Host Innate Immune Response of Geese Infected with Clade 2.3.4.4 H5N6 Highly Pathogenic Avian Influenza Viruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses
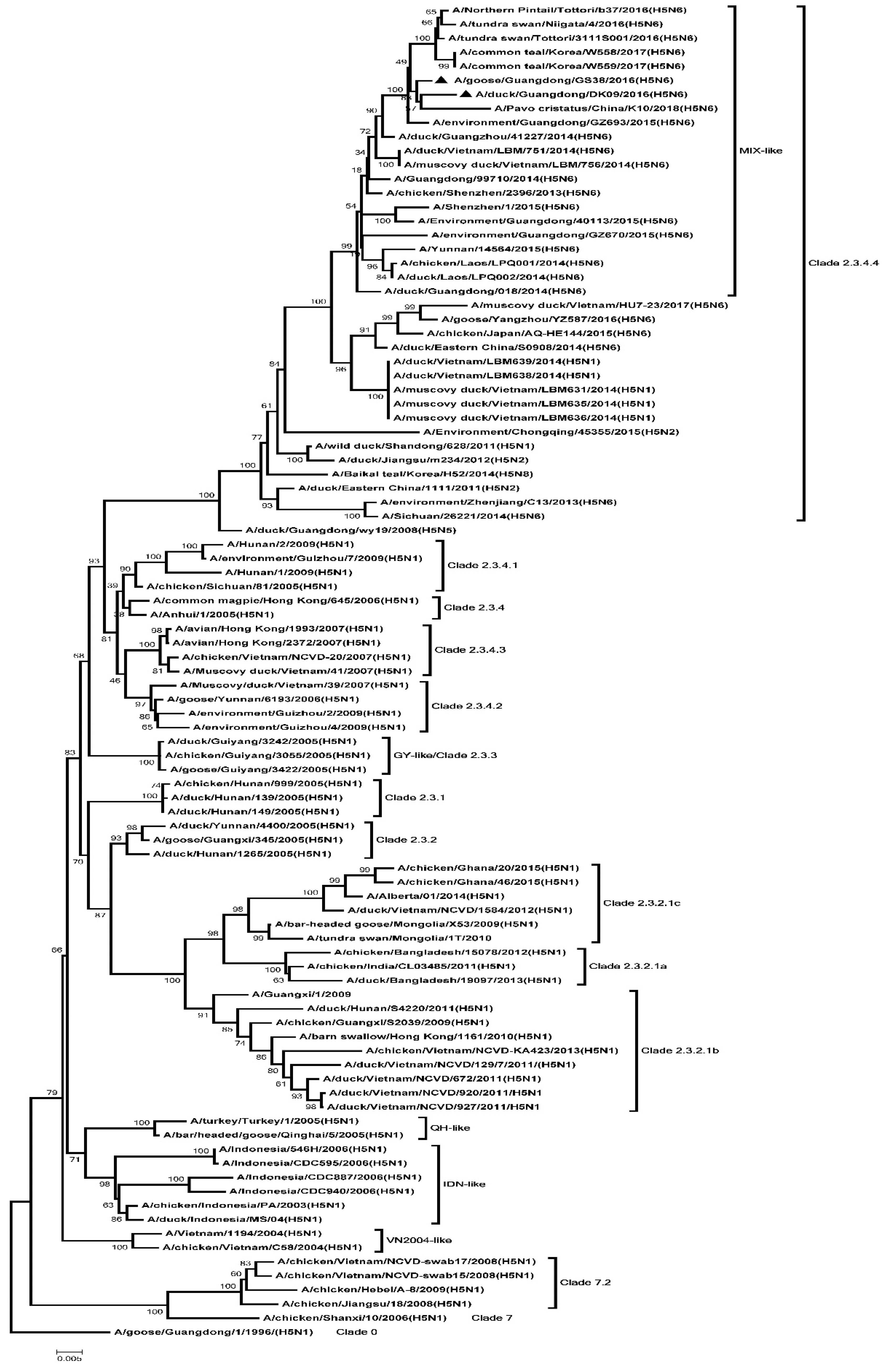
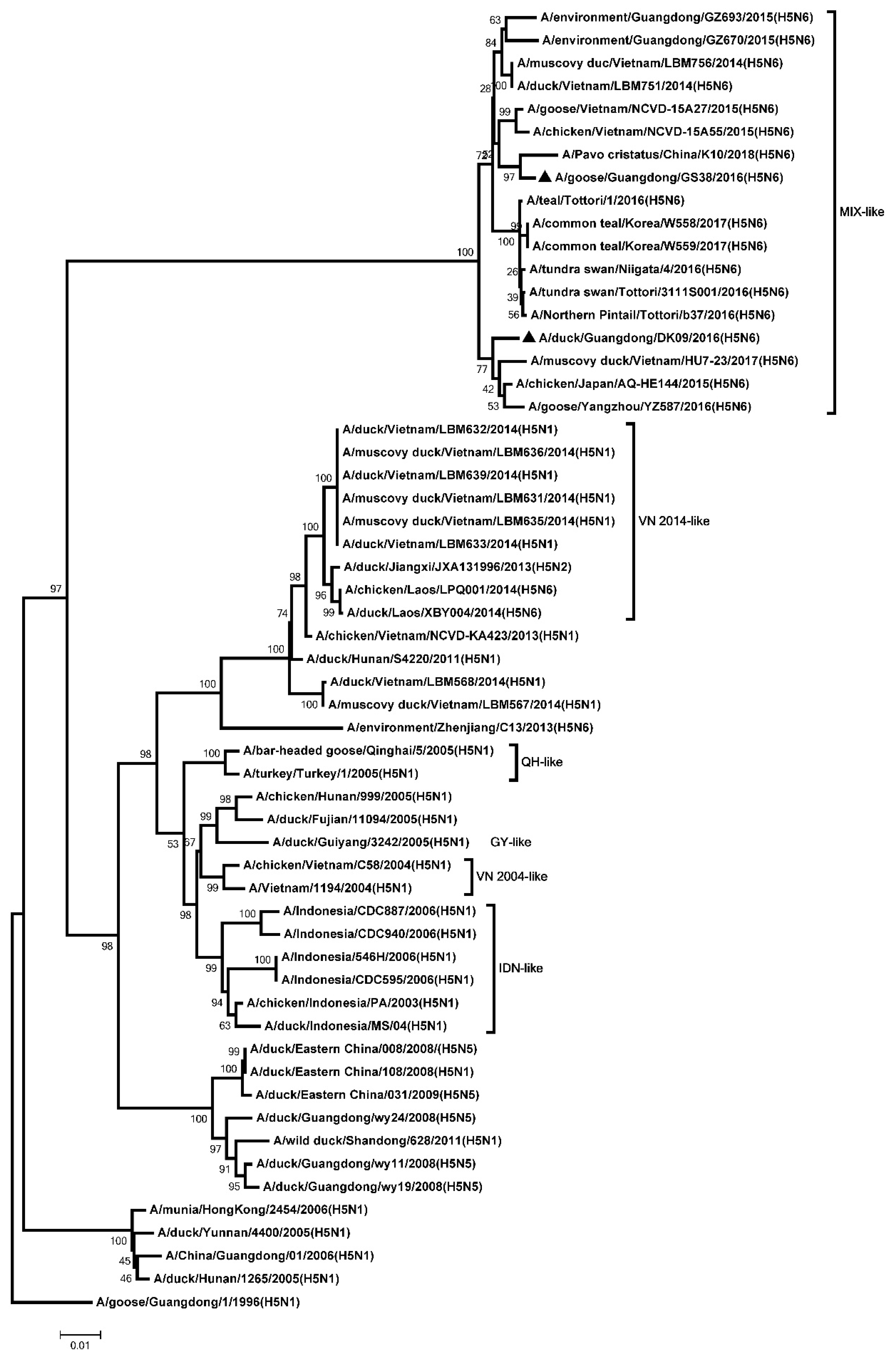
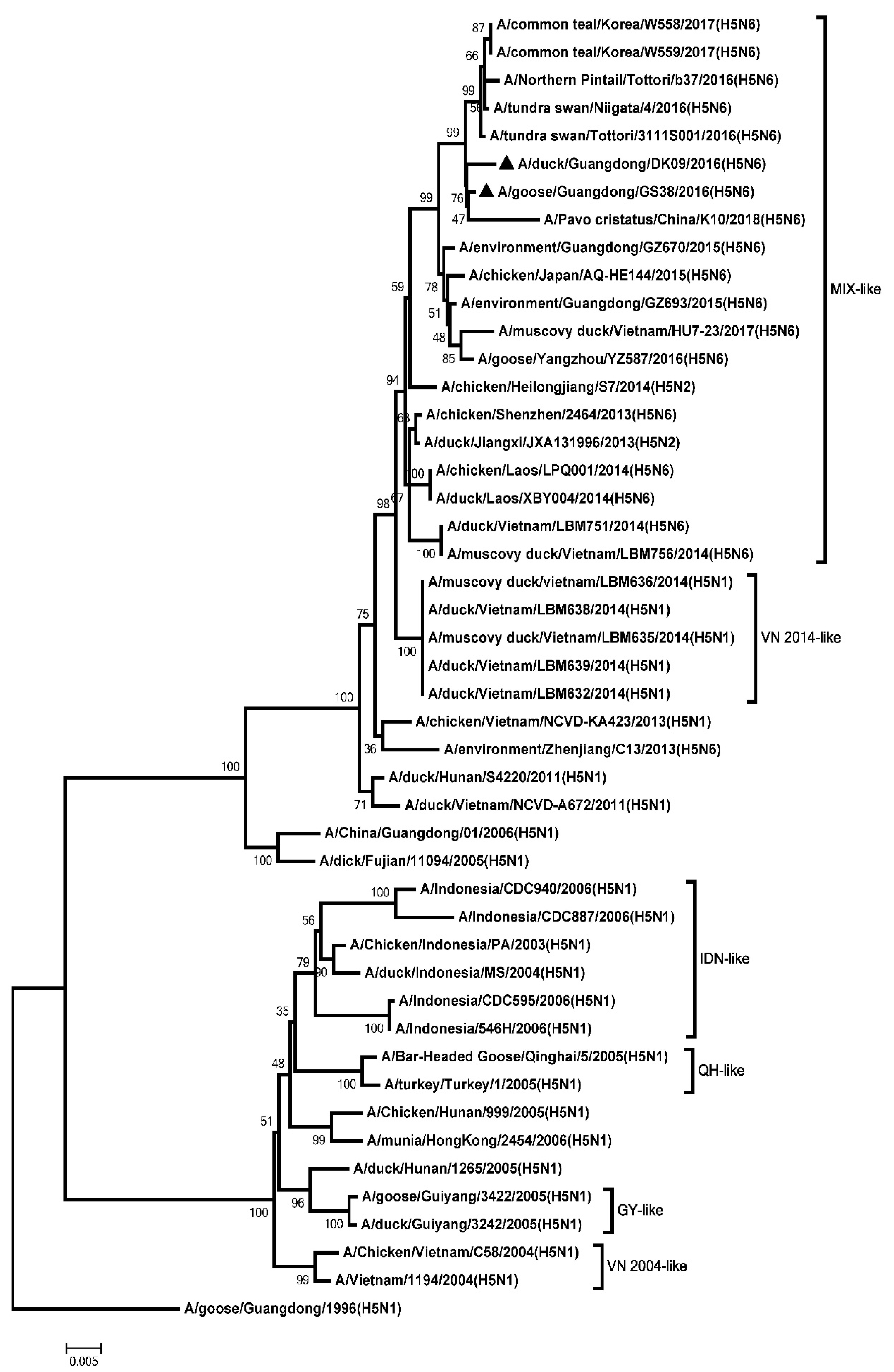
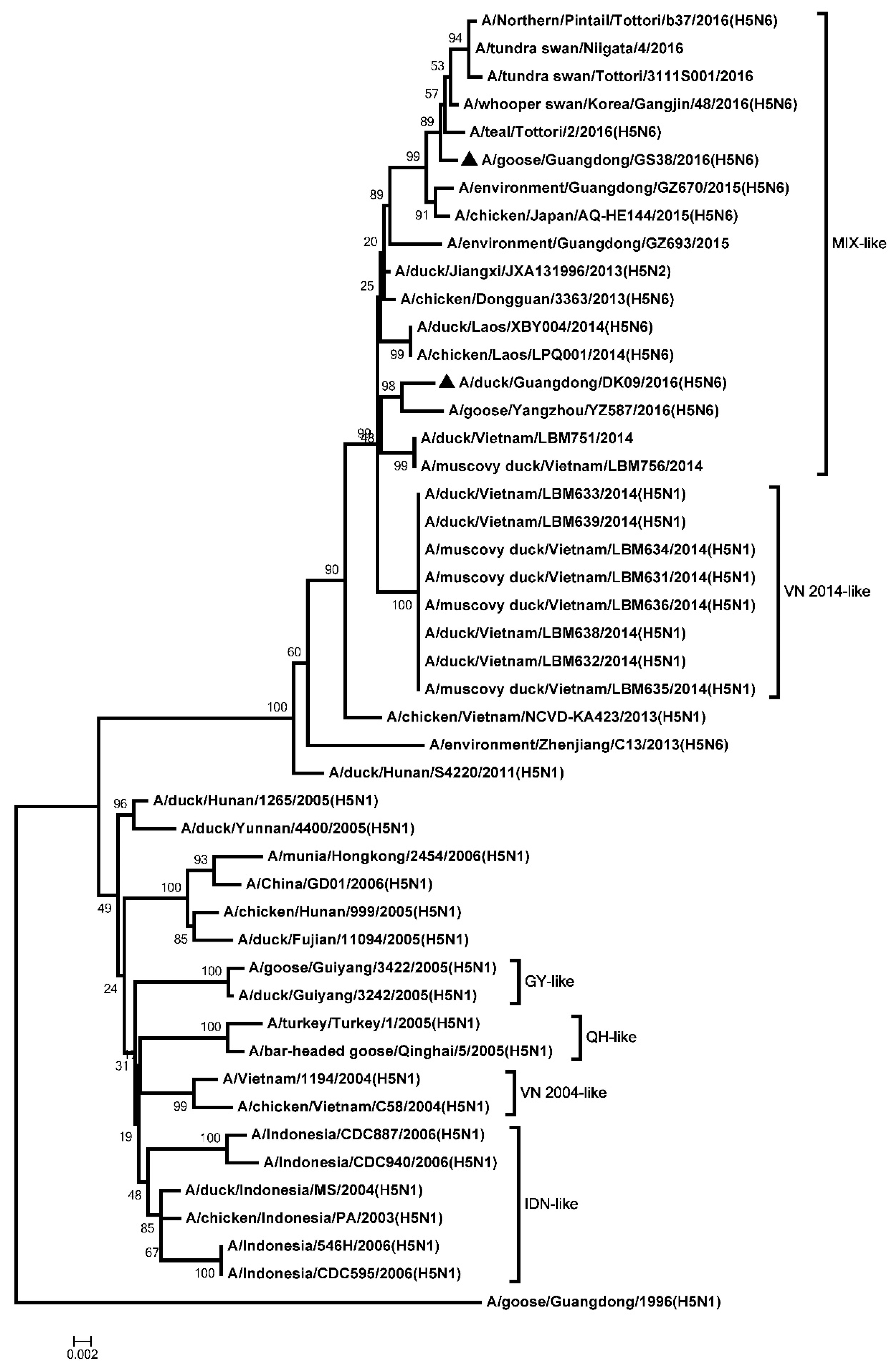
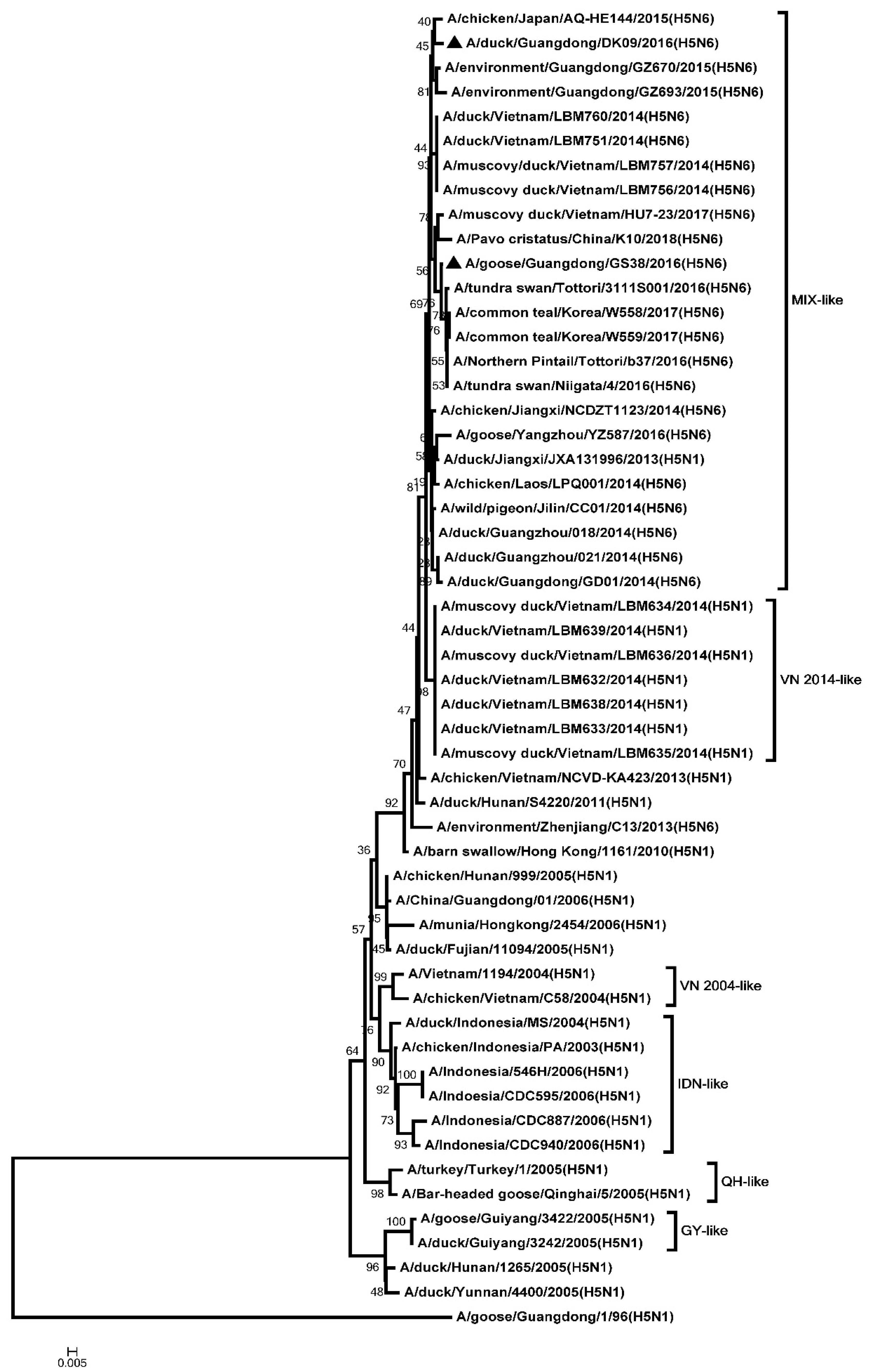
2.2. Phylogenetic Analysis
2.3. Animal Infection Study
2.4. Quantification of Innate Immune-Related Genes in H5N6 Infected Geese
2.5. Statistical Analysis
2.6. Ethics Statement
3. Results
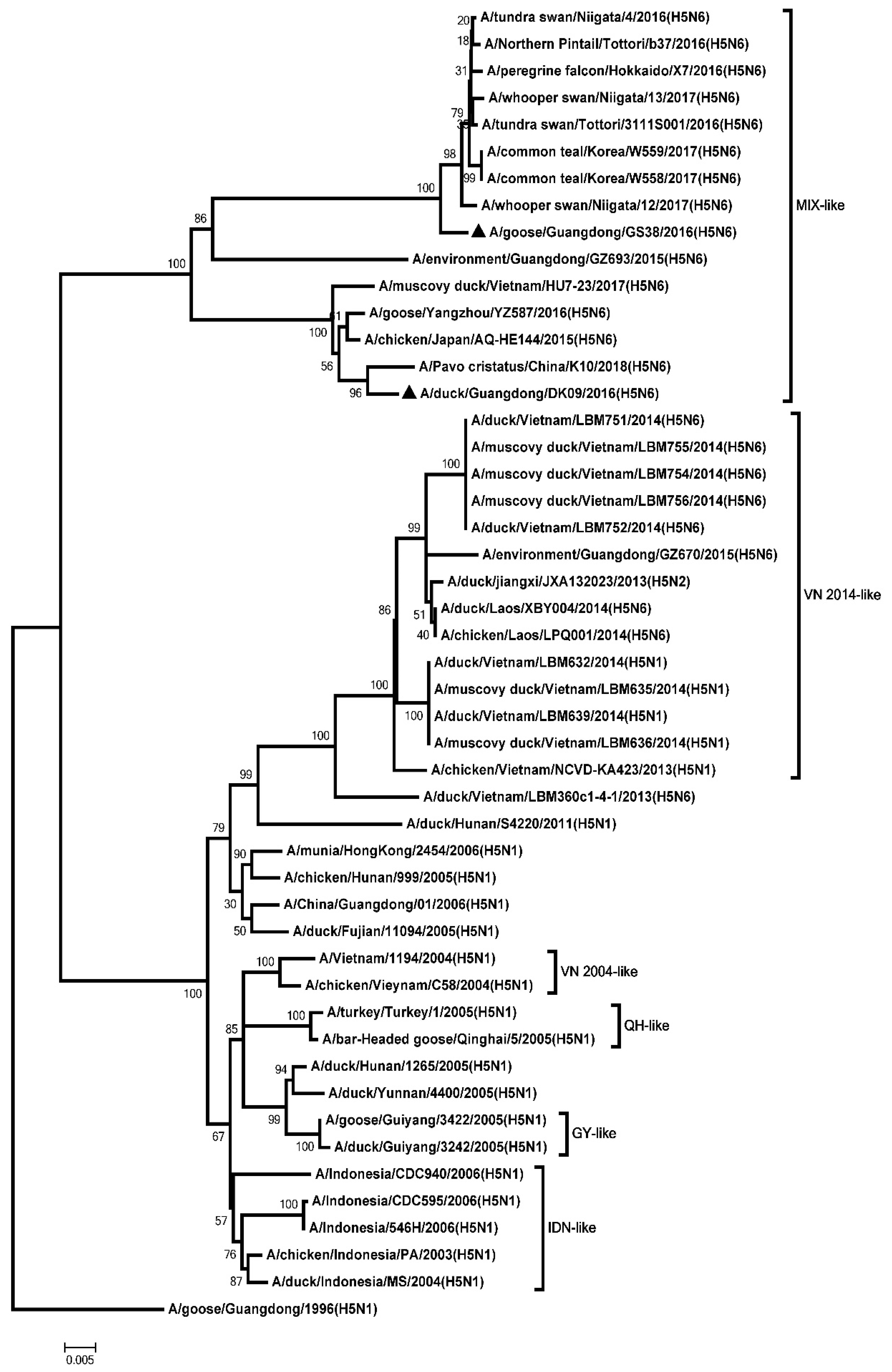
3.1. Genetic Analysis of the Two H5N6 Viruses Isolated from Southern China in 2016
3.2. Pathogenicity, Replication, and Shedding of the Two H5N6 Viruses in Geese
3.3. Transmission of the Two H5N6 Viruses in Geese
3.4. Quantification of PRRs, IFNs, and ISGs in the Lungs of the H5N6 Viruses Infected Geese
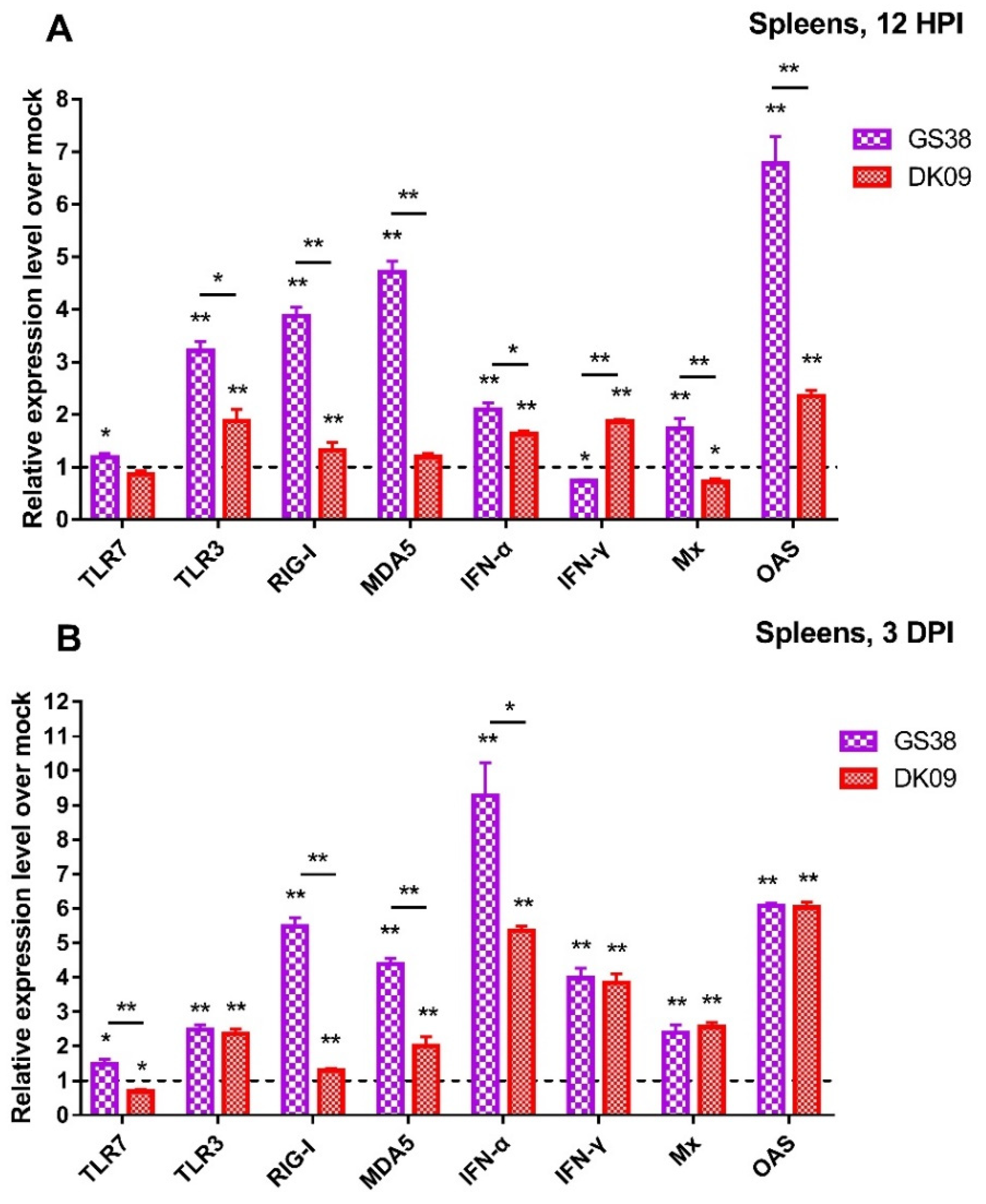
3.5. Quantification of PRRs, IFNs, and ISGs in the Spleens of H5N6 Viruses Infected Geese
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.; Chen, L.M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269–4274. [Google Scholar] [CrossRef]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza A viruses. Plos Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, Y.; Li, Z.; Shi, J.; Shinya, K.; Deng, G.; Qi, Q.; Tian, G.; Fan, S.; Zhao, H.; et al. Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J. Virol. 2006, 80, 5976–5983. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Deng, G.; Kong, H.; Gu, C.; Ma, S.; Yin, X.; Zeng, X.; Cui, P.; Chen, Y.; Yang, H.; et al. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res. 2017, 27, 1409–1421. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.Z.; Deng, G.H.; Ma, S.J.; Zeng, X.Y.; Yin, X.; Li, M.; Zhang, B.; Cui, P.F.; Chen, Y.; Yang, H.L.; et al. Rapid Evolution of H7N9 Highly Pathogenic Viruses that Emerged in China in 2017. Cell Host Microbe. 2018, 24, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Subbarao, K.; Cox, N.J.; Guo, Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 1999, 261, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.S. Influenza A (H5N1) in Hong Kong: an overview. Vaccine 2002, 20 (Suppl. 2), S77–S81. [Google Scholar] [CrossRef]
