Diversity and Bioactive Potential of Actinobacteria from Unexplored Regions of Western Ghats, India
Abstract
1. Introduction
2. Materials and Methods
2.1. Site, Sampling, Pre-Treatment, and Selective Isolation
2.2. Morphological Characterization of Isolates
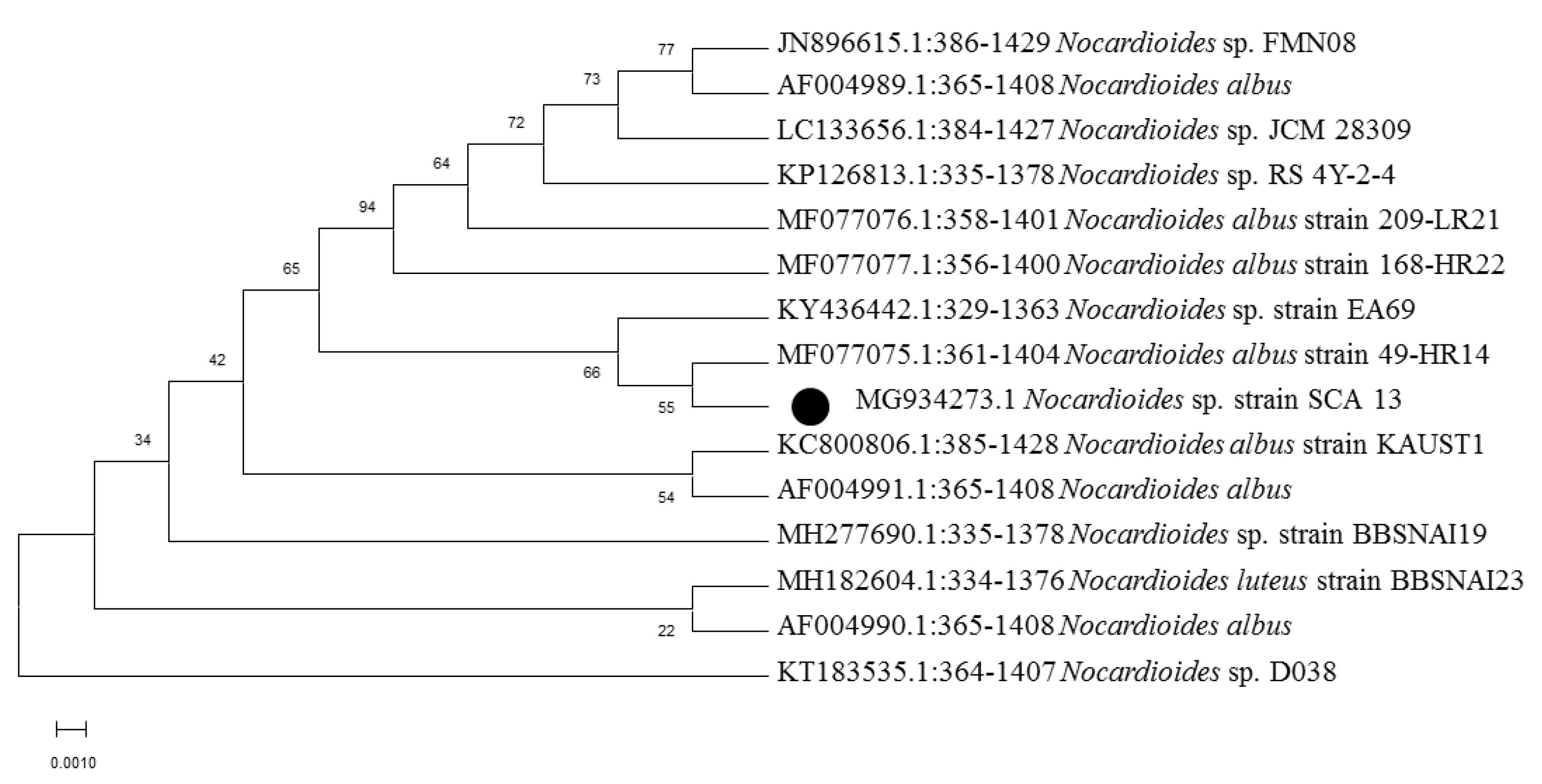
2.3. Molecular Identification and Phylogenetic Analysis
2.4. Isolates Cultivation and Metabolites Extraction
2.5. Antimicrobial Susceptibility Test
2.5.1. Disc Diffusion Assay
2.5.2. Minimum Inhibitory Concentration (MIC)
2.6. Antioxidant Assays
3. Results and Discussion
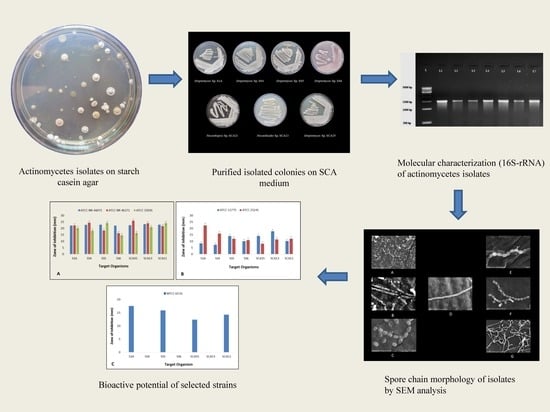
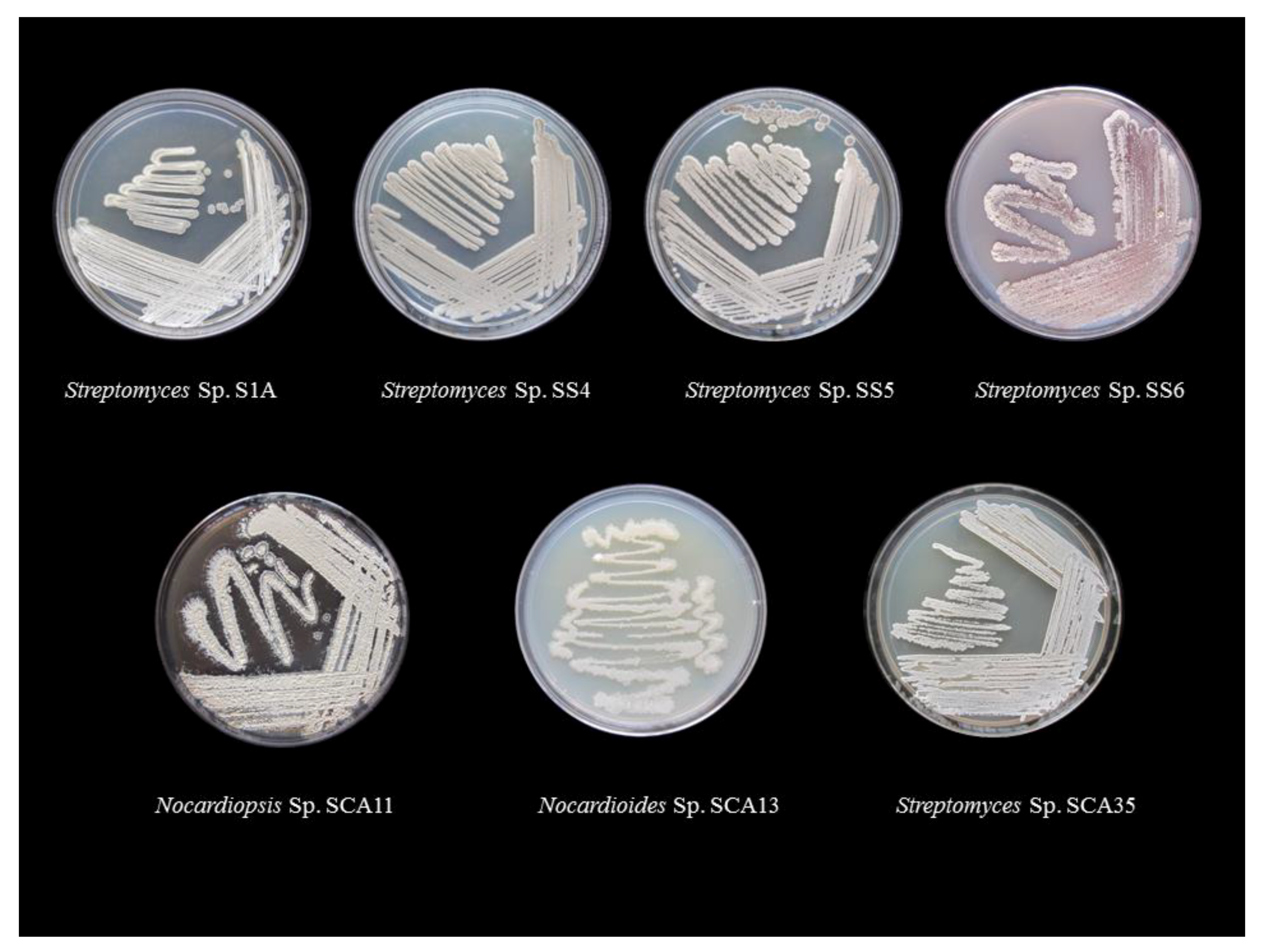
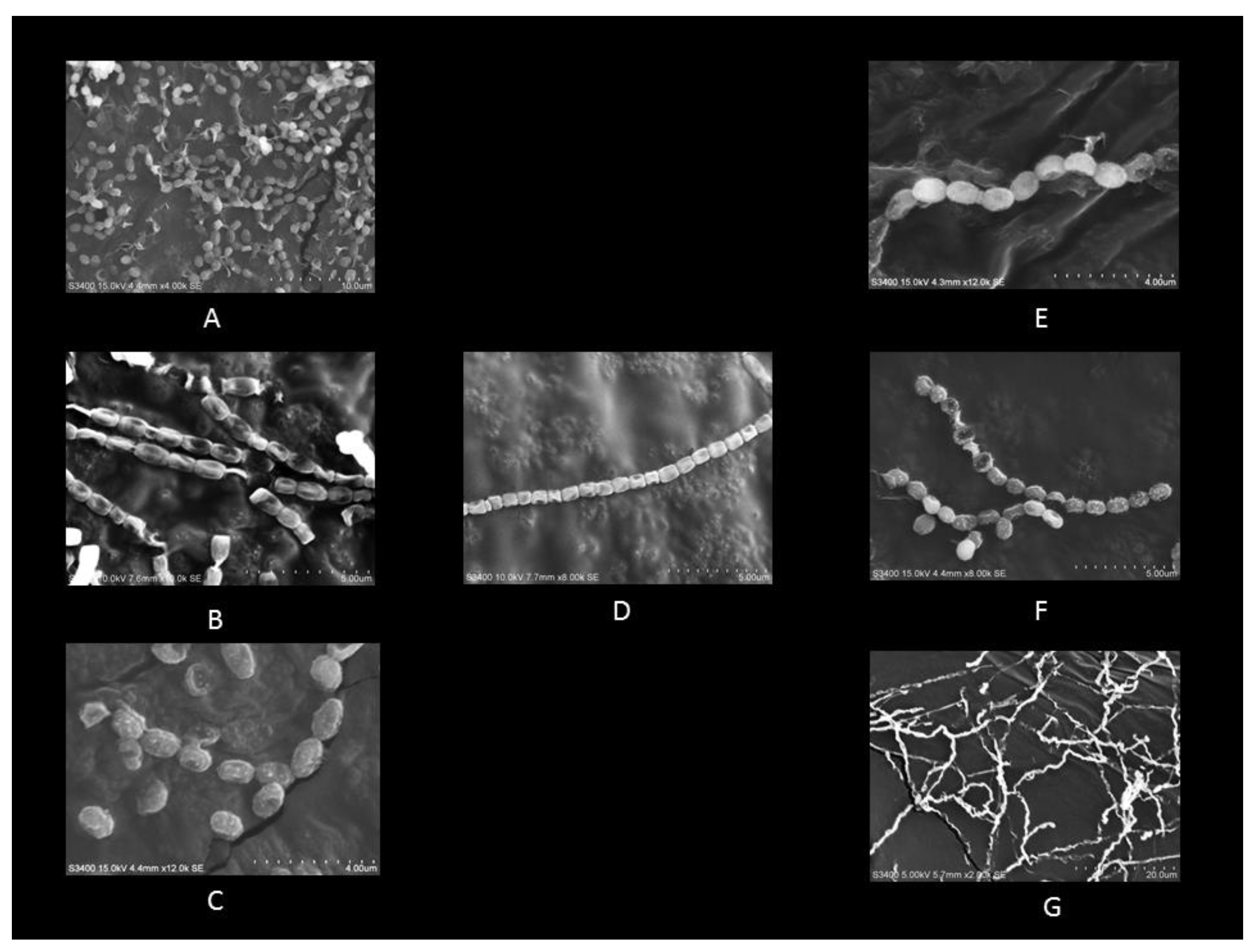
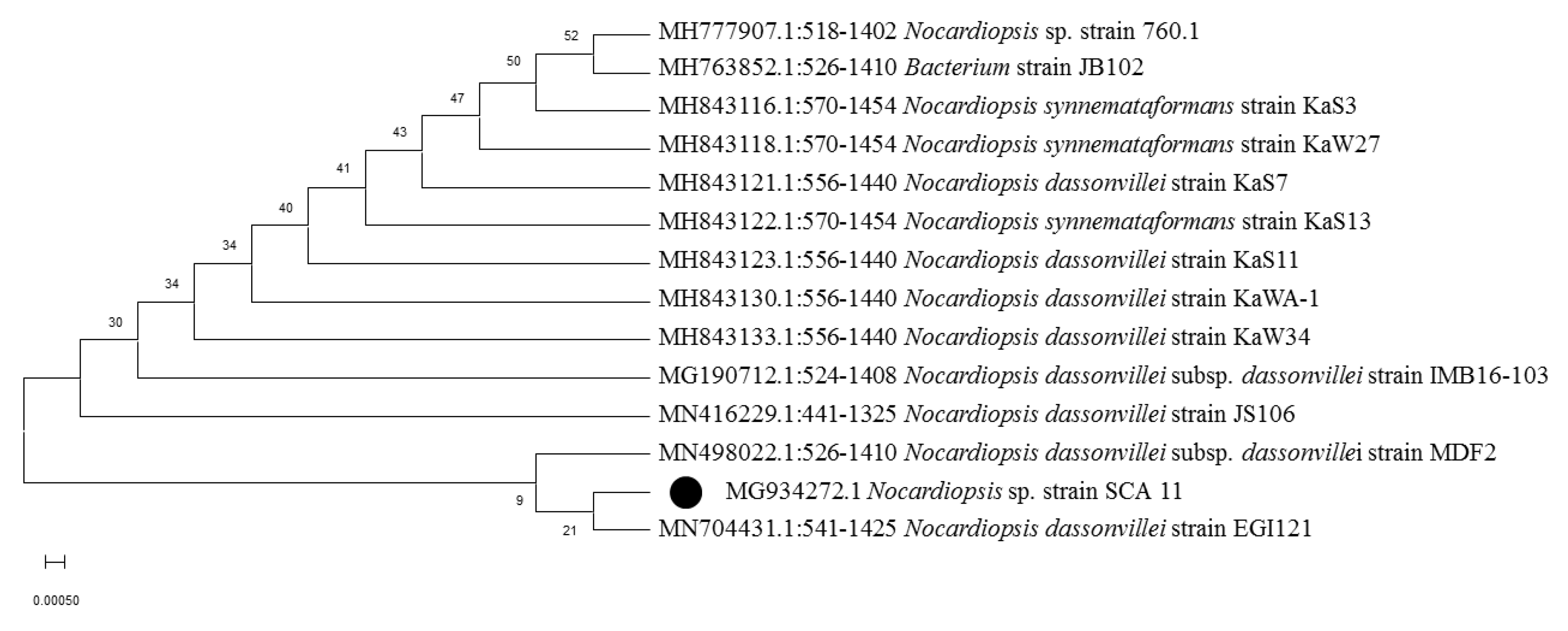
3.1. Isolation, Morphological, and Molecular Characterization of Actinobacterial Isolates
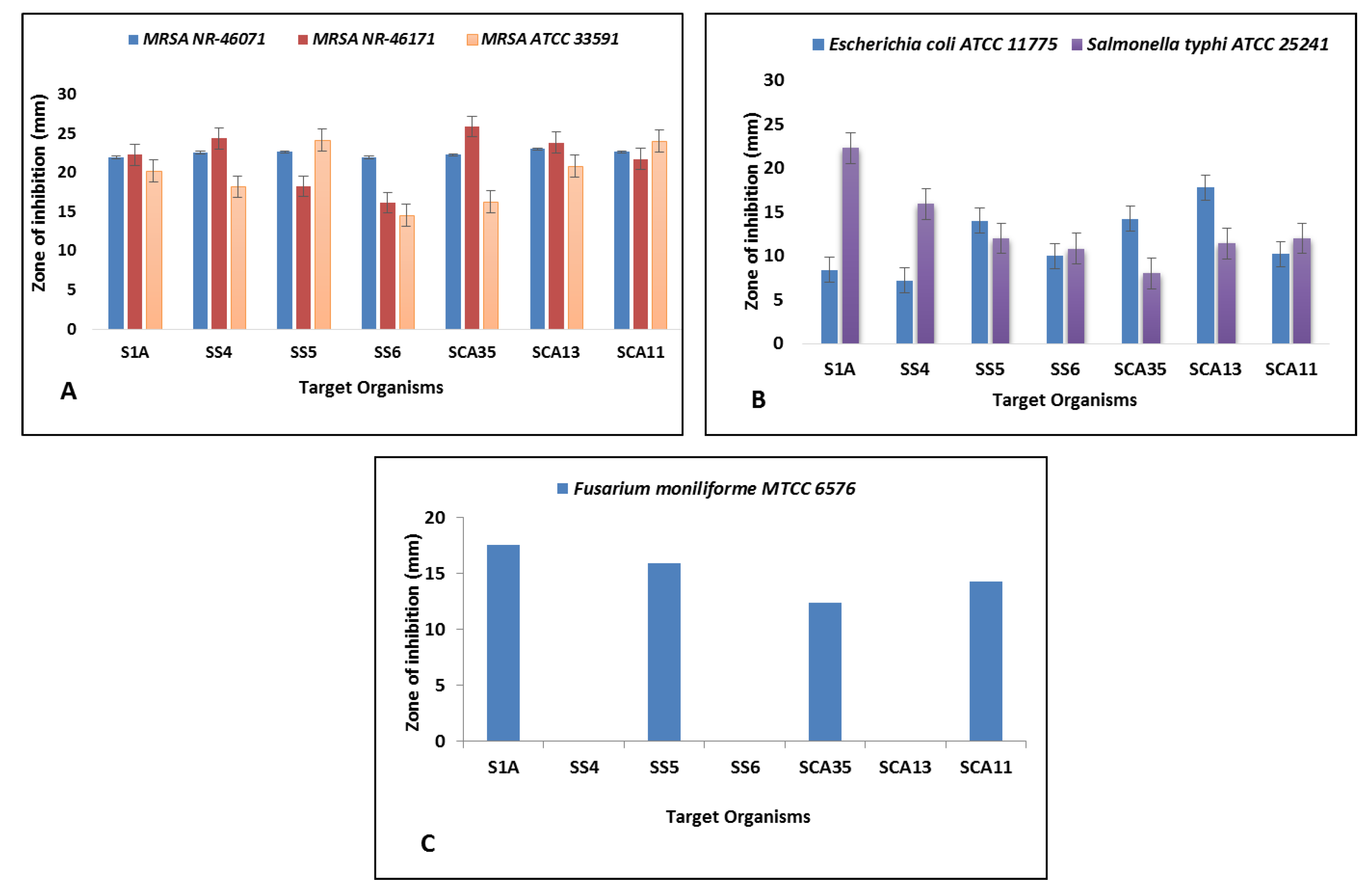
3.2. Antimicrobial and Antioxidant Potential of Isolates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, H.; Xiao, L.; Wei, J.; Schmitz, J.C.; Liu, M.; Wang, C.; Cheng, L.; Wu, N.; Chen, L.; Zhang, Y.; et al. Identification of Streptomyces sp. nov. WH26 producing cytotoxic compounds isolated from marine solar saltern in China. World J. Microbiol. Biotechnol. 2013, 29, 1271–1278. [Google Scholar] [CrossRef]
- David, M.Z.; Daum, R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, A.G.; Dhanarajan, G.; Nema, S.; Sen, R.; Roy, U. An antimicrobial metabolite from Bacillus sp.: Significant activity against pathogenic bacteria including multidrug-resistant clinical strains. Front. Microbiol. 2015, 6, 1335. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Jones, J.D.; Dangl, J.L. Reactive oxygen species signalling in response to pathogens. Plant. Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Giacco, F.; Brownlee, M. Oxidative stress and diabetic complications. Circ. Res. 2010, 107, 1058–1070. [Google Scholar] [CrossRef] [PubMed]
