Discrimination of Meat Produced by Bos taurus and Bos indicus Finished under an Intensive or Extensive System
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Management
2.2. Experimental Procedures
2.3. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mennecke, B.E.; Townsend, A.M.; Hayes, D.J.; Lonergan, S.M. A study of the factors that influence consumer attitudes toward beef products using the conjoint market analysis tool 1. J. Anim. Sci. 2007, 85, 2639–2659. [Google Scholar] [CrossRef]
- Nuernberg, K.; Dannenberger, D.; Nuernberg, G.; Ender, K.; Voigt, J.; Scollan, N.D.; Wood, J.D.; Nute, G.R.; Richardson, R.I. Effect of a grass-based and a concentrate feeding system on meat quality characteristics and fatty acid composition of longissimus muscle in different cattle breeds. Livest. Prod. Sci. 2005, 94, 137–147. [Google Scholar] [CrossRef]
- Vasta, V.; Luciano, G.; Dimauro, C.; Röhrle, F.; Priolo, A.; Monahan, F.J.; Moloney, A.P. The volatile profile of longissimus dorsi muscle of heifers fed pasture, pasture silage or cereal concentrate: Implication for dietary discrimination. Meat Sci. 2011, 87, 282–289. [Google Scholar] [CrossRef]
- Font-i-Furnols, M.; Guerrero, L. Consumer preference, behavior and perception about meat and meat products: An overview. Meat Sci. 2014. [Google Scholar] [CrossRef]
- Stampa, E.; Schipmann-Schwarze, C.; Hamm, U. Consumer perceptions, preferences, and behavior regarding pasture-raised livestock products: A review. Food Qual. Prefer. 2020. [Google Scholar] [CrossRef]
- FAO. FAOSTAT. Available online: http://www.fao.org/faostat/en/#home (accessed on 19 March 2019).
- Ferraz, J.B.S.; de Felício, P.E. Production systems—An example from Brazil. Meat Sci. 2010, 84, 238–243. [Google Scholar] [CrossRef]
- Wheeler, T.L.; Cundiff, L.V.; Shackelford, S.D.; Koohmaraie, M. Characterization of biological types of cattle (Cycle V): Carcass traits and longissimus palatability. J. Anim. Sci. 2001, 79, 1209. [Google Scholar] [CrossRef]
- Elzo, M.A.; Johnson, D.D.; Wasdin, J.G.; Driver, J.D. Carcass and meat palatability breed differences and heterosis effects in an Angus-Brahman multibreed population. Meat Sci. 2012, 90, 87–92. [Google Scholar] [CrossRef]