- WHO. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Available online: https://www.who.int/influenza/vaccines/virus/201909_zoonotic_vaccinevirusupdate.pdf?ua=1 (accessed on 30 September 2019).
- Zhao, G.; Gu, X.; Lu, X.; Pan, J.; Duan, Z.; Zhao, K.; Gu, M.; Liu, Q.; He, L.; Chen, J.; et al. Novel reassortant highly pathogenic H5N2 avian influenza viruses in poultry in China. Plos One 2012, 7, e46183. [Google Scholar] [CrossRef]
- Gu, M.; Liu, W.; Cao, Y.; Peng, D.; Wang, X.; Wan, H.; Zhao, G.; Xu, Q.; Zhang, W.; Song, Q.; et al. Novel Reassortant Highly Pathogenic Avian Influenza (H5N5) Viruses in Domestic Ducks, China. Emerg. Infect. Dis. 2011, 17, 1060–1063. [Google Scholar] [CrossRef]
- Jiao, P.; Cui, J.; Song, Y.; Song, H.; Zhao, Z.; Wu, S.; Qu, N.; Wang, N.; Ouyang, G.; Liao, M. New Reassortant H5N6 Highly Pathogenic Avian Influenza Viruses in Southern China, 2014. Front. Microbiol. 2016, 7, 754. [Google Scholar] [CrossRef]
- Song, Y.; Cui, J.; Song, H.; Ye, J.; Zhao, Z.; Wu, S.; Xu, C.; Jiao, P.; Liao, M. New reassortant H5N8 highly pathogenic avian influenza virus from waterfowl in Southern China. Front. Microbiol. 2015, 6, 1170. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Chen, Q.; Wang, Q.; Chen, J.; Jin, T.; Wong, G.; Quan, C.; Liu, J.; Wu, J.; Yin, R.; et al. Genesis, Evolution and Prevalence of H5N6 Avian Influenza Viruses in China. Cell Host Microbe. 2016, 20, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, S.; Kawaoka, Y. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr. Opin. Immunol. 2011, 23, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Sesma, A.; Marukian, S.; Ebersole, B.J.; Kaminski, D.; Park, M.S.; Yuen, T.; Sealfon, S.C.; Garcia-Sastre, A.; Moran, T.M. Influenza virus evades innate and adaptive immunity via the NS1 protein. J. Virol. 2006, 80, 6295–6304. [Google Scholar] [CrossRef]
- Szretter, K.J.; Gangappa, S.; Lu, X.; Smith, C.; Shieh, W.J.; Zaki, S.R.; Sambhara, S.; Tumpey, T.M.; Katz, J.M. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J. Virol. 2007, 81, 2736–2744. [Google Scholar] [CrossRef]
- Cameron, C.M.; Cameron, M.J.; Bermejo-Martin, J.F.; Ran, L.; Xu, L.; Turner, P.V.; Ran, R.; Danesh, A.; Fang, Y.; Chan, P.K.; et al. Gene expression analysis of host innate immune responses during Lethal H5N1 infection in ferrets. J. Virol. 2008, 82, 11308–11317. [Google Scholar] [CrossRef]
