Abstract
Background: The bacterial community responses to oil spill events are key elements to predict the fate of hydrocarbon pollution in receiving aquatic environments. In polar systems, cold temperatures and low irradiance levels can limit the effectiveness of contamination removal processes. In this study, the effects of a simulated acute oil spillage on bacterial communities from polar sediments were investigated, by assessing the role of hydrocarbon mixture, incubation time and source bacterial community in selecting oil-degrading bacterial phylotypes. Methods: The bacterial hydrocarbon degradation was evaluated by gas chromatography. Flow cytometric and fingerprinting profiles were used to assess the bacterial community dynamics over the experimental incubation time. Results: Direct responses to the simulated oil spill event were found from both Arctic and Antarctic settings, with recurrent bacterial community traits and diversity profiles, especially in crude oil enrichment. Along with the dominance of Pseudomonas spp., members of the well-known hydrocarbon degraders Granulosicoccus spp. and Cycloclasticus spp. were retrieved from both sediments. Conclusions: Our findings indicated that polar bacterial populations are able to respond to the detrimental effects of simulated hydrocarbon pollution, by developing into a more specialized active oil degrading community.
1. Introduction
Petroleum hydrocarbons are the main pollution source in polar ecosystems [1,2]. Although many petroleum products are used, stored, transported, and spilled in localized regions across the poles, the environmental contamination levels are hardly assessed or retrieved from country-specific documentations. In the Arctic, the risks of petroleum hydrocarbon pollution are increasingly because of the rising exploration activities, and a growing interest in developing the Northern Sea Route (NSR) as an alternative transportation route for oil and gas from Russia to Europe and other markets [3]. In the Antarctic, oil spills are known and registered events, but the spatial and temporal contamination patterns were not determined specifically, though wastes are managed under the common international environmental protocol (Madrid Protocol 1991).
Studies focused on Arctic and Antarctic seawater and ice samples showed that the introduction of hydrocarbons stimulated the selective growth of specialized hydrocarbon-degrading microorganisms [4,5]. They include mostly Proteobacteria members, mainly affiliated to Pseudoalteromonas, Pseudomonas, Psychrobacter spp. [6,7], Marinobacter [3,8], Oleispira [9], Shewanella [10], but also Bacteroidetes (e.g., Cytophaga spp.) and Actinobacteria (e.g., Rhodococcus spp.) [7,11,12,13,14]. The percentage of hydrocarbon-degrading bacteria were reported to increase up to ten folds, passing from 0.1 to 1% of the total heterotrophic bacteria in uncontaminated polar areas to 1–10% after an event of oil spill [12]. Moreover, consistent shift in the bacterial assemblage structure were detected immediately after the oil release into the environment [3]. In fact, multispecies microbial consortia can be more effective in oil degradation, through retaining a broader suite of enzymatic capacity than single microbial species, which grow on a limited range of carbon substrates [15].
The microbially driven mineralization processes can fundamentally contribute to hydrocarbon degradation and the natural attenuation of detrimental effects caused by oil spill [2]. The bacterial biodegradation potential is of utmost importance particularly in polar areas, since cold temperatures and light–dark cycles can limit the effectiveness of abiotic processes for contaminant removal [16].
Along with numerous field observations of microbial community responses to hydrocarbon contamination, the relative simplicity of experimental model systems is actively contributing to elucidate the bacterial potential for petroleum biodegradation in polar environments [17,18]. However, only few experimental studies analyzed the bacterial community patterns following oil spill of different composition [6], and the microbial responses to hydrocarbon contamination have not yet been evaluated concurrently in sediments from different polar regions.
This study was entailed to explore the effects of a hydrocarbon addition on the structural characteristics of bacterial communities in Arctic and Antarctic sediments. More specifically, we aimed at assessing (i) the aliphatic biodegradation patterns in sediments amended with different oil mixtures, (ii) the changes in bacterial community abundance (i.e., flow cytometry) and diversity profiles (i.e., Terminal Restriction Fragment Length Polymorphism analysis (T-RFLP)), and (iii) the occurrence of dominant bacterial species (i.e., Denaturing Gradient Gel Electrophoresis (DGGE)). By assuming a relevant impact of hydrocarbon amendments, we hypothesized direct responses from Arctic and Antarctic sediment microbial communities over the incubation time, with possible recurrent community traits that will occur because of the experimental stress factors.
2. Materials and Methods
2.1. Sampling Sites and Sediment Collection
Arctic samples were taken from the glacial open fjord Kongsfjorden, at the Research Village in Ny-Alesund (coordinates: 78°55′606′′N, 11°56′377′′E; Svalbard Archipelago, Arctic Norway) [7]. Antarctic samples were collected from the Byers Peninsula (Antarctic Specially Protected Area No. 126), at the mouth of the stream Petreles (coordinates: 62°40′09.9”S, 61°09′04”W; Livingston Island, Antarctica).
In each site of the north and south polar areas, during their respective summer periods in 2009, approximately 2 kg of sediment samples were collected between depths of 0 and 10 cm, together with seawater samples, by using sterile polycarbonate bottles, and stored in sterile polycarbonate bottles at 4 °C until processing. After arrival at the laboratory sediment samples were divided in subsamples used to set up microcosms. At the sampling days, seawater temperature and salinity were 7.9 °C and 22.8‰ and 6.0 °C and 30.0‰ in the Arctic and Antarctic sites, respectively.
2.2. Microcosm Set-Up
Hydrocarbon-enriched microcosms were set up in triplicates with Arctic and Antarctic samples by filling 250 mL glass beakers, covered with a sterile glass Petri dish cover to avoid external contamination during the incubation, with 150 g of sediment and 150 mL of filter-sterilized seawater from the same sampling site, as previously described [2]. Briefly, filter-sterilized crude oil (Arabian light) or commercial diesel oil (Q8 Italy) was added to each microcosm and mixed with a sterile glass stick (final concentration 1.5%, v/v). Hydrocarbon-free microcosms were used as control. All microcosms were incubated at 4 °C for 160 days. At established time intervals, microcosms were gently shaken and then left to settle. Finally, sediment subsamples (10 g) were aseptically collected with a sterile spoon from each microcosm for chemical and microbiological analyses.