- Fearon, I.M.; Faux, S.P. Oxidative stress and cardiovascular disease: Novel tools give (free) radical insight. J. Mol. Cell. Cardiol. 2009, 47, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Barnham, K.J.; Masters, C.L.; Bush, A.I. Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 2004, 3, 205–214. [Google Scholar] [CrossRef]
- Zhang, H.; Tsao, R. Dietary polyphenols, oxidative stress and antioxidant and anti-inflammatory effects. Curr. Opin. Food Sci. 2016, 8, 33–42. [Google Scholar] [CrossRef]
- Carocho, M.; Ferreira, I.C. A review on antioxidants, prooxidants and related controversy: Natural and synthetic compounds, screening and analysis methodologies and future perspectives. Food Chem. Toxicol. 2013, 51, 15–25. [Google Scholar] [CrossRef]
- Thompson, C.C.; Kruger, R.H.; Thompson, F.L. Unlocking marine biotechnology in the developing world. Trends Biotechnol. 2017, 35, 1119–1121. [Google Scholar] [CrossRef]
- Berdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xing, K.; Jiang, J.H.; Xu, L.H.; Li, W.J. Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 2011, 89, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol. Res. 2013, 169, 262–278. [Google Scholar] [CrossRef] [PubMed]
- Nagpure, A.; Choudhary, B.; Kumar, S.; Gupta, R.K. Isolation and characterization of chitinolytic Streptomyces sp. MT7 and its antagonism towards wood-rotting fungi. Ann. Microbiol. 2014, 64, 531–541. [Google Scholar] [CrossRef]