- Wright, S.A.; Ramos, P.; Johnson, D.D.; Scheffler, J.M.; Elzo, M.A.; Mateescu, R.G.; Bass, A.L.; Carr, C.C.; Scheffler, T.L. Brahman genetics influence muscle fiber properties, protein degradation, and tenderness in an Angus-Brahman multibreed herd. Meat Sci. 2018, 135, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Bressan, M.C.; Rodrigues, E.C.; Rossato, L.V.; Ramos, E.M.; da Gama, L.T. Physicochemical properties of meat from Bos taurus and Bos indicus. Rev. Bras. Zootec. 2011, 40, 1250–1259. [Google Scholar] [CrossRef][Green Version]
- Bressan, M.C.; Rossato, L.V.; Rodrigues, E.C.; Alves, S.P.; Bessa, R.J.B.; Ramos, E.M.; Gama, L.T. Genotype × environment interactions for fatty acid profiles in Bos indicus and Bos taurus finished on pasture or grain. J. Anim. Sci. 2011, 89, 221–232. [Google Scholar] [CrossRef] [PubMed]
- EBLEX. Quality Standard Mark Scheme for Beef and Lamb; Agriculture and Horticulture Development Board: Warwickshire, UK, 2012. [Google Scholar]
- Vlachos, A.; Arvanitoyannis, I.S.; Tserkezou, P. An Updated Review of Meat Authenticity Methods and Applications. Crit. Rev. Food Sci. Nutr. 2016, 56, 1061–1096. [Google Scholar] [CrossRef] [PubMed]
- Troy, D.J.; Kerry, J.P. Consumer perception and the role of science in the meat industry. Meat Sci. 2010, 86, 214–226. [Google Scholar]
- Tabachnick, B.G.; Fidell, L.S. Using Multivariate Statistics, 5th ed.; Pearson Education: Boston, MA, USA, 2007. [Google Scholar]
- Destefanis, G.; Barge, M.T.; Brugiapaglia, A.; Tassone, S. The use of principal component analysis (PCA) to characterize beef. Meat Sci. 2000, 56, 255–259. [Google Scholar] [CrossRef]
- Insausti, K.; Beriain, M.; Lizaso, G.; Carr, T.; Purroy, A. Multivariate study of different beef quality traits from local Spanish cattle breeds. Animal 2008, 2, 447–458. [Google Scholar] [PubMed]
- Domingo, G.; Iglesias, A.; Monserrat, L.; Sanchez, L.; Cantalapiedra, J.; Lorenzo, J.M. Effect of crossbreeding with Limousine, Rubia Gallega and Belgium Blue on meat quality and fatty acid profile of Holstein calves. Anim. Sci. J. 2015, 86, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Gagaoua, M.; Terlouw, E.M.C.; Micol, D.; Hocquette, J.F.; Moloney, A.P.; Nuernberg, K.; Bauchart, D.; Boudjellal, A.; Scollan, N.D.; Richardson, R.I.; et al. Sensory quality of meat from eight different types of cattle in relation with their biochemical characteristics. J. Integr. Agric. 2016, 15, 1550–1563. [Google Scholar] [CrossRef]