2.3. Analysis of Residual Hydrocarbons in Arctic and Antarctic Sediments
The biodegradation efficiency was monitored by gas chromatographic analysis to achieve a qualitative/quantitative estimation of residual hydrocarbons in each microcosm. Residual hydrocarbons from sediment subsamples (100 g) at the experiment beginning (0 days), after 90 days of incubation and end (160 days), were extracted using methyl-tertiary-butyl-ether:hexane (20:80), a modified version than the reference standard EPA (SW-846 3550B-Ultrasonic Extraction). Time 0 samples were used as control for abiotic losses. The ultrasound-based process guaranteed a strong adhesion of the sample matrix to the solvent extraction. The entire sample was transferred in a dark glass bottle, using sodium sulfate (5 g) as drying agent. The liquid layer of solvent was poured through a funnel built with filter (Whatman N° 2) Na2SO4 in a 250 mL pre-equilibrate evaporation flask. The extract was concentrated to a small volume by evaporation under reduced pressure in a rotavapor, and 1 mL was pipetted into glass vials with Teflon cap and refrigerated until analysis by gas chromatography. Immediately before the extraction, butylhydroxytoluene (BHT) was added as a surrogate standard. The standards used to monitor the biodegradation of crude oil and diesel oil were a mixture containing from C7 to C30 fractions and a mixture containing fractions from C13 to C18, respectively. The characterization of petroleum product was made after solvent extraction methods followed by gas chromatography analysis with a Flame Ionization Detector (FID). A gas chromatograph GC Series 17-A (Shimadzu Ltd., Kagoshima, Japan) equipped with Electron Capture detector (ECD) and FID and AOC-5000 autosampler (CombiPAL) was used for analysis. Analyses were conducted using Petrocol™ column (Supelco, St. Louis, MO, USA): 100 m long with inner diameter of 0.25 mm and the carrier material with diameter of 0.5 µm. The temperature of the injector and detector were set at 280 °C. We used a temperature program, which provides a better separation of various components with different boiling temperatures. The initial temperature of the gas chromatograph oven temperature control was maintained at 35 °C for ~15 min, and then the temperature was increased by 2 °C per minute to a temperature of 200 °C. The last step in temperature was set at 300 °C with an increase in temperature of 10 °C per minute and maintained for 2.5 min for a total analysis in 110 min. Helium was used as carrier gas at about 20 cm s−1, nitrogen as make-up gas at 30 mL min−1. This method allowed the separation of all the components of the matrix with particular reference to petroleum. The retention time was determinate by applying defined standard containing a mixture of compounds from C7 to C30 (Supelco) at a concentration of 1 mg mL−1 for each component in hexane solvent. The calibration curve for the quantitative analysis was conducted using five standards and increasing the concentration by using the gas chromatographic procedure management software provided by Shimadzu Class VP 7 Ltd. All lines obtained had a correlation coefficient greater than 0.95. Control of retention times was also conducted with the calculation of linear retention indices (LRI).
2.4. Microbial Community Characterization
2.4.1. Flow Cytometry
Total cell counts were assessed at regular time intervals for both treatments (i.e., 0, 30, 60, 80, 90, 120, and 160 days of incubation). Prokaryotic cells were detached from sediments by mixing sample aliquots (1 g) with phosphate buffered saline (PBS: 120 mM NaCl, 2.7 mM KCl in 10 mM phosphate buffer, pH 7.6; 9 mL), sodium pyrophosphate (0.1%), and tween 80 (10 µL). The mixture was fixed with formalin solution (final concentration 2%, v/v), shaken vigorously for 20 min, and stored at 4 °C until processing [19]. The total prokaryotic abundance was estimated by using the Flow Cytometer A-50 (Apogee Flow System, Hertfordshire, England), as described in Amalfitano et al. [20]. The Apogee Histogram Software v89.0, Apogee Flow System, Hertfordshire, England) was used for data elaboration, which were expressed as cells per gram of wet sediment (cells g−1).
2.4.2. DNA Extraction and Fingerprinting Analyses
The total DNA was extracted from 500 mg of starting material using a bead beating kit (FastDNA® SPIN Kit for Soil, Qbiogene, Heidelberg, Germany) following the manufacturer’s instructions. Two fingerprinting approaches were used for microbial community characterization on selected sampling times: at the microcosm set up (0 days), after 30 days, 90 days, and 160 days of incubation. As hydrocarbons degradation and emulsification rate appeared optically conspicuous in Arctic microcosms, an additional intermediate sampling was performed after 60 days of incubation.
The Terminal Restriction Fragment Length Polymorphism analysis (T-RFLP) was used to evaluate the diversity profiles of bacterial communities over time. The amplification of genes coding for 16S rRNA was carried out by PCR, with reaction mixture and conditions as previously described [12]. PCR reactions have been performed with a PTC-100 Peltier Thermal Cycler (MJ Research Celbio). Amplification products of three parallel PCR reactions were combined and purified with the Wizard SV Gel and PCR Clean-Up system (Promega, Durham, NC, USA) prior to perform T-RFLP analysis, as described in the following sections.
Approximately 300 ng of amplified 16S rRNA genes from each DNA sample were digested in duplicate with 10U of AluI restriction enzyme (Fermentas). Denaturation of reaction mixtures were carried out by heating at 95 °C for 3 min, and then incubated at 0 °C for 5 min. Samples were then sent to an outsourcing sequencing service (BMR-Genomics, Italy). The reaction outputs (electropherograms) were processed using methods as described by Luna et al. [21]. The “Abundance percentage” (Ap) of each T-RF was calculated as indicated by Lokow et al. [22]. The size of each T-RF was estimated in reference to an internal standard. All subsequent processing phases of T-RFLP data have been performed as suggested by Smith et al. [23] and Baldi et al. [24], to characterize the microbial community diversity patterns. After enzymatic digestion of PCR amplicons, each T-RF can be defined as an operational taxonomic unit (OTU) within a community [25]. Statistical analysis have been computed by considering the number of peaks as indicator of the species number (phylotype/genotype richness) and the band intensity peak height as the relative abundance of each bacterial species [26].
