; et al. The estrogen receptor locus is associated with a major gene influencing litter size in pigs. Proc. Natl. Acad. Sci. USA 1996, 93, 201. [Google Scholar] [CrossRef]
- Zhao, P.; Yu, Y.; Feng, W.; Du, H.; Yu, J.; Kang, H.; Zheng, X.; Wang, Z.; Liu, G.E.; Ernst, C.W.; et al. Evidence of evolutionary history and selective sweeps in the genome of Meishan pig reveals its genetic and phenotypic characterization. GigaScience 2018, 7, giy058. [Google Scholar] [CrossRef]
- Li, W.-T.; Zhang, M.-M.; Li, Q.-G.; Tang, H.; Zhang, L.-F.; Wang, K.-J.; Zhu, M.-Z.; Lu, Y.-F.; Bao, H.-G.; Zhang, Y.-M.; et al. Whole-genome resequencing reveals candidate mutations for pig prolificacy. Proc. R. Soc. B Biol. Sci. 2017, 284, 20172437. [Google Scholar] [CrossRef] [PubMed]
- Koketsu, Y.; Tani, S.; Iida, R. Factors for improving reproductive performance of sows and herd productivity in commercial breeding herds. Porc. Health Manag. 2017, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Soede, N.M.; Langendijk, P.; Kemp, B. Reproductive cycles in pigs. Anim. Reprod. Sci. 2011, 124, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Han, J.; Cao, R.; Zhang, J.; Li, B.; Liu, Z.; Liu, K.; Li, Q.; Pan, Z.; Chen, J.; et al. Sequence and regulation of the porcine FSHR gene promoter. Anim. Reprod. Sci. 2015, 154, 95–104. [Google Scholar] [CrossRef]
- LaVoie, H.A. Transcriptional control of genes mediating ovarian follicular growth, differentiation, and steroidogenesis in pigs. Mol. Reprod. Dev. 2017, 84, 788–801. [Google Scholar] [CrossRef]
- Fernandez-Rodriguez, A.; Munoz, M.; Fernandez, A.; Pena, R.N.; Tomas, A.; Noguera, J.L.; Ovilo, C.; Fernandez, A.I. Differential gene expression in ovaries of pregnant pigs with high and low prolificacy levels and identification of candidate genes for litter size. Biol. Reprod. 2011, 84, 299–307. [Google Scholar] [CrossRef]
- Che, L.; Xu, M.; Yang, Z.; Xu, S.; Che, L.; Lin, Y.; Fang, Z.; Feng, B.; Li, J.; Chen, D.; et al. Detection of Placental Proteomes at Different Uterine Positions in Large White and Meishan Gilts on Gestational Day 90. PLoS ONE 2016, 11, e0167799. [Google Scholar] [CrossRef]
- Tung, E.; Roberts, C.T.; Heinemann, G.K.; De Blasio, M.J.; Kind, K.L.; van Wettere, W.H.; Owens, J.A.; Gatford, K.L. Increased placental nutrient transporter expression at midgestation after maternal growth hormone treatment in pigs: A placental mechanism for increased fetal growth. Biol. Reprod. 2012, 87, 126. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Yang, Y.; Niu, G.; Tang, Z.; Li, K. Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 2017, 24, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Panda, A.C.; De, S.; Grammatikakis, I.; Kim, J.; Ding, J.; Noh, J.H.; Kim, K.M.; Mattison, J.A.; de Cabo, R.; et al. Circular RNAs in monkey muscle: Age-dependent changes. Aging 2015, 7, 903–910. [Google Scholar] [CrossRef]
- Gruner, H.; Cortés-López, M.; Cooper, D.A.; Bauer, M.; Miura, P. CircRNA accumulation in the aging mouse brain. Sci. Rep. 2016, 6, 38907. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, L.; Wu, T.; Feng, Y.; Ding, Y.; Ye, P.; Yin, Z. Transcriptomic analysis of ovaries from pigs with high and low litter size. PLoS ONE 2015, 10, e0139514. [Google Scholar] [CrossRef]