- Santos-Silva, J.; Bessa, R.J.B.; Santos-Silva, F. Effect of genotype, feeding system and slaughter weight on the quality of light lambs. II. Fatty acid composition of meat. Livest. Prod. Sci. 2002, 77, 187–194. [Google Scholar] [CrossRef]
- Moreno, T.; Varela, A.; Oliete, B.; Carballo, J.A.; Sánchez, L.; Montserrat, L. Nutritional characteristics of veal from weaned and unweaned calves: Discriminatory ability of the fat profile. Meat Sci. 2006, 73, 209–217. [Google Scholar] [CrossRef]
- Acciaro, M.; Dimauro, C.; Decandia, M.; Sitzia, M.; Manca, C.; Giovanetti, V.; Cabiddu, A.; Addis, M.; Rassu, S.; Molle, G. Discriminant analysis to identify ruminant meat produced from pasture or stall-fed animals. In Proceedings of the Grassland resources for extensive farming systems in marginal lands: Major drivers and future scenarios. 19th Symposium of the European Grassland Society, Alghero, Italy, 7–10 May 2017; pp. 88–90. [Google Scholar]
- Horcada, A.; López, A.; Polvillo, O.; Pino, R.; Cubiles-de-la-Vega, D.; Tejerina, D.; García-Torres, S. Fatty acid profile as a tool to trace the origin of beef in pasture- and grain-fed young bulls of Retinta breed. Spanish J. Agric. Res. 2017, 15. [Google Scholar] [CrossRef]
- Costa, P.; Lemos, J.P.; Lopes, P.A.; Alfaia, C.M.; Costa, A.S.H.; Bessa, R.J.B.; Prates, J.A.M. Effect of low- and high-forage diets on meat quality and fatty acid composition of Alentejana and Barrosã beef breeds. Animal 2012, 6, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Serra, X.; Guerrero, L.; Guàrdia, M.D.; Gil, M.; Sañudo, C.; Panea, B.; Campo, M.M.; Olleta, J.L.; García-Cachán, M.D.; Piedrafita, J.; et al. Eating quality of young bulls from three Spanish beef breed-production systems and its relationships with chemical and instrumental meat quality. Meat Sci. 2008, 79, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Bessa, R.J.B.; Alves, S.P.; Jerónimo, E.; Alfaia, C.M.; Prates, J.A.M.; Santos-Silva, J. Effect of lipid supplements on ruminal biohydrogenation intermediates and muscle fatty acids in lambs. Eur. J. Lipid Sci. Technol. 2007, 109, 868–878. [Google Scholar] [CrossRef]
- Wood, J.D.; Enser, M.; Fisher, A.V.; Nute, G.R.; Sheard, P.R.; Richardson, R.I.; Hughes, S.I.; Whittington, F.M. Fat deposition, fatty acid composition and meat quality: A review. Meat Sci. 2008, 78, 343–358. [Google Scholar] [CrossRef]
- Wood, J.D.; Enser, M. Manipulating the Fatty Acid Composition of Meat to Improve Nutritional Value and Meat Quality; Elsevier Ltd.: Amsterdand, The Netherlands, 2017; ISBN 9780081005934. [Google Scholar]