Denaturing Gradient Gel Electrophoresis (DGGE) was used to promptly identify the dominant bacterial species occurring in contaminated Arctic and Antarctic sediments over the experimental incubation time. The variable V3 region of the 16S rDNA of bacteria was amplified by using universal primers (27F, 5′-AGA GTT TGA TC(AC) TGG CTC AG-3′ with GC-clamp in position 5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC G-5′ spanning Escherichia coli position 8-27; 518R, 5′-ATT ACC GCG GCT GCT GG-3′ spanning E.coli position 518-534; - Biomers.net GmbH - Ulm, Germany). PCR was performed using a thermocycler (Mastercycler, GeneAmp PCR-System 9700, Applied Biosystems, Foster City, CA 94404, USA), as described by Gerçe et al. [27], and the same reference was used also for all DGGE conditions. A presence/absence matrix of bands was constructed for DGGE gel analysis (Alpha Imager 2.200; Biozym Scientific GmbH, Oldendorf, Germany).
Selected DGGE bands were carefully cut out under UV lamp with sterile scalpels, eluted overnight in 50 μL water at 4 °C (DNA-free PCR water, Molzym GmbH and Co. KG, Bremen, Germany), and PCR re-amplified using the DNA diffuses into the water as starting material in a PCR. PCR conditions were the same as described before with some modifications [27]. The NCBI GenBank database (http://www.ncbi.nlm.nih.gov) was used for comparing the sequences to 16S ribosomal RNA (rRNA) gene sequences by using the Basic Local Alignment Search Tool (BLASTN) algorithm [28].
2.5. Statistical Analyses
To examine the differences in bacterial community profiles over time, fingerprint data were analyzed by presence⁄absence and relative abundance-based matrices using semimetric Bray–Curtis distance measures. Bray–Curtis similarities were calculated on both T-RFLP and DGGE data and used for Cluster analysis and visualized in a lower dimensional space by applying non-metric multidimensional scaling (nMDS). Statistical differences between bacterial community structures in the multivariate dataset by carrying out the Analysis of SIMilarities (ANOSIM). All calculation were carried out by the software Primer 6 (v. 6β R6, PRIMER-E Ltd., UK). Two-way ANOVA was performed to establish the effect of incubation time, treatment (diesel oil and crude oil), and sediment origin (Arctic and Antarctic) on the bacterial community diversity.
3. Results
3.1. Residual Hydrocarbons in Arctic and Antarctic Microcosms
The Arctic community was more efficient in the degradation of small chain hydrocarbons (except C-10) in the experiment setting with addition of crude oil, as it was demonstrated by the removal of hydrocarbons with C-10 and C-12 (Figure 1a), with biodegradation rates of 99% and 95% (Figure 1c). The fractions between C-16 and C-30 were optimally degraded but with a minor extent, except for the hydrocarbons with C-21 and C-22 chains, which were almost present also at the end of the experiment. Sediments in the microcosm enriched with diesel oil showed a very efficient community in the biodegradation of longer chain hydrocarbons (from C-21 to C-25), reaching up to 75% of their removal (Figure 1b). The Figure 1c shows more clearly the overall biodegradation rate for crude oil and diesel oil during the experiment.

Figure 1.
Residual hydrocarbons in Arctic microcosms enriched with crude oil (a) and diesel oil (b) over the incubation time (0, 90, and 160 days of incubation). Overall biodegradation of hydrocarbon mixtures (c).
A relevant reduction of long chain hydrocarbons was found in Antarctic sediments (Figure 2a–c). In particular, at the end of the experiment the hydrocarbon chains from C-20 to C-30 in crude oil (Figure 2a), and chains from C-23 to C-30 in diesel oil (Figure 2b) were totally removed.

Figure 2.
Residual hydrocarbons in Antarctic microcosms enriched with crude oil (a) and diesel oil (b) over the incubation time (0, 90, and 160 days of incubation). Overall biodegradation of hydrocarbon mixtures (c).
3.2. Patterns of Microbial Cell Abundance in Arctic and Antarctic Contaminated Sediments
In Arctic sediments, the microbial abundance varied between 0.2 × 106 cells g−1 and 3.1 × 106 cells g−1, with values increasing over time only in presence of crude oil. In Antarctic sediments, cell abundance reached values up to 14.4 and 9.1 × 106 cells g−1 (in crude oil and diesel, respectively), and likely decreased along the incubation time.
The fold increase of microbial abundance with respect to the relative control treatments (i.e., treatment-to-control ratio) was plotted to show the effects of oil contamination over time (Figure 3). As indicated by values below 1, a detrimental impact by oil contamination was evident at day 30 in all contaminated sediments, also reflecting the highest abundance values found in both control sediments (i.e., 79.1 × 106 cells g−1 and 18.7 × 106 cells g−1 in Arctic and Antarctic sediments, respectively). In Arctic sediments, the detrimental effects of diesel contamination were evident throughout the entire incubation period. From day 60 in sediments with crude oil, fold increase values exceeded 1, thus indicating a possible hydrocarbon-stimulated increase of microbial abundance. A similar pattern was found in Antarctic sediments contaminated by diesel oil.

Figure 3.
Patterns of the total prokaryotic cell counts as assessed by flow cytometry in Arctic and Antarctic microcosms. Data are presented as fold increase of microbial abundance in contaminated sediments with respect to the control treatments.
3.3. Community Diversity Profiles in Contaminated Sediments over the Incubation Time
A number of t-RFs ranging from 23 to 63 in the bacterial community profiles were obtained from T-RFLP analysis. Most t-RFs were identified at the Arctic sediments after 0 days of incubation in crude oil treatment, and in the Antarctic sediments after 30 days of incubation both in diesel oil and crude oil treatment. The minimum number was detected in Arctic sediments at the end of the experiments in both the microcosm settings. No statistically significant differences were found between treatments (i.e., diesel oil or crude oil at each time), while significant differences occurred between sampling times (Figure 4). In detail, for the Arctic microcosm the t-RFs after 160 days of incubation were significantly different from those detected after 30 and 90 days of incubation. In the case of Antarctic microcosm, significant differences occurred between all sampling times, namely 30, 90 and 160 days.
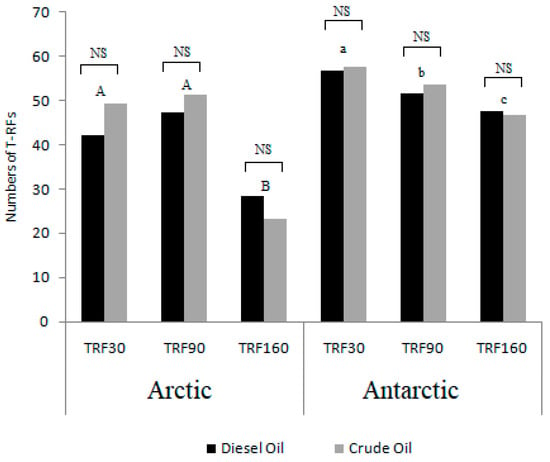
Figure 4.