- Che, Q.; Zhu, T.; Qi, X.; Mandi, A.; Mo, X.; Li, J. Hybrid isoprenoids from a reed rhizosphere soil derived actinomycete Streptomyces sp. CHQ-64. Org. Lett. 2012, 14, 3438–3441. [Google Scholar] [CrossRef]
- Ibeyaima, A.; Rana, J.; Dwivedi, A.K.; Saini, N.; Gupta, S.; Sarethy, I.P. Pseudonocardiaceae sp. TD-015 from the Thar Desert, India: Antimicrobial activity and identification of antimicrobial compounds. Curr. Bioact. Compd. 2017, 14, 112–118. [Google Scholar] [CrossRef]
- Gautham, S.A.; Shobha, K.S.; Onkarappa, R.; Kekuda, P.T.R. Isolation, characterization and antimicrobial potential of Streptomyces species from Western Ghats of Karnataka, India. Res. J. Pharm. Technol. 2012, 5, 233–238. [Google Scholar]
- Nampoothiri, M.K.; Ramkumar, B.; Pandey, A. Western Ghats of India: Rich source of microbial diversity. J. Sci. Ind. Res. 2013, 72, 617–623. [Google Scholar]
- Ganesan, P.; Anand, S.; Sivanandhan, S.; David, R.H.A.; Paulraj, M.G.; Al-Dhabi, N.A.; Ignacimuthu, S. Larvicidal, ovicidal and repellent activities of Streptomyces enissocaesilis (S12-17) isolated from Western Ghats of Tamil Nadu, India. J. Entomol. 2018, 6, 1828–1835. [Google Scholar]
- Ganesan, P.; Reegan, A.D.; David, R.H.A.; Gandhi, M.R.; Paulraj, M.G.; Al-Dhabi, N.A.; Ignacimuthu, S. Antimicrobial activity of some actinomycetes from Western Ghats of Tamil Nadu, India. Alexandria J. 2017, 53, 101–110. [Google Scholar] [CrossRef]
- Siddharth, S.; Vittal, R.R. Evaluation of antimicrobial, enzyme inhibitory, antioxidant and cytotoxic activities of partially purified volatile metabolites of marine Streptomyces sp. S2A. Microorganisms 2018, 6, 72. [Google Scholar] [CrossRef]