- Monteiro, A.C.G.; Gomes, E.; Barreto, A.S.; Silva, M.F.; Fontes, M.A.; Bessa, R.J.B.; Lemos, J.P.C. Eating quality of “Vitela Tradicional do Montado”-PGI veal and Mertolenga-PDO veal and beef. Meat Sci. 2013, 94, 63–68. [Google Scholar] [CrossRef]
- French, P.; O’Riordan, E.G.; Monahan, F.J.; Caffrey, P.J.; Vidal, M.; Mooney, M.T.; Troy, D.J.; Moloney, A.P. Meat quality of steers finished on autumn grass, grass silage or concentrate-based diets. Meat Sci. 2000, 56, 173–180. [Google Scholar] [CrossRef]
- Bressan, M.C.; Rodrigues, E.C.; de Paula, M.d.L.; Ramos, E.M.; Portugal, P.V.; Silva, J.S.; Bessa, R.B.; da Gama, L.T. Differences in intramuscular fatty acid profiles among Bos indicus and crossbred Bos taurus × Bos indicus bulls finished on pasture or with concentrate feed in Brazil. Ital. J. Anim. Sci. 2016, 15, 10–21. [Google Scholar] [CrossRef]
- Nian, Y.; Allen, P.; Harrison, S.M.; Brunton, N.P.; Prendiville, R.; Kerry, J.P. Fatty acid composition of young dairy bull beef as affected by breed type, production treatment, and relationship to sensory characteristics. Anim. Prod. Sci. 2019, 59, 1360. [Google Scholar] [CrossRef]
- Moloney, A.P.; O’riordan, E.G.; Schmidt, O.; Monahan, F.J. The fatty acid profile and stable isotope ratios of C and N of muscle from cattle that grazed grass or grass/clover pastures before slaughter and their discriminatory potential. Irish J. Agric. Food Res. 2018, 57, 84–94. [Google Scholar] [CrossRef]
- Prache, S.; Kondjoyan, N.; Delfosse, O.; Chauveau-Duriot, B.; Andueza, D.; Cornu, A. Discrimination of pasture-fed lambs from lambs fed dehydrated alfalfa indoors using different compounds measured in the fat, meat and plasma. Animal 2009, 3, 598–605. [Google Scholar] [CrossRef]
- Monahan, F.J.; Schmidt, O.; Moloney, A.P. Meat provenance: Authentication of geographical origin and dietary background of meat. Meat Sci. 2018, 144, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Negrini, R.; Nicoloso, L.; Crepaldi, P.; Milanesi, E.; Colli, L.; Chegdani, F.; Pariset, L.; Dunner, S.; Leveziel, H.; Williams, J.L.; et al. Assessing SNP markers for assigning individuals to cattle populations. Anim. Genet. 