- Huang, L.; Yin, Z.J.; Feng, Y.F.; Zhang, X.D.; Wu, T.; Ding, Y.Y.; Ye, P.F.; Fu, K.; Zhang, M.Q. Identification and differential expression of microRNAs in the ovaries of pigs (Sus scrofa) with high and low litter sizes. Anim. Genet. 2016, 47, 543–551. [Google Scholar] [CrossRef]
- Carletti, M.Z.; Christenson, L.K. MicroRNA in the ovary and female reproductive tract1. J. Anim. Sci. 2009, 87, E29–E38. [Google Scholar] [CrossRef]
- Knapczyk-Stwora, K.; Nynca, A.; Ciereszko, R.E.; Paukszto, L.; Jastrzebski, J.P.; Czaja, E.; Witek, P.; Koziorowski, M.; Slomczynska, M. Flutamide-induced alterations in transcriptional profiling of neonatal porcine ovaries. J. Anim. Sci. Biotechnol. 2019, 10, 35. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.-L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 2018. [Google Scholar] [CrossRef]
- Bonizzato, A.; Gaffo, E.; Te Kronnie, G.; Bortoluzzi, S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016, 6, e483. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef]
- Thadani, R.; Tammi, M.T. MicroTar: Predicting microRNA targets from RNA duplexes. BMC Bioinform. 2006, 18, S20. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Huang, J.; Liu, R.; Su, L.; Xiao, Q.; Yu, M. Transcriptome analysis revealed the embryo-induced gene expression patterns in the endometrium from Meishan and Yorkshire pigs. Int. J. Mol. Sci. 2015, 16, 22692–22710. [Google Scholar] [CrossRef]
- Yang, S.; Zhou, X.; Pei, Y.; Wang, H.; He, K.; Zhao, A. Identification of differentially expressed genes in porcine ovaries at proestrus and estrus stages using RNA-Seq technique. Biomed Res. Int. 2018, 2018, 9150723. [Google Scholar] [CrossRef] [PubMed]
- Zi, X.D.; Xu, H.W.; Wang, Y. Variation in sequences and mRNA expression levels of inhibin subunits alpha (INHA) and betaA (INHBA) genes between prolific and nonprolific goat breeds. Mol. Reprod. Dev. 2012, 79, 238. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Ponsuksili, S.; Tholen, E.; Jennen, D.G.J.; Schellander, K.; Wimmers, K. Candidate gene markers for sperm quality and fertility of boar. Anim. Reprod. Sci. 2006, 92, 349–363. [Google Scholar] [CrossRef]
- Giesecke, K.; Sieme, H.; Distl, O. Infertility and candidate gene markers for fertility in stallions: A review. Vet. J. 2010, 185, 265–271. [Google Scholar] [CrossRef]
- Chumnarnsilpa, S.; Robinson, R.C.; Grimes, J.M.; Leyrat, C. Calcium-controlled conformational choreography in the N-terminal half of adseverin. Nat. Commun. 2015, 6, 8254. [Google Scholar] [CrossRef]
- Ghoshdastider, U.; Popp, D.; Burtnick, L.D.; Robinson, R.C. The expanding superfamily of gelsolin homology domain proteins. Cytoskeleton 2013, 70, 775–795. [Google Scholar] [CrossRef]
- Pelletier, R.; Trifaro, J.M.; Carbajal, M.E.; Okawara, Y.; Vitale, M.L. Calcium-dependent actin filament-severing protein scinderin levels and localization in bovine testis, epididymis, and spermatozoa. Biol. Reprod. 1999, 60, 1128–1136. [Google Scholar] [CrossRef]
- Haim, B.; Gili, C.; Sara, R. Role of actin cytoskeleton in mammalian sperm capacitation and the acrosome reaction. Reproduction 2005, 129, 263–268. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, W.; Wang, X.; Wang, T.; Chen, Y.; Chen, B.; Liu, R.; Bai, P.; Xing, J. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol. Med. (Camb. Mass.) 2018, 24, 40. [Google Scholar] [CrossRef]