The bacterial richness expressed as the number of T-RFs (T-RFLP) detected in the microcosms samples from two experimental treatments (crude oil and diesel oil) in three sampling times (TRF30: 30 days of incubation; TRF60: 60 days of incubation; TRF160: 160 days of incubation). The different simple and capital letters denote statistically significant differences among each sampling times in Arctic and microcosms; NS—not significant different (ANOVA, p = 0.05).
As calculated from the T-RFLP data, the highest genetic diversity and evenness were detected in Arctic sediments with crude oil after 90 days of incubation and Antarctic sediments with diesel oil after 30 days of incubation (Figure S1). In both Arctic and Antarctic microcosms, richness globally decreased over time after the addition of hydrocarbons.
In Arctic sediments, the T-RFLP analysis revealed that the bacterial community structure had differences over time in the treatment with crude oil, as demonstrated by the low Bray–Curtis similarity. The sediments enriched with crude oil exhibited the higher dissimilarity rate over time with 32.4% of similarity between the different time samplings. The community appeared more similar over time in the microcosm supplemented with diesel oil and in the control setting, showing a similarity of 42.3% and 56.9% between sampling times, respectively. Similarity percentage of 35.0% (ANOSIM Global R > 0.4; P > 0.8) was detected when crude oil and diesel oil settings were compared, while a similarity of 47.1% (ANOSIM Global R > 0.2; P > 0.01) was recorded in case of comparison with the control experiment (Figure S2a).
Similarly, the T-RFLP analysis suggest an influence of hydrocarbon introduction on the Antarctic bacterial community structure (ANOSIM Global R > 0.4; P > 0.08). Results obtained for the Antarctic sediments enriched with crude oil showed an average similarity of 61.8% between sample aliquots collected during the time course experiment, and 59.4% (ANOSIM Global R > 0.7; P > 0.2) in comparison with the control. Differently, the bacterial community of microcosm supplemented with diesel oil showed a similarity of 81.0% between aliquots collected at different sampling times, and a value of 68.7% (ANOSIM Global R > 0.6; P > 0.5) compared with the control (Figure S2b).
3.4. Occurrence of Dominant Bacterial Species across the Experimental Conditions
The DGGE gel obtained from the Arctic sediments revealed the presence of a total of 30 dominant phylotypes and a banding pattern of 11 to 18 bands (Figure S3). A total of 77 bands were excised from gel and analyzed for taxonomical affiliation by 16S rRNA gene sequencing. Among them, 30 were considered as phylotype representatives (Table 1 and Figure S3). The phylogenetic analysis of partial 16S rRNA gene sequences from selected DGGE bands revealed that Arctic bacteria were related to the Proteobacteria (Alpha-, Beta-, Delta-, and Gammaproteobacteria), Actinobacteria, Firmicutes, and CF group of Bacteroidetes. Pseudomonas frederiksbergensis (DGGE_band_114), Pseudomonas costantinii (DGGE_band_123), and Magnetospirillum gryphiswaldense (DGGE_band_131) were observed only in the presence of diesel oil (from 30 to 160 days of incubation, and from 60 to 160 days of incubation, respectively), whereas Magnetospirillum magnetotacticum (DGGE_band_95) only in the presence of crude oil (from 30 to 90 days of incubation). A number of sequences were retrieved both in presence of crude oil and diesel oil at almost all sampling times, and shared with the control setting: Kofleria flava (DGGE_band_62), Granulosicoccus antarcticus (DGGE_band_66), Marinobacter antarcticus (DGGE_band_69), Rhodoferax fermentans (DGGE_band_70), Pibocella ponti (DGGE_band_115), Pseudomonas congelans (DGGE_band_121), and Pseudomonas sabulinigri (DGGE_band_129).

Table 1.
16S rRNA gene sequence affiliation of selected DGGE bands to their closest phylogenetic neighbors. Sequences that were found in both microcosms are in bold.
Other phylotypes were shared between the control experiment and the crude oil treatment or diesel oil treatment. The occurrence of sequences observed only in the absence of hydrocarbons was also verified (Figure 5).

Figure 5.
Venn diagrams showing phylotypes distribution in Arctic and Antarctic microcosms detected by denaturing gradient gel electrophoresis (DGGE) analysis.
The DGGE gel obtained from the Antarctic sediments revealed the presence of 23 dominant phylotypes and a banding pattern of 12 to 14 bands (Figure S4). A total of 53 bands were excised from DGGE gels and characterized by 16S rRNA gene sequence analysis, resulting in 15 phylotype representatives that gave sequencing results (Table 1 and Figure S4). The phylogenetic analysis of partial 16S rRNA gene sequences from selected DGGE bands revealed that bacteria in the Antarctic microcosm were related to the Proteobacteria (Alpha-, Beta-, Gamma-, and Deltaproteobacteria), Actinobacteria, and CF group of Bacteroidetes. A shift in diversity and the emergence of a small number of dominant bands compared to the corresponding control treatment were observed. The crude oil and diesel oil addition during incubation resulted in a shift towards the appearance or disappearance of specialized hydrocarbon degraders. For example, Oleispira lenta (DGGE_band_50) sequences were retrieved only in Antarctic microcosm supplemented with diesel oil after 90 days of incubation. The same was true for Rhodococcus qingshengii (DGGE_band_43) in Antarctic microcosms with crude oil, even if it was observed also after 160 days of incubation. Cycloclasticus pugetii (DGGE_band_42) became visible only at the end of the experiment with addition of crude oil (160 days), while Hoeflea sp. (DGGE_band_30) was observed only in the same treatment after 30 days, but then it disappeared.
As for Arctic sediments, a number of sequences were retrieved in both hydrocarbon-enriched microcosms at all sampling times, and shared with the control. The betaproteobacterium Sideroxydans lithotrophicus (DGGE_band_22) co-occurred in control and crude oil treatments, but it was absent in the diesel oil treatment. Marinimicrobium agarilyticum (DGGE_band_18) appeared in the control only at the end of the incubation. Finally, sequences from Pseudomonas grimontii (DGGE_band_51) were retrieved from all microcosms (including the control) after 160 days. Salinibacterium amurskyense (DGGE_band_8), visible at 0 days of incubation, disappeared after hydrocarbon addition.