2009, 40, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Hulsegge, B.; Calus, M.P.L.; Windig, J.J.; Hoving-Bolink, A.H.; Maurice-van Eijndhoven, M.H.T.; Hiemstra, S.J. Selection of SNP from 50K and 777K arrays to predict breed of origin in cattle. J. Anim. Sci. 2013, 91, 5128–5134. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L. Meat Authenticity and Traceability. In Lawrie’s Meat Science: Eighth Edition; Elsevier: Amsterdand, The Netherlands, 2017; pp. 585–633. ISBN 9780081006979. [Google Scholar]
- Gama, L.T.; Bressan, M.C.; Rodrigues, E.C.; Rossato, L.V.; Moreira, O.C.; Alves, S.P.; Bessa, R.J.B. Heterosis for meat quality and fatty acid profiles in crosses among Bos indicus and Bos taurus finished on pasture or grain. Meat Sci. 2013, 93, 98–104. [Google Scholar] [CrossRef] [PubMed]
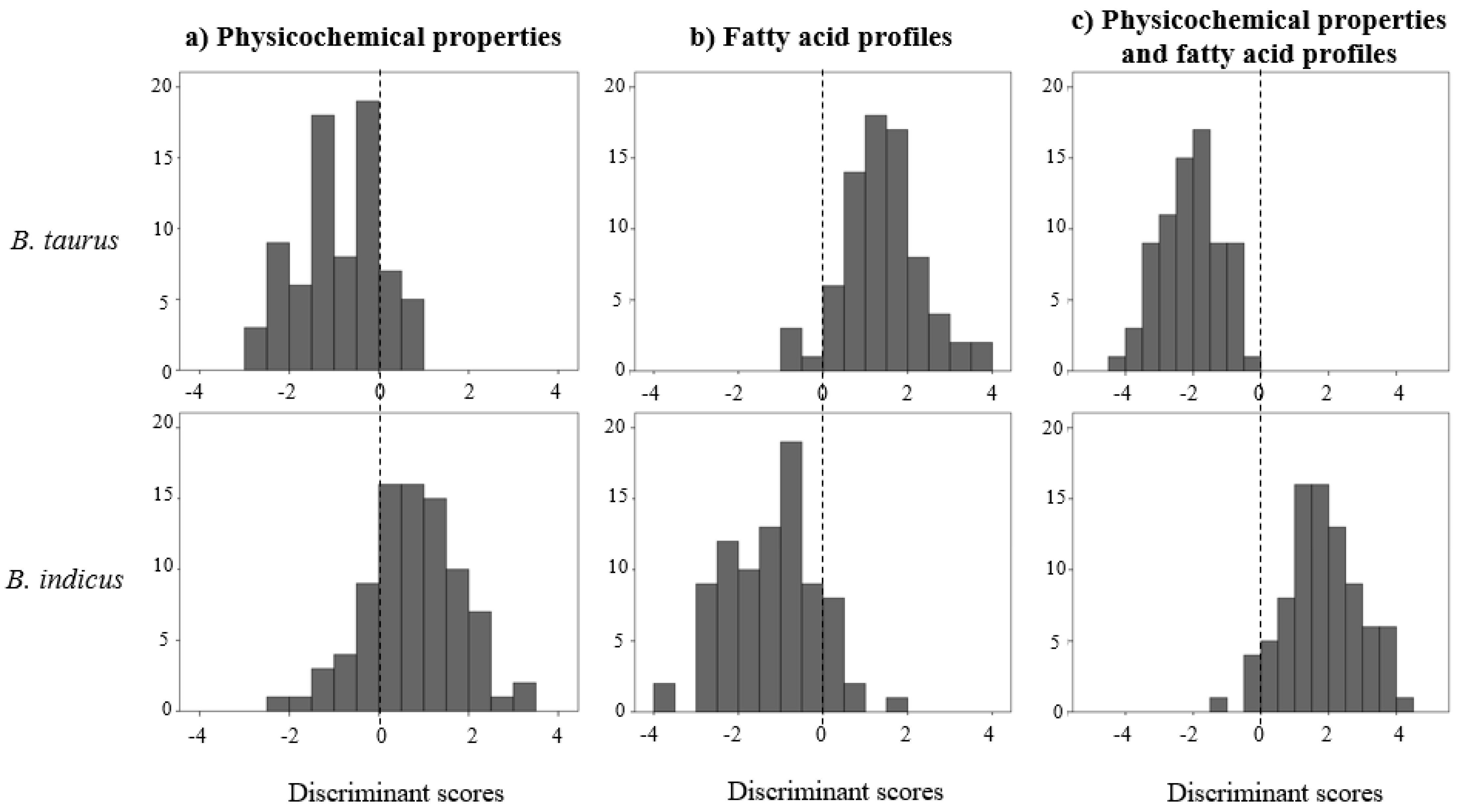
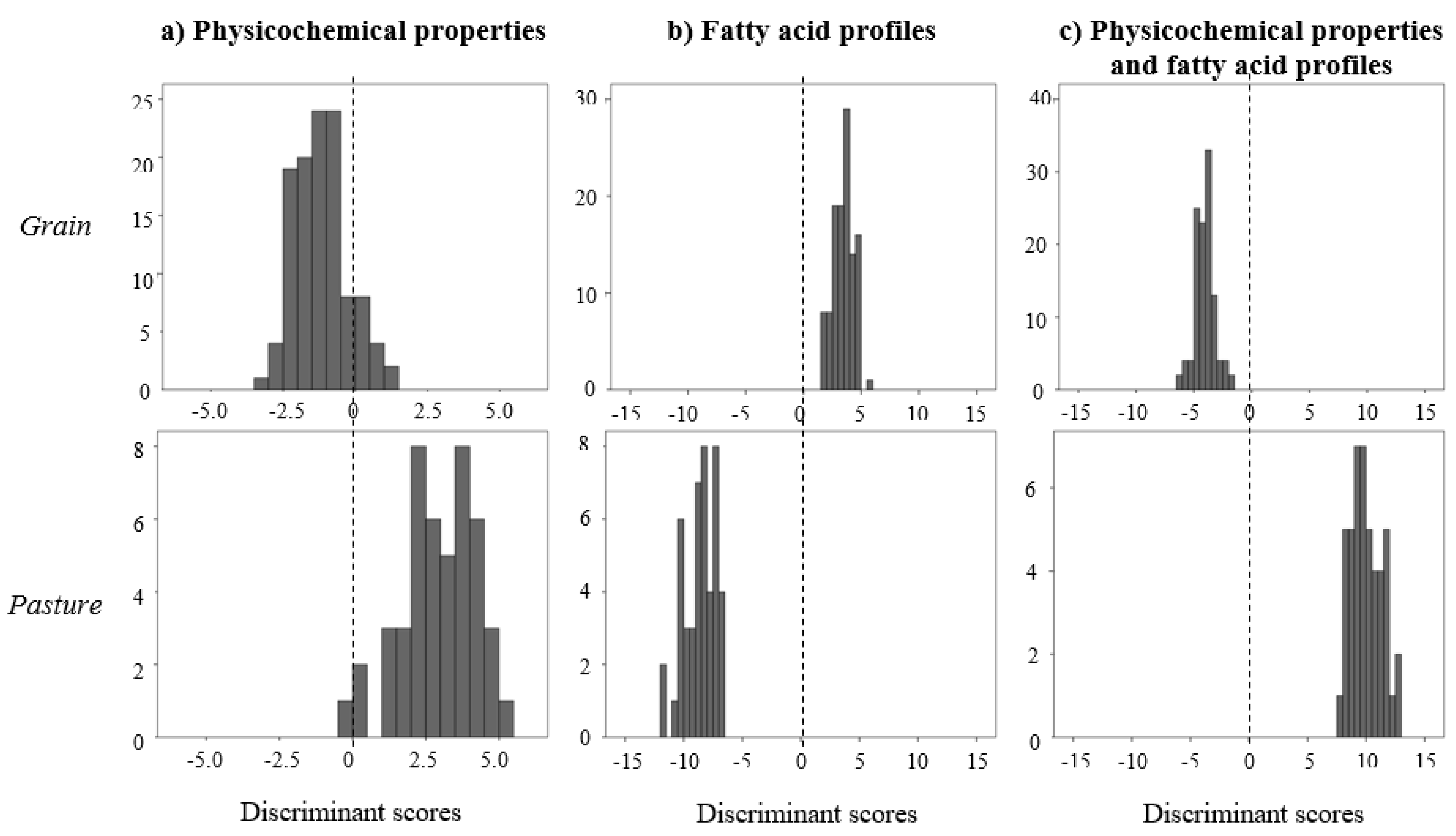

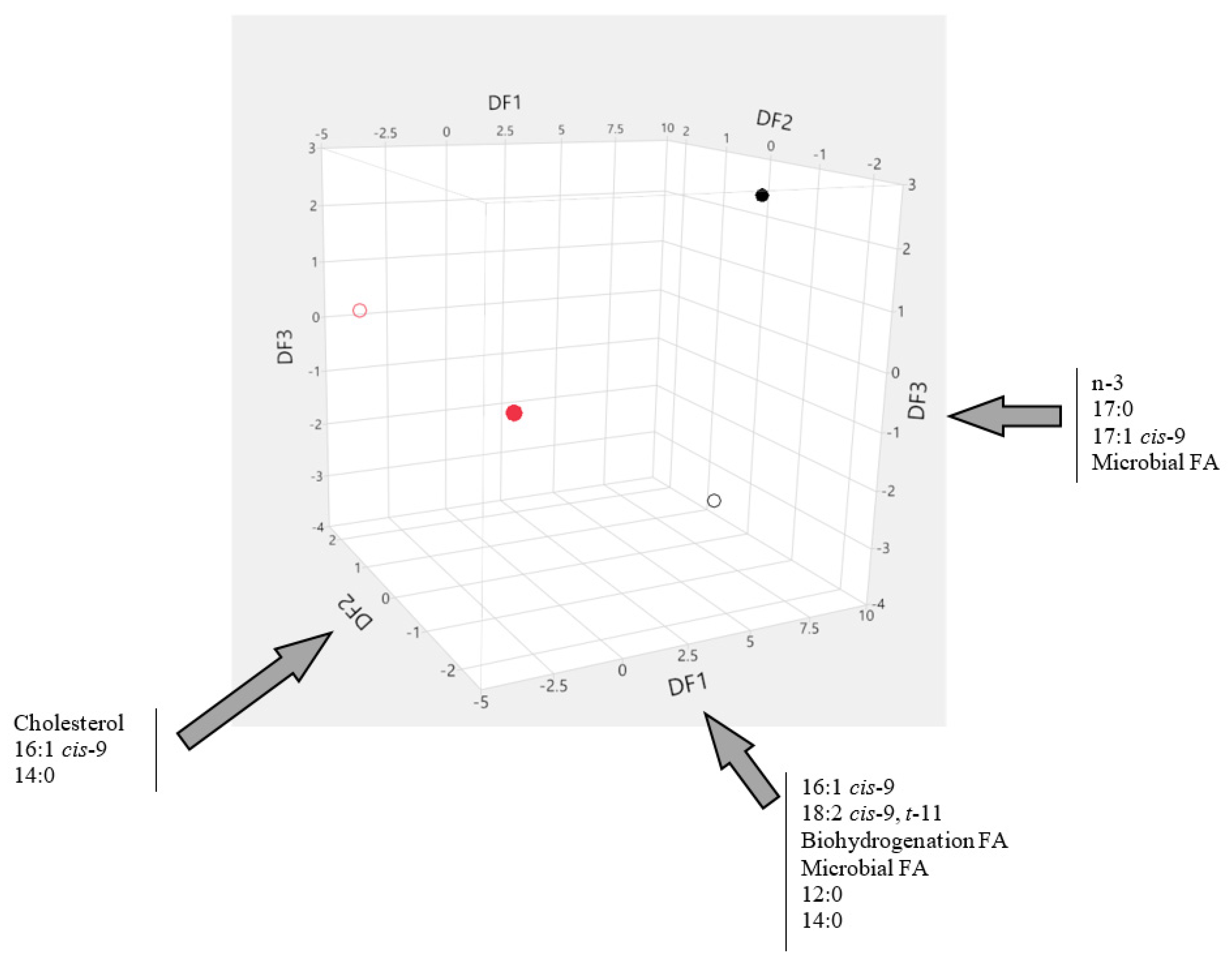
| Group | Variable | Mean | CV (%) |
|---|---|---|---|
| Meat chemical composition | Moisture (%) | 72.63 | 2.8 |
| Protein (%) | 19.08 | 8.9 | |
| Ash (%) | 0.90 | 20.0 | |
| Fat (%) | 6.39 | 49.0 | |
| Cholesterol (mg/100 g) | 49.27 | 32.5 | |
| Meat colour parameters | L* 24 h | 32.76 | 7.4 |
| a* 24 h | 20.00 | 7.8 | |
| b* 24 h | 4.23 | 27.4 | |
| L* 10 d | 33.61 | 7.9 | |
| a* 10 d | 16.51 | 12.6 | |
| b* 10 d | 4.87 | 28.5 | |
| Meat physical properties | Cooking loss 24 h (%) | 29.06 | 16.0 |
| Shear Force 24 h (kg) | 7.98 | 21.2 | |
| Cooking loss 10 d (%) | 30.20 | 17.1 | |
| Shear Force 10 d (kg) | 5.64 | 27.0 | |
| pH 24 h | 5.85 | 3.1 | |
| Fatty acids (FA, %) | Microbial FA | 2.26 | 21.2 |
| Biohydrogenation FA | 4.06 | 23.2 | |
| n-3 FA | 1.22 | 93.4 | |
| n-6 FA | 3.49 | 40.4 | |
| 12:0 | 0.07 | 42.9 | |
| 14:0 | 3.04 | 21.1 | |
| 14:1c-9 | 0.49 | 36.7 | |
| 16:0 | 24.62 | 8.0 | |
| 16:2 | 0.34 | 26.5 | |
| 16:1c-9 | 2.79 | 23.3 | |
| 17:0 | 1.21 | 13.2 | |
| 17:1c-9 | 1.03 | 72.8 | |
| 18:0 | 20.42 | 17.8 | |
| 18:1c-9 | 32.64 | 8.9 | |
| 18:1c-11 | 1.20 | 26.7 | |
| 20:0 | 0.18 | 27.8 | |
| 20:1-c13 | 0.28 | 60.7 | |
| 18:2-c9,t-11 | 0.48 | 27.1 |
| Discriminant Factors | Genetic Groups | Finishing Systems | Genetic Group × Finishing System Combinations |
|---|---|---|---|
| Physicochemical (PC) properties | 0.582 | 0.222 | 0.070 |
| Fatty acid (FA) profiles | 0.363 | 0.032 | 0.004 |
| PC properties and FA profiles | 0.208 | 0.024 | 0.002 |
| Discrimination Criteria | Discriminant Function | ||
|---|---|---|---|
| Variable | Unstandardized Coefficients | Standardized Coefficients | |