- Suzuki, K.; Okada, H.; Itoh, T.; Tada, T.; Mase, M.; Nakamura, K.; Kubo, M.; Tsukamoto, K. Association of increased pathogenicity of Asian H5N1 highly pathogenic avian influenza viruses in chickens with highly efficient viral replication accompanied by early destruction of innate immune responses. J. Virol. 2009, 83, 7475–7486. [Google Scholar] [CrossRef]
- Wei, L.; Jiao, P.; Song, Y.; Cao, L.; Yuan, R.; Gong, L.; Cui, J.; Zhang, S.; Qi, W.; Yang, S.; et al. Host immune responses of ducks infected with H5N1 highly pathogenic avian influenza viruses of different pathogenicities. Vet. Microbiol. 2013, 166, 386–393. [Google Scholar] [CrossRef]
- Hayashi, T.; Hiromoto, Y.; Chaichoune, K.; Patchimasiri, T.; Chakritbudsabong, W.; Prayoonwong, N.; Chaisilp, N.; Wiriyarat, W.; Parchariyanon, S.; Ratanakorn, P.; et al. Host cytokine responses of pigeons infected with highly pathogenic Thai avian influenza viruses of subtype H5N1 isolated from wild birds. Plos One 2011, 6, e23103. [Google Scholar] [CrossRef]
- Jiao, P.R.; Song, Y.F.; Huang, J.N.; Xiang, C.W.; Cui, J.; Wu, S.Y.; Qu, N.; Wang, N.C.; Ouyang, G.W.; Liao, M. H7N9 Avian Influenza Virus Is Efficiently Transmissible and Induces an Antibody Response in Chickens. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Hulse-Post, D.J.; Sturm-Ramirez, K.M.; Humberd, J.; Seiler, P.; Govorkova, E.A.; Krauss, S.; Scholtissek, C.; Puthavathana, P.; Buranathai, C.; Nguyen, T.D.; et al. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc. Natl. Acad. Sci. USA 2005, 102, 10682–10687. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shi, J.; Zhong, G.; Deng, G.; Tian, G.; Ge, J.; Zeng, X.; Song, J.; Zhao, D.; Liu, L.; et al. Continued evolution of H5N1 influenza viruses in wild birds, domestic poultry, and humans in China from 2004 to 2009. J. Virol. 2010, 84, 8389–8397. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Havers, F.; Chen, E.; Yuan, Z.; Yuan, H.; Ou, J.; Shang, M.; Kang, K.; Liao, K.; Liu, F.; et al. Risk factors for influenza A(H7N9) disease--China, 2013. Clin. Infect. Dis. 2014, 59, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.X.; Cheng, W.; Yu, Z.; Liu, S.L.; Mao, H.Y.; Chen, E.F. Risk factors for avian influenza virus in backyard poultry flocks and environments in Zhejiang Province, China: a cross-sectional study. Infect. Dis. Poverty 2018, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- OIE. Immediate notifications and follow-up reports of highly pathogenic avian influenza (types H5 and H7). Available online: https://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/2020/ (accessed on 24 January 2020).