Comparison of the Bray–Curtis dissimilarity matrices showed a similarity within Antarctic microcosms communities (both crude oil and diesel oil treatments) of 91% and 98%, respectively. Antarctic-crude oil and Antarctic-diesel oil communities showed similar values of similarity (88.5% and 88.7%, respectively) with the control experiment (Figure 6a). With regard to the Arctic sediments, this analysis revealed that the communities grouped differently in dependence of the hydrocarbon enrichment. The similarity within the different communities over the set time was always higher than 80%, but it drastically decrease when compared to the community from the control microcosm with 57 and 47% of similarity to Arctic-crude oil and Arctic-diesel oil, respectively (ANOSIM Global R = 0.9; P < 0.03) (Figure 6b). The nMDS analysis computed on DGGE matrix results clearly showed the different clustering of polar communities in response to hydrocarbon addition, by highlighting the occurrence of a separate cluster grouping all samples from Antarctic microcosms, whereas Arctic microcosm samples formed three main clusters, one for crude oil treatment samples, a second for diesel oil treatment samples, and the last one for control samples (Figure 7).

Figure 6.
Non-metric multidimensional scaling (nMDS) computed on Bray–Curtis similarities calculated presence/absence matrix obtained from DGGE analysis for (a) Arctic and (b) Antarctic microcosms.

Figure 7.
Non-metric multidimensional scaling (nMDS) computed on Bray–Curtis similarities calculated from DGGE analysis results, plotted by clustering data according to sediment origin.
4. Discussion
This study aimed to investigate the effects of an acute oil spillage on microbial communities from polar sediments, also verifying the possible analogies/differences in the occurring possible specific responses related to the community origin.
A different response was highlighted in the biodegradation processes on the two hydrocarbon mixtures. Generally, the trend in hydrocarbon degradation involves a faster removal of short and medium chain length alkanes than the long chain length alkanes. Despite this, different patterns have been reported. For example, Mason et al. [29] detected a faster degradation of dodecane than phenanthrene and toluene, whereas Yergeau at al. [30] and Bacosa et al. [31] reported faster polycyclic aromatic hydrocarbon degradation within 30 days of incubation. In our case, the gas chromatographic analysis on residual aliphatic hydrocarbons at the end of incubation showed that the Arctic community was able to better utilize hydrocarbons from C13 to C30 in crude oil. On the other hand, the Antarctic community was more efficient (up to 100% per single compound removed) in the biodegradation of longer chain hydrocarbons (crude oil from C7 to C19; diesel oil from C7 to C22). This finding might rely on the different composition of the source microbial community that naturally occurred at the diverse levels of pollution reported from the sampling Arctic and Antarctic areas, as it was confirmed by the differences encountered between DDGE results. However, the bacterial community dynamics showed similar patterns in the two microcosm settings in terms of bacterial abundance. The hydrocarbon contamination was likely limiting the microbial cell growth at the first month from the simulated oil spill event. A possible growth-stimulating effect was found afterwards, with higher cell abundance values found in contaminated sediments than in the control treatment. The first biodegradative processes, and the consequent appearance of hydrocarbon intermediates from reactions, could in fact contribute to the development of a bacterial community specialized in the degradation of hydrocarbons, with increase of cell abundance. The sediment bacterial communities can develop into active oil degrading consortia after oil contamination, despite hydrocarbon-degrading bacteria are recurrently reported even in uncontaminated areas [13]. Obligate hydrocarbonoclastic bacteria may occur immediately after the release of oil into the environment, by becoming dominant component of the microbial community [32,33,34].
Although an increase in abundance of the cultivable bacterial fraction is also reported in response to hydrocarbons, the bacterial diversity can dramatically decrease [35]. Our findings pointed to higher specialized communities at the end of the experiment in terms of diversity profiles.
Despite recent advances in genomics and sequencing technologies, molecular approaches have been scantly applied to similar experimental study about effects of a simulated oil spill event on polar bacterial communities. Here, T-RFLP analysis was chosen as a semiquantitative and highly reproducible method [36,37], and it was a useful tool for investigating the species diversity and evenness, despite the phylogenetic characterization is limited to the observation and comparison of terminal restriction fragment (t-RF) [38,39]. This technique was coupled with DGGE, which is less sensitive than T-RFLP [40,41,42], but can better reflect the community composition, by evidencing potential dominant phylotypes.
Further similarities between Arctic and Antarctic communities were underlined by the changes in diversity profiles and by the appearance of specialized phylotypes over time. Both communities were affected by the hydrocarbon addition if compared with the control microcosm, and changes in bacterial diversity were also detected also in the time course between the different sampling times. The harsher crude oil effect on diversity profiles represented a common point in the two polar microcosms, as proven also by the similarity average calculated by the Bray–Curtis analysis, in comparison with the control and the diesel oil treatment, thus suggesting a more disturbing effect of crude oil on microcosms. Indeed, the similarity in crude oil treatment was lower, and this was particularly true for Arctic microcosm, as demonstrated by the low Bray–Curtis similarity (32.4% and 61.9% for Arctic and Antarctic, respectively).
The appearance of bacterial species specialized in the hydrocarbon degradation supports the detrimental effect of crude oil and diesel oil on microbial communities structure. Some species appeared in relation to the addition of hydrocarbon substrates, not only related to aliphatic hydrocarbon isoforms. For example, Oleispira lenta was found exclusively within the diesel oil enriched Antarctic community, whereas Rhodococcus qingshengii and Cycloclasticus pugetii (well-known degraders of polycyclic aromatic hydrocarbons) were both retrieved only in crude oil-enriched microcosm. Rhodococci are known as dominant alkane degraders in polar environment [35,43,44], despite a less pronounced cold adaptation than, for example, Oleispira spp. strains, obligate hydrocarbonoclastic [45,46]. Previous investigations in microcosm at low temperatures highlighted the bloom of organisms related to Oleispira sp. oil-degrading microbial communities [47,48]. Conversely, Pseudomonas grimontii sequences were obtained from both crude oil and diesel oil enriched Antarctic microcosms. The appearance of such bacterial sequences suggests a great versatility of the genus members, able to use as carbon source hydrocarbon fractions present in both substrates. For many years, members of the genera Pseudomonas and Cycloclasticus have been investigated for their hydrocarbon degradative properties, and were found in cold environments [2,49,50]. The different time of appearance confirmed that the several members of hydrocarbon-degrading community tend to occupy distinct trophic niches, and generally the aliphatic-degraders bloom first, followed by degraders of more complex and less bioavailable hydrocarbons [51].