- Sirotkin, A.V.; Ovcharenko, D.; Grossmann, R.; Lauková, M.; Mlynček, M. Identification of MicroRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. J. Cell. Physiol. 2009, 219, 415–420. [Google Scholar] [CrossRef]
- Toms, D.; Pan, B.; Li, J. Endocrine regulation in the ovary by MicroRNA during the estrous cycle. Front. Endocrinol. 2018, 8. [Google Scholar] [CrossRef]
- Song, Y.-N.; Shi, L.-L.; Liu, Z.-Q.; Qiu, G.-F. Global analysis of the ovarian microRNA transcriptome: Implication for miR-2 and miR-133 regulation of oocyte meiosis in the Chinese mitten crab, Eriocheir sinensis (Crustacea:Decapoda). BMC Genom. 2014, 15, 547. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.; Cheng, Q.; Zhou, C.; Ding, Z. Role of microRNA-133a in epithelial ovarian cancer pathogenesis and progression. Oncol. Lett. 2014, 7, 1043–1048. [Google Scholar] [CrossRef]
- Guo, J.; Xia, B.; Meng, F.; Lou, G. miR-133a suppresses ovarian cancer cell proliferation by directly targeting insulin-like growth factor 1 receptor. Tumor Biol. 2014, 35, 1557–1564. [Google Scholar] [CrossRef]
- Yao, N.; Yang, B.Q.; Liu, Y.; Tan, X.Y.; Lu, C.L.; Yuan, X.H.; Ma, X. Follicle-stimulating hormone regulation of microRNA expression on progesterone production in cultured rat granulosa cells. Endocrine 2010, 38, 158–166. [Google Scholar] [CrossRef]
- Dai, A.; Sun, H.; Fang, T.; Zhang, Q.; Wu, S.; Jiang, Y.; Ding, L.; Yan, G.; Hu, Y. MicroRNA-133b stimulates ovarian estradiol synthesis by targeting Foxl2. FEBS Lett. 2013, 587, 2474–2482. [Google Scholar] [CrossRef]
- Li, Y.; Deng, X.; Zeng, X.; Peng, X. The Role of Mir-148a in Cancer. J. Cancer 2016, 7, 1233–1241. [Google Scholar] [CrossRef]
- Friedrich, M.; Pracht, K.; Mashreghi, M.-F.; Jäck, H.-M.; Radbruch, A.; Seliger, B. The role of the miR-148/-152 family in physiology and disease. Eur. J. Immunol. 2017, 47, 2026–2038. [Google Scholar] [CrossRef]
- Ghosh-Choudhury, T.; Xiao, W.; Gunaratne, P.; Anderson, M. Loss of miR-148a/b expression promotes ovarian cancer by targeting Erbb3 and MYB. Gynecol. Oncol. 2012, 125, S98. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, G.; Yan, J.; Guo, J.; Tang, Z. Identification of Ovarian Circular RNAs and Differential Expression Analysis between MeiShan and Large White Pigs. Animals 2020, 10, 1114. https://doi.org/10.3390/ani10071114
Liang G, Yan J, Guo J, Tang Z. Identification of Ovarian Circular RNAs and Differential Expression Analysis between MeiShan and Large White Pigs. Animals. 2020; 10(7):1114. https://doi.org/10.3390/ani10071114
Chicago/Turabian StyleLiang, Guoming, Junyu Yan, Jin Guo, and Zhonglin Tang. 2020. "Identification of Ovarian Circular RNAs and Differential Expression Analysis between MeiShan and Large White Pigs" Animals 10, no. 7: 1114. https://doi.org/10.3390/ani10071114
APA StyleLiang, G., Yan, J., Guo, J., & Tang, Z. (2020). Identification of Ovarian Circular RNAs and Differential Expression Analysis between MeiShan and Large White Pigs. Animals, 10(7), 1114. https://doi.org/10.3390/ani10071114