- Steinhauer, D.A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 1999, 258, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Gambaryan, A.S.; Teneberg, S.; Piskarev, V.E.; Yamnikova, S.S.; Lvov, D.K.; Robertson, J.S.; Karlsson, K.A. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 1997, 233, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Chutinimitkul, S.; Van Riel, D.; Munster, V.J.; Van den Brand, J.M.; Rimmelzwaan, G.F.; Kuiken, T.; Osterhaus, A.D.; Fouchier, R.A.; De Wit, E. In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J. Virol. 2010, 84, 6825–6833. [Google Scholar] [CrossRef]
- Yang, Z.Y.; Wei, C.J.; Kong, W.P.; Wu, L.; Xu, L.; Smith, D.F.; Nabel, G.J. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 2007, 317, 825–828. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lu, B.; Zhou, H.; Suguitan, A.L., Jr.; Cheng, X.; Subbarao, K.; Kemble, G.; Jin, H. Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J. Virol. 2010, 84, 6570–6577. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, Y.; Shinya, K.; Deng, G.; Jiang, Y.; Li, Z.; Guan, Y.; Tian, G.; Li, Y.; Shi, J.; et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. Plos Pathog. 2009, 5, e1000709. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Swayne, D.E.; Thomas, C.; Rameix-Welti, M.A.; Naffakh, N.; Warnes, C.; Altholtz, M.; Donis, R.; Subbarao, K. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J. Virol. 2009, 83, 4704–4708. [Google Scholar] [CrossRef] [PubMed]
- Hurt, A.C.; Selleck, P.; Komadina, N.; Shaw, R.; Brown, L.; Barr, I.G. Susceptibility of highly pathogenic A(H5N1) avian influenza viruses to the neuraminidase inhibitors and adamantanes. Antivir. Res. 2007, 73, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Baek, Y.H.; Song, M.S.; Lee, E.Y.; Kim, Y.I.; Kim, E.H.; Park, S.J.; Park, K.J.; Kwon, H.I.; Pascua, P.N.; Lim, G.J.; et al. Profiling and characterization of influenza virus N1 strains potentially resistant to multiple neuraminidase inhibitors. J. Virol. 2015, 89, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Ilyushina, N.A.; Seiler, J.P.; Rehg, J.E.; Webster, R.G.; Govorkova, E.A. Effect of neuraminidase inhibitor-resistant mutations on pathogenicity of clade 2.2 A/Turkey/15/06 (H5N1) influenza virus in ferrets. Plos Pathog. 2010, 6, e1000933. [Google Scholar] [CrossRef]