- Siddharth, S.; Vittal, R.R. Isolation and characterization of bioactive compounds with antibacterial, antioxidant and enzyme inhibitory activities from marine-derived rare actinobacteria, Nocardiopsis sp. SCA21. Microb. Pathog. 2019, 103775. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.L.; Tan, L.T.; Palanisamy, U.D.; Abd Malek, S.N.; Yin, W.F.; Chan, K.G. Streptomyces antioxidans sp. nov., a novel mangrove soil actinobacterium with antioxidative and neuroprotective potentials. Front. Microbiol. 2016, 7, 899. [Google Scholar] [CrossRef] [PubMed]
- Ceylan, O.; Okmen, G.; Ugur, A. Isolation of soil Streptomyces as source of antibiotics active against antibiotic-resistant bacteria. EurAsian J. Biosci. 2008, 2, 73–82. [Google Scholar]
- Junaid, S.; Dileep, N.; Rakesh, K.N.; Kekuda, P.T.R. Antimicrobial and antioxidant efficacy of Streptomyces species SRDP-TK-07 isolated from a soil of Talakaveri, Karnataka, India. Pharmanest 2013, 4, 736–750. [Google Scholar]
- Bennur, T.; Kumar, A.R.; Zinjarde, S.; Javdekar, V. Nocardiopsis species as potential sources of diverse and novel extracellular enzymes. Appl Microbiol Biotechnol. 2014, 98, 9173–9185. [Google Scholar] [CrossRef]
- Fiedler, H.P.; Bruntner, C.; Bull, A.T. Marine actinomycetes as a source of novel secondary metabolites. Ant. V. Leeuw. 2004, 87, 37–42. [Google Scholar] [CrossRef]
- Sengupta, S.; Pramanik, A.; Ghosh, A.; Bhattacharyya, M. Antimicrobial activities of actinomycetes isolated from unexplored regions of Sundarbans mangrove ecosystem. BMC Microbiol. 2015, 15, 170. [Google Scholar] [CrossRef]
- Satheeja, S.; Jebakumar, S.R.D. Phylogenetic analysis and antimicrobial activities of Streptomyces isolates from mangrove sediment. J. Basic Microbiol. 2011, 51, 71–79. [Google Scholar] [CrossRef]