| Physicochemical properties | Cholesterol | 0.057 | 0.781 |
| Shear Force 24 h | 0.340 | 0.525 | |
| b* 10 d | −0.312 | −0.413 | |
| (Constant) | −3.981 | - | |
| Fatty acid profiles | Biohydrogenation FA | −1.195 | −0.919 |
| n-3 | −1.382 | −1.570 | |
| 12:0 | 20.192 | 0.659 | |
| 14:0 | −1.429 | −0.892 | |
| 16:0 | 0.322 | 0.614 | |
| 16:1c-7 | 11.810 | 1.119 | |
| 17:0 | −3.027 | −0.484 | |
| 17:1c-9 | 1.260 | 0.952 | |
| 20:1c-13 | 3.545 | 0.599 | |
| (Constant) | −1.187 | - | |
| Physicochemical properties and fatty acid profiles | Microbial FA | −2.589 | −1.240 |
| Biohydrogenation FA | 0.586 | 0.451 | |
| n-3 | 0.893 | 1.014 | |
| 12:0 | −19.301 | −0.630 | |
| 14:0 | 0.908 | 0.567 | |
| 14:1c-9 | 2.662 | 0.473 | |
| 17:0 | 4.638 | 0.741 | |
| 18:0 | 0.255 | 0.863 | |
| 18:2c-9,t-11 | 5.147 | 0.635 | |
| Moisture | 0.156 | 0.319 | |
| Ash | −1.286 | −0.222 | |
| Cholesterol | 0.056 | 0.768 | |
| Shear Force 24 h | 0.171 | 0.265 | |
| b* 10d | −0.244 | −0.322 | |
| (Constant) | −26.590 | - | |
| Discrimination Criteria | Discriminant Function | ||
|---|---|---|---|
| Variable | Unstandardized Coefficients | Standardized Coefficients | |
| Physicochemical properties | Protein | 1.074 | 0.953 |
| Cholesterol | −0.024 | −0.366 | |
| a* 24 h | −0.239 | −0.357 | |
| b* 24 h | 0.201 | 0.233 | |
| Shear Force 24 h | 0.114 | 0.189 | |
| (Constant) | −16.266 | - | |
| Fatty acid profiles | Microbial FA | −1.977 | −0.792 |
| n-3 | −0.983 | −0.564 | |
| n-6 | 0.546 | 0.656 | |
| 12:0 | 30.608 | 0.623 | |
| 14:0 | −0.941 | −0.503 | |
| 16:1c-7 | −6.184 | −0.320 | |
| 16:1c-9 | 2.630 | 1.531 | |
| 17:0 | 2.404 | 0.386 | |
| 17:1c-9 | −2.088 | −0.683 | |
| 18:0 | 0.195 | 0.668 | |
| 18:1c-11 | −2.508 | −0.528 | |
| 20:0 | 7.111 | 0.334 | |
| 20:1c-13 | 4.135 | 0.432 | |
| (Constant) | −5.044 | - | |
| Physicochemical properties and fatty acid profiles | Moisture | 0.357 | 0.691 |
| Fat | 0.435 | 1.015 | |
| L* 24 h | −0.088 | −0.214 | |
| Microbial FA | −1.798 | −0.721 | |
| Biohydrogenation FA | 0.779 | 0.695 | |
| n-3 | −0.844 | −0.484 | |
| n-6 | 0.535 | 0.642 | |
| 12:0 | 26.423 | 0.538 | |
| 14:0 | −1.020 | −0.545 | |
| 16:0 | −0.210 | −0.368 | |
| 16:2 | −8.253 | −0.427 | |
| 16:1c-9 | 2.830 | 1.648 | |
| 17:0 | 3.196 | 0.512 | |
| 17:1c-9 | −2.438 | −0.797 | |
| 18:1-c11 | −2.025 | −0.426 | |
| 20:0 | 7.619 | 0.358 | |