- Hatta, M.; Gao, P.; Halfmann, P.; Kawaoka, Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 2001, 293, 1840–1842. [Google Scholar] [CrossRef]
- Taft, A.S.; Ozawa, M.; Fitch, A.; Depasse, J.V.; Halfmann, P.J.; Hill-Batorski, L.; Hatta, M.; Friedrich, T.C.; Lopes, T.J.; Maher, E.A.; et al. Identification of mammalian-adapting mutations in the polymerase complex of an avian H5N1 influenza virus. Nat. Commun. 2015, 6, 7491. [Google Scholar] [CrossRef]
- Yamada, S.; Hatta, M.; Staker, B.L.; Watanabe, S.; Imai, M.; Shinya, K.; Sakai-Tagawa, Y.; Ito, M.; Ozawa, M.; Watanabe, T.; et al. Biological and structural characterization of a host-adapting amino acid in influenza virus. Plos Pathog. 2010, 6, e1001034. [Google Scholar] [CrossRef]
- Li, Z.; Chen, H.; Jiao, P.; Deng, G.; Tian, G.; Li, Y.; Hoffmann, E.; Webster, R.G.; Matsuoka, Y.; Yu, K. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 2005, 79, 12058–12064. [Google Scholar] [CrossRef]
- Le, Q.M.; Sakai-Tagawa, Y.; Ozawa, M.; Ito, M.; Kawaoka, Y. Selection of H5N1 influenza virus PB2 during replication in humans. J. Virol. 2009, 83, 5278–5281. [Google Scholar] [CrossRef]
- Feng, X.X.; Wang, Z.; Shi, J.Z.; Deng, G.H.; Kong, H.H.; Tao, S.Y.; Li, C.Y.; Liu, L.L.; Guan, Y.T.; Chen, H.L. Glycine at Position 622 in PB1 Contributes to the Virulence of H5N1 Avian Influenza Virus in Mice. J. Virol. 2016, 90, 1872–1879. [Google Scholar] [CrossRef]
- Lan, Y.; Zhang, Y.; Dong, L.B.; Wang, D.Y.; Huang, W.J.; Xin, L.; Yang, L.M.; Zhao, X.A.; Li, Z.; Wang, W.; et al. A comprehensive surveillance of adamantane resistance among human influenza A virus isolated from mainland China between 1956 and 2009. Antivir 2010, 15, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.H.; Hoffmann, E.; Webster, R.G. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 2002, 8, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.R.; Tian, G.B.; Li, Y.B.; Deng, G.H.; Jiang, Y.P.; Liu, C.; Liu, W.L.; Bu, Z.G.; Kawaoka, Y.; Chen, H.L. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice (vol 82, pg 1146, 2008). J. Virol. 2008, 82, 4190. [Google Scholar] [CrossRef]