- Vu, H.T.; Nguyen, D.T.; Nguyen, H.Q. Antimicrobial and cytotoxic properties of bioactive metabolites produced by Streptomyces cavourensis YBQ59 isolated from Cinnamomum cassia Prels in Yen Bai province of Vietnam. Curr. Microbiol. 2018, 75, 1247–1255. [Google Scholar] [CrossRef]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.R. Production of induced secondary metabolites by a co-culture of sponge-associated actinomycetes, Actinokineospora sp. EG49 and Nocardiopsis sp. RV163. Mar. Drugs 2014, 12, 3046–3059. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, V.; Martín, J.; Sarmiento-Vizcaíno, A. Anthracimycin B, a potent Antibiotic against gram-positive bacteria isolated from cultures of the deep-sea actinomycetes Streptomyces cyaneofuscatus M-169. Mar. Drugs 2018, 16, 406. [Google Scholar] [CrossRef] [PubMed]
- Shobha, K.S.; Onkarappa, R. In vitro susceptibility of C. albicans and C. neoformens to potential metabolites from Streptomycetes. Indian J. Microbiol. 2011, 51, 445–449. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Manasa, M.; Poornima, G.; Abhipsa, V.; Rekha, C.; Prashith, K.T.R.; Onkarappa, R.; Mukunda, S. Antimicrobial and antioxidant potential of Streptomyces sp. RAMPP-065 isolated from Kudremukh soil, Karnataka, India. Sci. Technol. Arts Res. J. 2012, 1, 39–44. [Google Scholar] [CrossRef][Green Version]
- Gautham, S.A.; Onkarappa, R. Pharmacological activities of metabolite from Streptomyces fradiae strain GOS1. Int. J. Chem. 2013, 11, 583–590. [Google Scholar]
- Chang, H.B.; Kim, J.-H. Antioxidant properties of dihydroherbimycin A from a newly isolated Streptomyces sp. Biotechnol. Lett. 2007, 29, 599–603. [Google Scholar] [CrossRef]