Similarly to the experimental conditions here observed, observations in various marine habitats have shown increases in the occurrence of Gammaproteobacteria after oil contamination [15], even if the Alphaproteobacteria component resulted also well represented in the Arctic microcosm. The changes observed in the Arctic community in terms of taxonomic composition highlighted differences between the control and the contaminated microcosms. The species Magnetospirillum magnetotacticum, Sediminicola luteus, Microbulbifer pacificus, Sphingopyxis flavimaris, and Thiobacillus thioparus were found only in sediments with crude oil, whereas Cycloclasticus pugetii, Novosphingobium nitrogenifigens, Pibocella ponti, Magnetospirillum gryphiswaldense, and Pseudomonas spp. appeared in sediments with diesel oil. These taxa are known as aromatic hydrocarbon biodegraders [52,53]. Interestingly, the species of Microbulbifer, Thiobacillus, and Novosphingobium were never reported in cold environment, and only some references are available for Magnetospirillum, Sphingopyxis, and Sediminicola [4,54,55].
5. Conclusions
In this study, a simulated acute oil spillage had direct effects on the microbial community structure from polar sediments, showing notable analogies regardless the Arctic and Antarctic community origin. Both communities showed a similar bacterial abundance variation, with the occurrence of specialized phylotypes. Major changes in bacterial diversity were observed in relation to the incubation time rather than to hydrocarbon content and sediment origin.
The study outcomes provided an environmental relevant contribute by highlighting the selection of microbial consortia with higher potential in case of oil spills in polar areas, thus contributing to pave the way for novel biotechnological applications of sediment polar communities in bioremediation processes.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2076-2607/7/12/632/s1.
Author Contributions
Conceptualization and design of the work, R.M., A.L.G., L.M., and V.B.; methodology and experiments, R.M., B.G., C.S., and R.H.; analysis and curation of the data, R.M., C.R., S.A.; software and graphical elaboration, C.R. and M.P.; writing, draft, and critical analysis of the manuscript, C.R., R.M., A.L.G., and S.A.; final approval, all authors.
Funding
This research was supported by grants from the project PolyArc “Occurrence of PCB-degrading bacteria in relation to PCB contamination in coastal marine environments in Ny-Ålesund (Svalbard, Spitsbergen)”, which was financed by the European Centre for Arctic Environmental Research in Ny-Ålesund (ARCFAC V; FP6) [grant no. ARCFAC-026129-2008-70].
Acknowledgments
The authors wish to thank the colleagues Stefania Giannarelli and Sandro Francesconi (University of Pisa, Italy) for assistance with sample collection in the Arctic and Antonio Camacho (University of Valencia, Spain) for sampling in Antarctica.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Cripps, G.C.; Shears, J. The fate in the marine environment of a minor diesel fuel spill from an Antarctic research station. Environ. Monit. Assess. 1997, 46, 221–232. [Google Scholar] [CrossRef]
- Malavenda, R.; Rizzo, C.; Michaud, L.; Gerce, B.; Bruni, V.; Syldatk, C.; Hausmann, R.; Lo Giudice, A. Biosurfactant production by Arctic and Antarctic bacteria growing on hydrocarbons. Polar Biol. 2015, 38, 1565–1574. [Google Scholar] [CrossRef]
- Gerdes, B.; Brinkmeyer, R.; Dieckmann, G.; Helmke, E. Influence of crude oil on changes of bacterial community in Arctic sea-ice. FEMS Microbiol. Ecol. 2005, 53, 129–139. [Google Scholar] [CrossRef]
- Delille, D.; Pelletier, E. Natural attenuation of diesel-oil contamination in a subantarctic soil (Crozet Island). Polar Biol. 2002, 25, 682–687. [Google Scholar]
- Powell, S.M.; Snapeb, I.; Bowmana, J.P.; Thompsona, B.; Starkb, J.; McCammon, S.A.; Riddleb, M.J. A comparison of the short term effects of diesel fuel and lubricant oils on Antarctic benthic microbial communities. J. Exp. Mar. Biol. Ecol. 2005, 322, 53–65. [Google Scholar] [CrossRef]
- Deppe, U.; Richnow, H.H.; Michaelis, W.; Antranikian, G. Degradation of crude oil by an Arctic microbial consortium. Extremophiles 2005, 9, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Conte, A.; Papale, M.; Amalfitano, S.; Mikkonen, A.; Rizzo, C.; De Domenico, E.; Michaud, L.; Lo Giudice, A. Bacterial community structure along the subtidal sandy sediment belt of a high Arctic fjord (Kongsfjorden, Svalbard Islands). Sci. Total Environ. 2018, 619–620, 203–211. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Bruni, V.; De Domenico, M.; Michaud, L. Cold-Adapted hydrocarbon-Degrading microorganisms. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin, Germany, 2010; pp. 1897–1922. [Google Scholar]
- Crisafi, F.; Giuliano, L.; Yakimov, M.M.; Azzaro, M.; Denaro, R. Isolation and degradation potential of a cold-adapted oil/PAHdegrading marine bacterial consortium from Kongsfjorden (Arctic region). Rend. Lincei Sci. Fis. E Nat. 2016, 27, 261–270. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Giuliano, L.; Gentile, G.; Crisafi, E.; Chernikova, T.N.; Abraham, W.R.; Lunsdorf, H.; Timmis, K.N.; Golyshin, P.N. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int. J. Syst. Evol. Microbiol. 2003, 53, 779–785. [Google Scholar] [CrossRef]
- Gentile, G.; Bonasera, V.; D’Amico, C.; Giuliano, L.; Yakimov, M.M. Shewanella sp. GA-22, a psychrophilic hydrocarbonoclastic Antarctic bacterium producing polyunsaturated fatty acids. J. Appl. Microbiol. 2003, 95, 1124–1133. [Google Scholar] [CrossRef]