- Yuan, R.Y.; Cui, J.; Zhang, S.; Cao, L.; Liu, X.K.; Kang, Y.F.; Song, Y.F.; Gong, L.; Jiao, P.R.; Liao, M. Pathogenicity and transmission of H5N1 avian influenza viruses in different birds. Vet. Microbiol. 2014, 168, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.M.; Jiao, P.R.; Yuan, R.Y.; Song, Y.F.; Cui, P.F.; Guo, X.C.; Zheng, B.F.; Jia, W.X.; Qi, W.B.; Ren, T.; et al. Goose Toll-like receptor 7 (TLR7), myeloid differentiation factor 88 (MyD88) and antiviral molecules involved in anti-H5N1 highly pathogenic avian influenza virus response. Vet. Immunol. Immunop. 2013, 153, 99–106. [Google Scholar] [CrossRef]
- Berhane, Y.; Kobasa, D.; Embury-Hyatt, C.; Pickering, B.; Babiuk, S.; Joseph, T.; Bowes, V.; Suderman, M.; Leung, A.; Cottam-Birt, C.; et al. Pathobiological Characterization of a Novel Reassortant Highly Pathogenic H5N1 Virus Isolated in British Columbia, Canada, 2015. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Gao, W.; Zu, Z.; Liu, J.; Song, J.; Wang, X.; Wang, C.; Liu, L.; Tong, Q.; Wang, M.; Sun, H.; et al. Prevailing I292V PB2 mutation in avian influenza H9N2 virus increases viral polymerase function and attenuates IFN-beta induction in human cells. J. Gen. Virol. 2019, 100, 1273–1281. [Google Scholar] [CrossRef]
- Xiao, C.; Ma, W.; Sun, N.; Huang, L.; Li, Y.; Zeng, Z.; Wen, Y.; Zhang, Z.; Li, H.; Li, Q.; et al. PB2-588 V promotes the mammalian adaptation of H10N8, H7N9 and H9N2 avian influenza viruses. Sci. Rep. 2016, 6, 19474. [Google Scholar] [CrossRef]
- Qu, N.; Zhao, B.; Chen, Z.; He, Z.; Li, W.; Liu, Z.; Wang, X.; Huang, J.; Zhang, Y.; He, W.; et al. Genetic characteristics, pathogenicity and transmission of H5N6 highly pathogenic avian influenza viruses in Southern China. Transbound Emerg. Dis. 2019, 66, 2411–2425. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, J.; Huang, J.; Li, W.; Liu, Z.; He, Z.; Chen, Z.; He, W.; Zhao, B.; Qin, Z.; et al. Immune-Related Gene Expression in Ducks Infected With Waterfowl-Origin H5N6 Highly Pathogenic Avian Influenza Viruses. Front. Microbiol. 2019, 10, 1782. [Google Scholar] [CrossRef]
- Wei, L.M.; Jiao, P.R.; Song, Y.F.; Han, F.; Cao, L.; Yang, F.; Ren, T.; Liao, M. Identification and expression profiling analysis of goose melanoma differentiation associated gene 5 (MDA5) gene. Poult. Sci. 2013, 92, 2618–2624. [Google Scholar] [CrossRef] [PubMed]