- Saurav, K.; Kannabiran, K. Cytotoxicity and antioxidant activity of 5-(2, 4-dimethylbenzyl) pyrrolidin-2-one extracted from marine Streptomyces VITSVK5 spp. Saudi J. Biol. Sci. 2012, 19, 81–86. [Google Scholar] [CrossRef]
- Ser, H.L.; Palanisamy, U.D.; Yin, W.F.; Malek, S.N.A.; Chan, K.G.; Goh, B.H. Presence of antioxidative agent, Pyrrolo[1,2-a] pyrazine-1,4-dione, hexahydro-in newly isolated Streptomyces mangrovisoli sp. nov. Front. Microbiol. 2015, 6, 854. [Google Scholar] [CrossRef]
- Narendhran, S.; Rajiv, P.; Vanathi, P.; Sivaraj, R. Spectroscopic analysis of bioactive compounds from Streptomyces cavouresis kuv39: Evaluation of antioxidant and cytotoxicity activity. Int. J. Pharm. Pharm. Sci. 2014, 6, 319–322. [Google Scholar]
- Tian, S.Z.; Pu, X.; Luo, G.; Zhao, L.X.; Xu, L.H.; Li, W.J.; Luo, Y. Isolation and characterization of new p-Terphenyls with antifungal, antibacterial, and antioxidant activities from halophilic actinomycete Nocardiopsis gilva YIM 90087. J. Agric. Food Chem. 2013, 61, 3006–3012. [Google Scholar] [CrossRef]

| Name | Sample Code | Sampling Area | Latitude (N) | Longitude (E) | Elevation (m) |
|---|---|---|---|---|---|
| Streptomyces Sp. | S1A | Bisle Ghat, Hassan District | 12°71′88.04″ | 75°68′70.02″ | 802 |
| Streptomyces Sp. | SS4 | Bisle Ghat, Hassan District | 12°72′00.89″ | 75°68′41.42″ | 752 |
| Streptomyces Sp. | SS5 | Bisle Ghat, Hassan District | 12°71′24.30″ | 75°68′04.74″ | 710 |
| Streptomyces Sp. | SS6 | Virajpet, Madikeri District | 12°19′75.83″ | 75°79′52.93″ | 885 |
| Streptomyces Sp. | SCA35 | Virajpet, Madikeri District | 12°18′83.38″ | 75°83′06.79″ | 864 |
| Nocardiopsis Sp. | SCA11 | Virajpet, Madikeri District | 12°21′21.64″ | 75°80′24.84″ | 830 |