- Michaud, L.; Lo Giudice, A.; Saitta, M.; De Domenico, M.; Bruni, V. The biodegradation efficiency on diesel oil by two psychrotrophic Antarctic marine bacteria during a two-month-long experiment. Mar. Pollut. Bull. 2004, 49, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Yakimov, M.M.; Gentile, G.; Bruni, V.; Cappello, S.; D’Auria, G.; Golyshin, P.N.; Giuliano, L. Crude oil-induced structural shift of coastal bacterial communities of Rod Bay (Terra Nova Bay, Ross Sea) and characterization of cultured cold-adapted hydro-carbonoclastic bacteria. FEMS Microbiol. Ecol. 2004, 49, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Pini, F.; Grossi, C.; Nereo, S.; Michaud, L.; Lo Giudice, A.; Bruni, V.; Baldi, F.; Fani, R. Molecular and physiological characterisation of psychotrophic hydrocarbon-degrading bacteria isolated from Terra Nova Bay (Antarctica). Eur. J. Soil Biol. 2007, 43, 368–379. [Google Scholar] [CrossRef]
- Röling, W.F.M.; Milner, M.G.; Jones, D.M.; Lee, K.; Daniel, F.; Swannell, R.P.J.; Head, I.M. Robust hydrocarbon degradation during nutrient-enhanced oil spill bioremediation. Appl. Environ. Microbiol. 2002, 68, 5537–5548. [Google Scholar] [CrossRef]
- Das, N.; Chandran, P. Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnol. Res. Int. 2011, 1–13. [Google Scholar] [CrossRef]
- Bell, T.H.; Yergeau, E.; Maynard, C.; Juck, D.; Whyte, L.G.; Greer, C.W. Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J. 2013, 7, 1200–1210. [Google Scholar] [CrossRef]
- Garneau, M.; Michel, C.; Meisterhans, G.; Fortin, N.; King, T.; Greer, C.; Lee, K. Hydrocarbon biodegradation by Arctic sea-ice and sub-ice microbial communities during microcosm experiments, Northwest Passage (Nunavut, Canada). FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef]
- Amalfitano, S.; Fazi, S. Recovery and quantification of bacterial cells associated with streambed sediments. J. Microbiol. Methods 2008, 75, 237–243. [Google Scholar] [CrossRef]
- Amalfitano, S.; Fazi, S.; Ejarque, E.; Freixa, A.; Romaní, A.M.; Butturini, A. Deconvolution model to resolve cytometric microbial community patterns in flowing waters. Cytom. Part A 2018, 93, 194–200. [Google Scholar] [CrossRef]
- Luna, G.M.; Dell’Anno, A.; Danovaro, R. DNA extraction procedure: A critical issue for bacterial diversity assessment in marine sediments. Environ. Microbiol. 2006, 8, 308–320. [Google Scholar] [CrossRef]
- Lukow, T.; Dunfield, P.F.; Liesack, W. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 2000, 32, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.J.; Danilowicz, B.S.; Clear, A.K.; Costello, F.J.; Wilson, B.; Meijer, W.G.T. Align, a web-based tool for comparison of multiple terminal restriction fragment length polymorphism profiles. FEMS Microbiol. Ecol. 2005, 54, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Baldi, F.; Marchetto, D.; Pini, F.; Fani, R.; Michaud, L.; Lo Giudice, A.; Berto, D.; Giani, M. Biochemical and microbial features of shallow marine sediments along the Terra Nova Bay (Ross Sea, Antarctica). Cont. Shelf Res. 2010, 15, 1614–1625. [Google Scholar] [CrossRef]
- Engebretson, J.J.; Moyer, C.L. Fidelity of select restriction endonucleases in determining microbial diversity by terminal-restriction fragment length polymorphism. Appl. Environ. Microbiol. 2003, 69, 4823–4829. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Danovaro, R.; Luna, G.M.; Dell’Anno, A.; Pietrangeli, B. Comparison of two fingerprinting techniques, terminal restriction fragment length polymorphism and automated ribosomal intergenic spacer analysis, for determination of bacterial diversity in aquatic environments. Appl. Environ. Microbiol. 2006, 72, 5982–5989. [Google Scholar] [CrossRef] [PubMed]
- Gerçe, B.; Schwartz, T.; Syldatk, C.; Hausmann, R. Differences between bacterial communities associated with the surface or tissue of Mediterranean sponge species. Microb. Ecol. 2011, 61, 769–782. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biolology 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Mason, O.U.; Hazen, T.C.; Borglin, S.; Chain, P.S.G.; Dubinsky, E.A.; Fortney, J.L.; Han, J.; Holman, H.Y.; Hultman, J.; Lamendella, R.; et al. Metagenomics, metatranscriptomics and single cell sequencing reveal bacterial response to the Gulf oil spill. ISME J. 2012, 6, 1715–1727. [Google Scholar] [CrossRef]
- Yergeau, E.; Maynard, C.; Sanschagrin, S.; Champagne, J.; Juck, D.; Lee, K.; Greer, C.W. Microbial community composition, functions, and activities in the Gulf of Mexico 1 year after the Deepwater Horizon accident. Appl. Environ. Microbiol. 2015, 81, 5855–5866. [Google Scholar] [CrossRef]
- Bacosa, H.P.; Erdner, D.L.; Rosenheim, B.E.; Shetty, P.; Seitz, K.W.; Baker, B.J.; Liu, Z. Hydrocarbon degradation and response of seafloor sediment bacterial community in the northern Gulf of Mexico to light Louisiana sweet crude oil. ISME J. 2018, 12, 2532–2543. [Google Scholar] [CrossRef]
- Acosta-González, A.; Martirani-von Abercron, S.M.; Rosselló-Móra, R.; Wittich, R.M.; Marqués, S. The effect of oil spills on the bacterial diversity and catabolic function in coastal sediments: A case study on the Prestige oil spill. Environ. Sci. Pollut. Res. 2015, 22, 15200–15214. [Google Scholar] [CrossRef] [PubMed]