- Barber, M.R.; Aldridge, J.R., Jr.; Webster, R.G.; Magor, K.E. Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci. USA 2010, 107, 5913–5918. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.J.; Ding, N.; Ding, S.S.; Yu, S.Q.; Meng, C.H.; Chen, H.J.; Qiu, X.S.; Zhang, S.L.; Yu, Y.; Zhan, Y.; et al. Goose RIG-I functions in innate immunity against Newcastle disease virus infections. Mol. Immunol. 2013, 53, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, W.; Wu, Z.; Zhang, J.Y.; Wang, M.S.; Jia, R.Y.; Zhu, D.K.; Liu, M.F.; Sun, K.F.; Yang, Q.; et al. Goose Mx and OASL Play Vital Roles in the Antiviral Effects of Type I, II and III Interferon against Newly Emerging Avian Flavivirus. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, A.; Chen, S.; Wu, Z.; Zhang, J.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Yang, Q.; et al. Differential immune-related gene expression in the spleens of duck Tembusu virus-infected goslings. Vet. Microbiol. 2017, 212, 39–47. [Google Scholar] [CrossRef]
- Penski, N.; Hartle, S.; Rubbenstroth, D.; Krohmann, C.; Ruggli, N.; Schusser, B.; Pfann, M.; Reuter, A.; Gohrbandt, S.; Hundt, J.; et al. Highly Pathogenic Avian Influenza Viruses Do Not Inhibit Interferon Synthesis in Infected Chickens but Can Override the Interferon-Induced Antiviral State. J. Virol. 2011, 85, 7730–7741. [Google Scholar] [CrossRef]
- Gao, S.; Kang, Y.; Yuan, R.; Ma, H.; Xiang, B.; Wang, Z.; Dai, X.; Wang, F.; Xiao, J.; Liao, M.; et al. Immune Responses of Chickens Infected with Wild Bird-Origin H5N6 Avian Influenza Virus. Front. Microbiol. 2017, 8, 1081. [Google Scholar] [CrossRef]



| Genes | Forward Primer Sequences (5′–3′) | Reverse Primer Sequences (5′–3′) | Accession No. |
|---|---|---|---|
| TLR7 | CCTTGTATTCCCTGCCCCAA | ACTTCAAAAACTGAGCATCT | KJ022638 |
| TLR3 | CGGTCGGAAAGCAATGTCAA | AGCTGATTATGAGAAATGTC | KP238287 |
| RIG-I | AGCACCTGACAGCCAAAT | AGTGCGAGTCTGTGGGTT | JF804977 |
| MDA5 | TGCTGTAGTGGAGGATTTG | CTGCTCTGTCCCAGGTTT | JX976550 |
| IFN-α | CAGCACCACATCCACCAC | TACTTGTTGATGCCGAGGT | AY524422 |
| IFN-γ | TGAGCCAGATTGTTTCCC | CAGGTCCACGAGGTCTTT | AY524421 |
| Mx | TTCACAGCAATGGAAAGGGA | ATTAGTGTCGGGTCTGGGA | KU247604 |
| OASL | CTGGCGGTGAAGAATACGGT | CGTCTTGAAGACGCGGATTT | KU058695 |
| GAPDH | CATCTTCCAGGAGCGCGACC | AGACACCGGTGGACTCCACA | MG674174 |
| Strains | Group | Illness a | Death | MDT |
|---|---|---|---|---|
| GS38 | Inoculated b | 9/9 | 8/9 | 6.4 |
| Contact c | 4/5 | 4/5 | 8.8 | |
| DK09 | Inoculated | 9/9 | 8/9 | 10.4 |
| Contact | 3/5 | 3/5 | 12 |