| Nocardioides Sp. | SCA13 | Virajpet, Madikeri District | 12°18′47.27″ | 75°76′24.17″ | 798 |
| Isolate | Medium | Diffusible Pigment | Colony Morphology | Aerial Mycelium | Substrate Mycelium |
|---|---|---|---|---|---|
| Streptomyces Sp. S1A | SCA | None | Powdery | White | Cream |
| Streptomyces Sp. SS4 | SCA | None | Cottony | Dark Grey | Grey |
| Streptomyces Sp. SS5 | SCA | None | Cottony | Grey | Cream |
| Streptomyces Sp. SS6 | SCA | Pink | Rough | Pale red | Pink |
| Streptomyces Sp. SCA35 | SCA | None | Powdery | White | Cream |
| Nocardiopsis Sp. SCA11 | SCA | None | Powdery | Cream | Light brown |
| Nocardioides Sp. SCA13 | SCA | None | Raised | White | White |
| Source | Organism | GenBank Accession No | % Similarity |
|---|---|---|---|
| Soil | Streptomyces Sp. S1A | KU921223 | 98.32% |
| Soil | Streptomyces Sp. SS4 | MF668120 | 99.71% |
| Soil | Streptomyces Sp. SS5 | MF925722 | 99.70% |
| Soil | Streptomyces Sp. SS6 | MF925723 | 99.72% |
| Soil | Streptomyces Sp. SCA35 | MN176654 | 99.35% |
| Soil | Nocardiopsis Sp. SCA11 | MG934272 | 98.99% |
| Soil | Nocardioides Sp. SCA13 | MG934273 | 98.28% |
| Organisms | Minimum Inhibition Concentration (µg/mL) | |||||
|---|---|---|---|---|---|---|
| Test Organisms | ||||||
| MRSA ATCC NR-46071 | MRSA ATCC NR-46171 | MRSA ATCC 33591 | Salmonella typhi ATCC 25241 | Escherichia coli ATCC 11775 | Fusarium moniliforme MTCC 6576 | |
| Streptomyces Sp. S1A | 15.62 | 15.62 | 62.5 | 125 | >500 | 62.5 |
| Streptomyces Sp. SS4 | 62.5 | 31.25 | 125 | 250 | >500 | - |
| Streptomyces Sp. SS5 | 31.25 | 125 | 15.62 | 250 | 250 | 125 |
| Streptomyces Sp. SS6 | 31.25 | 62.5 | 62.5 | 250 | 125 | - |
| Streptomyces Sp. SCA35 | 62.5 | 15.62 | 125 | >500 | 125 | 250 |