| 20:1-c13 | 4.000 | 0.418 | |
| 18:2-c9,t-11 | −7.032 | −0.741 | |
| (Constant) | −22.621 | - | |
| Discrimination Criteria | Variable | Unstandardized Coefficients | Standardized Coefficients | ||||
|---|---|---|---|---|---|---|---|
| Function 1 | Function 2 | Function 3 | Function 1 | Function 2 | Function 3 | ||
| Physicochemical properties | Protein | 1.003 | 0.445 | 0.205 | 0.892 | 0.396 | 0.183 |
| Cholesterol | −0.069 | 0.075 | 0.024 | −0.665 | 0.716 | 0.228 | |
| a* 24 h | −0.124 | −0.189 | −0.123 | −0.187 | −0.283 | −0.185 | |
| Shear Force 24h | 0.028 | 0.256 | −0.278 | 0.042 | 0.388 | −0.421 | |
| b* 10 d | −0.011 | −0.029 | 0.625 | −0.015 | −0.037 | 0.812 | |
| (Constant) | −13.410 | −10.294 | −3.454 | - | - | - | |
| Fatty acid profiles | Microbial FA | 1.064 | −2.227 | −1.376 | 0.423 | −0.885 | −0.547 |
| Biohydrogenation FA | −0.906 | 0.775 | −0.244 | −0.636 | 0.544 | −0.171 | |
| n-3 | 1.132 | 3.052 | 1.786 | 0.602 | 1.622 | 0.949 | |
| n-6 | −0.481 | −0.165 | 0.127 | −0.574 | −0.197 | 0.151 | |
| 12:0 | −29.838 | −10.257 | 12.169 | −0.609 | −0.209 | 0.248 | |
| 14:0 | 1.402 | 1.132 | −1.063 | 0.721 | 0.582 | −0.547 | |
| 16:0 | 0.161 | −0.125 | 0.349 | 0.271 | −0.210 | 0.589 | |
| 16:2 | 9.052 | −1.963 | 11.573 | 0.465 | −0.101 | 0.595 | |
| 16:1c-9 | −2.851 | 0.818 | 1.860 | −1.341 | 0.384 | 0.874 | |
| 17:0 | −1.901 | 7.194 | 4.292 | −0.300 | 1.137 | 0.678 | |
| 17:1c-9 | 1.839 | −3.413 | −2.751 | 0.595 | −1.105 | −0.891 | |
| 18:1-c11 | 2.185 | −0.353 | 0.896 | 0.409 | −0.066 | 0.168 | |
| 20:1-c13 | −3.571 | −1.981 | 0.632 | −0.375 | −0.208 | 0.066 | |
| 18:2-c9,t-11 | 7.404 | 3.278 | −0.731 | 0.715 | 0.316 | −0.071 | |
| (Constant) | −4.327 | −8.274 | −17.221 | ||||
| Physicochemical properties and fatty acid profiles | Microbial FA | 1.615 | 0.009 | −2.469 | 0.642 | 0.004 | −0.981 |
| Biohydrogenation FA | −0.928 | −0.078 | 0.791 | −0.651 | −0.054 | 0.555 | |
| n-3 | 0.572 | −0.016 | 3.268 | 0.304 | −0.008 | 1.737 | |
| 12:0 | −31.774 | 24.073 | −2.326 | −0.649 | 0.491 | −0.047 | |
| 14:0 | 1.065 | −1.172 | 0.438 | 0.548 | −0.603 | 0.225 | |
| 16:0 | 0.232 | 0.267 | 0.015 | 0.391 | 0.450 | 0.025 | |
| 16:2 | 9.532 | 8.389 | 1.755 | 0.490 | 0.431 | 0.090 | |
| 16:1c-9 | −2.599 | 1.492 | 1.722 | −1.222 | 0.702 | 0.810 | |
| 17:0 | −1.841 | −0.122 | 8.369 | −0.291 | −0.019 | 1.323 | |
| 17:1c-9 | 1.638 | −0.419 | −4.283 | 0.530 | −0.136 | −1.386 | |