| Strains | Time | Virus Replication Titers in Organs of Inoculated Geese (log10EID50/0.1 mL) a | ||||||
|---|---|---|---|---|---|---|---|---|
| Liver | Spleen | Lung | Kidney | Intestine | Pancreas | Bursa of Fabricius | ||
| GS38 | 12HPI | 2.92 ± 0.58 | 1.58 ± 0.14 | 2.25 ± 0.75 | < 1.5 | 1.58 ± 0.14 | 1.75 ± 0.43 | < 1.5 |
| 3 DPI | 1.83 ± 0.58 | 4.17 ± 1.53 | 2.83 ± 1.28 | 3.92 ± 1.51 | 3.75 ± 1.98 | 2.83 ± 1.89 | 3.67 ± 1.23 | |
| DK09 | 12HPI | 1.58 ± 0.14 | <1.5 | 2.17 ± 1.15 | 2.33 ± 1.44 | 2.92 ± 0.38 | 1.92 ± 1.42 | 2 ± 0.87 |
| 3 DPI | 4.75 ± 2.95 | 3.92 ± 3.17 | 5.33 ± 1.80 | 2.08 ± 0.52 | 3.17 ± 2.89 | 2.75 ± 2.15 | 3.33 ± 1.67 | |
| Strain | Infection Sample | 3 DPI | 5 DPI | 7 DPI | 9 DPI | 11 DPI | 13 DPI | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T | C | T | C | T | C | T | C | T | C | ||
| GS38 | Inoculated a | 2/12 | 1/12 | 5/6 | 5/6 | 3/3 | 2/3 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Contacted b | 1/5 | 1/5 | 4/5 | 3/5 | 2/3 | 2/3 | 1/3 | 0/3 | 0/2 | 0/2 | 0/1 | 0/1 | |
| DK09 | Inoculated | 2/12 | 1/12 | 6/9 | 2/9 | 5/7 | 6/7 | 2/6 | 2/6 | 0/5 | 2/5 | 0/1 | 0/1 |
| Contacted | 0/5 | 0/5 | 1/5 | 1/5 | 2/5 | 1/5 | 3/5 | 2/5 | 0/4 | 0/4 | 0/2 | 0/2 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Huang, J.; Huang, Q.; Zhang, J.; Liu, J.; Xue, Q.; Li, W.; Liao, M.; Jiao, P. Host Innate Immune Response of Geese Infected with Clade 2.3.4.4 H5N6 Highly Pathogenic Avian Influenza Viruses. Microorganisms 2020, 8, 224. https://doi.org/10.3390/microorganisms8020224
Wu S, Huang J, Huang Q, Zhang J, Liu J, Xue Q, Li W, Liao M, Jiao P. Host Innate Immune Response of Geese Infected with Clade 2.3.4.4 H5N6 Highly Pathogenic Avian Influenza Viruses. Microorganisms. 2020; 8(2):224. https://doi.org/10.3390/microorganisms8020224
Chicago/Turabian StyleWu, Siyu, Jianni Huang, Qiwen Huang, Junsheng Zhang, Jing Liu, Qian Xue, Weiqiang Li, Ming Liao, and Peirong Jiao. 2020. "Host Innate Immune Response of Geese Infected with Clade 2.3.4.4 H5N6 Highly Pathogenic Avian Influenza Viruses" Microorganisms 8, no. 2: 224. https://doi.org/10.3390/microorganisms8020224
APA StyleWu, S., Huang, J., Huang, Q., Zhang, J., Liu, J., Xue, Q., Li, W., Liao, M., & Jiao, P. (2020). Host Innate Immune Response of Geese Infected with Clade 2.3.4.4 H5N6 Highly Pathogenic Avian Influenza Viruses. Microorganisms, 8(2), 224. https://doi.org/10.3390/microorganisms8020224