| Nocardiopsis Sp. SCA11 | 31.25 | 15.62 | 31.25 | 250 | >500 | 125 |
| Nocardioides Sp. SCA13 | 62.5 | 62.5 | 125 | 250 | 125 | - |
| Isolates | IC50 (µg/mL) Crude Extracts | |
|---|---|---|
| DPPH | ABTS | |
| Streptomyces Sp. S1A | 189.40 ± 0.12 | 156.81 ± 0.06 |
| Streptomyces Sp. SS4 | 98.29 ± 0.32 | 123.48 ± 0.13 |
| Streptomyces Sp. SS5 | 86.45 ± 0.04 | 114.87 ± 0.29 |
| Streptomyces Sp. SS6 | 114.15 ± 0.03 | 164.04 ± 0.07 |
| Streptomyces Sp. SCA35 | 65.86 ± 0.49 | 49.11 ± 0.73 |
| Nocardiopsis Sp. SCA11 | 30.91 ± 0.25 | 48.24 ± 0.30 |
| Nocardioides Sp. SCA13 | 42.30 ± 0.10 | 37.91 ± 0.17 |
| Trolox (Standard) | 11.07 ± 0.06 | 9.87 ± 0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddharth, S.; Vittal, R.R.; Wink, J.; Steinert, M. Diversity and Bioactive Potential of Actinobacteria from Unexplored Regions of Western Ghats, India. Microorganisms 2020, 8, 225. https://doi.org/10.3390/microorganisms8020225
Siddharth S, Vittal RR, Wink J, Steinert M. Diversity and Bioactive Potential of Actinobacteria from Unexplored Regions of Western Ghats, India. Microorganisms. 2020; 8(2):225. https://doi.org/10.3390/microorganisms8020225
Chicago/Turabian StyleSiddharth, Saket, Ravishankar Rai Vittal, Joachim Wink, and Michael Steinert. 2020. "Diversity and Bioactive Potential of Actinobacteria from Unexplored Regions of Western Ghats, India" Microorganisms 8, no. 2: 225. https://doi.org/10.3390/microorganisms8020225
APA StyleSiddharth, S., Vittal, R. R., Wink, J., & Steinert, M. (2020). Diversity and Bioactive Potential of Actinobacteria from Unexplored Regions of Western Ghats, India. Microorganisms, 8(2), 225. https://doi.org/10.3390/microorganisms8020225