| 20:0 | −8.541 | 0.954 | 0.464 | −0.381 | 0.043 | 0.021 | |
| 20:1-c13 | −4.655 | 0.668 | −2.159 | −0.489 | 0.070 | −0.227 | |
| 18:2-c9,t-11 | 8.447 | −3.425 | 2.122 | 0.815 | −0.331 | 0.205 | |
| Protein | 0.381 | 0.289 | 0.078 | 0.339 | 0.257 | 0.069 | |
| Cholesterol | −0.007 | −0.086 | −0.023 | −0.068 | −0.826 | −0.222 | |
| Shear Force 24 h | 0.047 | −0.165 | 0.180 | 0.071 | −0.251 | 0.272 | |
| (Constant) | −11.095 | −9.634 | −16.564 | ||||
| Source of Information | Genetic Group | Finishing System | Genetic Group × Finishing System | |||||
|---|---|---|---|---|---|---|---|---|
| Taurus | Indicus | Grain | Pasture | Indicus Pasture | Indicus Grain | Taurus Pasture | Taurus Grain | |
| Physicochemical (PC) properties | 81.3 (80.0) | 81.2 (78.8) | 98.2 (98.2) | 93.5 (93.5) | 69.2 (69.2) | 94.9 (94.9) | 75.0 (65.0) | 80.0 (80.0) |
| Fatty acid (FA) profiles | 93.3 (92.0) | 89.4 (87.1) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (96.2) | 93.2 (88.1) | 100.0 (100.0) | 92.7 (90.9) |
| PC and FA profiles | 100.0 (97.3) | 95.3 (94.1) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (96.2) | 100.0 (96.6) | 100.0 (100.0) | 98.2 (98.2) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bressan, M.C.; Rodrigues, E.C.; Rossato, L.V.; Neto-Fonseca, I.; Alves, S.P.; Bessa, R.J.B.; Gama, L.T. Discrimination of Meat Produced by Bos taurus and Bos indicus Finished under an Intensive or Extensive System. Animals 2020, 10, 1737. https://doi.org/10.3390/ani10101737
Bressan MC, Rodrigues EC, Rossato LV, Neto-Fonseca I, Alves SP, Bessa RJB, Gama LT. Discrimination of Meat Produced by Bos taurus and Bos indicus Finished under an Intensive or Extensive System. Animals. 2020; 10(10):1737. https://doi.org/10.3390/ani10101737
Chicago/Turabian StyleBressan, Maria C., Erika C. Rodrigues, Lizandra V. Rossato, Isabel Neto-Fonseca, Susana P. Alves, Rui J.B. Bessa, and Luis T. Gama. 2020. "Discrimination of Meat Produced by Bos taurus and Bos indicus Finished under an Intensive or Extensive System" Animals 10, no. 10: 1737. https://doi.org/10.3390/ani10101737
APA StyleBressan, M. C., Rodrigues, E. C., Rossato, L. V., Neto-Fonseca, I., Alves, S. P., Bessa, R. J. B., & Gama, L. T. (2020). Discrimination of Meat Produced by Bos taurus and Bos indicus Finished under an Intensive or Extensive System. Animals, 10(10), 1737. https://doi.org/10.3390/ani10101737