- Duran, R.; Cravo-Laureau, C. Role of environmental factors and microorganisms in determining the fate of polycyclic aromatic hydrocarbons in the marine environment. FEMS Microbiol. Rev. 2016, 40, 814–830. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Nigro, L.M.; Gutierrez, T.; D’Ambrosio, L.; Joye, S.B.; Highsmith, R.; Teske, A. Pulsed blooms and persistent oil-degrading bacterial populations in the water column during and after the Deepwater Horizon blowout. Deep Sea Res. Part II Top. Stud. Oceanogr. 2016, 129, 282–291. [Google Scholar] [CrossRef]
- Saul, D.J.; Aislabie, J.M.; Brown, C.E.; Harris, L.; Foght, J.M. Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol. Ecol. 2005, 53, 141–155. [Google Scholar] [CrossRef]
- Horz, H.P.; Tchawa Yimga, M.; Liesack, W. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 2001, 67, 4177–4185. [Google Scholar] [CrossRef]
- Sipos, R.; Székely, A.J.; Palatinszky, M.; Révész, S.; Márialigeti, K.; Nikolausz, M. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targeting bacterial community analysis. FEMS Microbiol. Ecol. 2007, 60, 341–350. [Google Scholar] [CrossRef]
- Tiquia, S.M.; Michel, F.C. Bacterial diversity in livestock manure composts as characterized by terminal restriction fragment length polymorphisms (T-RFLP) of PCR-amplified 16S rRNA and gene sequences. In Microbiology of Composting and Other Biodegradation Processes; Insam, H., Ed.; Springer: Heidelberg, Germany, 2002. [Google Scholar]
- Sanchez, J.I.; Rossetti, L.; Martinez, B.; Rodriguez, A.; Giraffa, G. Application of reverse transcriptase PCR-based T-RFLP to perform semi-quantitative analysis of metabolically active bacteria in dairy fermentations. J. Microbiol. Methods 2006, 65, 268–277. [Google Scholar] [CrossRef]
- Moeseneder, M.M.; Arrieta, J.M.; Muyzer, G.; Winter, C.; Herndl, G.J. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 1999, 65, 3518–3525. [Google Scholar]
- Nikolausz, M.; Sipos, R.; Revesz, S.; Szekely, A.; Marialigeti, K. Observation of bias associated with re-amplification of DNA isolated from denaturing gradient gels. FEMS Microbiol. Lett. 2005, 244, 385–390. [Google Scholar] [CrossRef]
- Nunan, N.; Daniell, T.J.; Singh, B.K.; Papert, A.; McNicol, J.W.; Prosser, J.I. Links between plant and rhizoplane bacterial communities in grassland soils, characterized using molecular techniques. Appl. Environ. Microbiol. 2005, 71, 6784–6792. [Google Scholar] [CrossRef]
- Eriksson, M.; Ka, J.O.; Mohn, W.W. Effects of low temperature and freeze-thaw cycles on hydrocarbon biodegradation in arctic tundra soil. Appl. Environ. Microbiol. 2001, 67, 5107–5112. [Google Scholar] [CrossRef] [PubMed]
- Whyte, L.G.; Smits, T.H.M.; Labbè, D.; Witholt, B.; Greer, C.W.; Van Beilen, J.B. Gene cloning and characterization of multiple alkane hydroxilase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl. Environ. Microbiol. 2002, 68, 5933–5942. [Google Scholar] [CrossRef] [PubMed]
- Yakimov, M.M.; Timmis, K.N.; Golyshin, P.N. Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 2007, 18, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Kube, M.; Chernikova, T.N.; Al-Ramahi, Y.; Beloqui, A.; Lopez-Cortez, N.; Guazzaroni, M.-A.; Heipieper, H.J.; Klages, S.; Kotsyurbenko, O.R.; Langer, I.; et al. Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica. Nat. Commun. 2013, 23, 2156. [Google Scholar] [CrossRef]
- Coulon, F.; McKew, B.A.; Osborn, A.M.; McGenity, T.J.; Timmis, K.N. Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ. Microbiol. 2007, 9, 177–186. [Google Scholar] [CrossRef]
- Gentile, G.; Bonsignore, M.; Santisi, S.; Catalfamo, M.; Giuliano, L.; Genovese, L.; Yakimov, M.M.; Denaro, R.; Genovese, M.; Cappello, S. Biodegradation potentiality of psychrophilic bacterial strain Oleispira antarctica RB-8T. Mar. Pollut. Bull. 2016, 105, 125–130. [Google Scholar] [CrossRef]
- Whyte, L.G.; Hawari, J.; Zhou, E.; Bourbonniére, L.; Inniss, W.E.; Greer, C.W. Biodegradation of variable chain-length alkanes at low temperatures by a psychrotrophic Rhodococcus sp. Appl. Environ. Microbiol. 1998, 64, 2578–2584. [Google Scholar]
- Grossman, M.J.; Prince, R.C.; Garrett, R.M.; Garrett, K.K.; Bare, R.E.; O’Neil, K.R.; Sowlay, M.R.; Hinton, S.M.; Lee, K.; Sergy, G.; et al. Microbial diversity in oiled and un-oiled shoreline sediments in the Norwegian Arctic. In Microbial Biosystems: New Frontiers Proceedings of the 8th International Symposium on Microbial Ecology; Bell, C.R., Brylinsky, M., Johnson-Green, P., Eds.; Atlantic Canada Society for Microbial Ecology: Halifax, Canada, 2000; pp. 775–787. [Google Scholar]
- Terrisse, F.; Cravo-Laureau, C.; Noël, C.; Cagnon, C.; Dumbrell, A.J.; McGenity, T.J.; Duran, R. Variation of oxygenation conditions on a hydrocarbonoclastic microbial community reveals Alcanivorax and Cycloclasticus ecotypes. Front. Microbiol. 2017, 8, 1549. [Google Scholar] [CrossRef]
- Brito, E.M.S.; Guyoneaud, R.; Goñi-Urriza, M.; Ranchou-Peyruse, A.; Verbaere, A.; Crapez, M.A.C.; Wasserman, J.C.A.; Duran, R. Characterization of hydrocarbonoclastic bacterial communities from mangrove sediments in Guanabara Bay, Brazil. Res. Microbiol. 2006, 157, 752–762. [Google Scholar] [CrossRef]
- LaRoe, S.L.; Wang, B.; Han, J.I. Isolation and characterization of a novel polycyclic aromatic hydrocarbon-degrading bacterium, Sphingopyxis sp strain M2R2, capable of passive spreading motility through soil. Environ. Eng. Sci. 2010, 27, 452–456. [Google Scholar] [CrossRef]
- Yergeau, E.; Sanschagrin, S.; Beaumier, D.; Greer, C.W. Metagenomic analysis of the bioremediation of diesel- contaminated Canadian high arctic soils. PLoS ONE 2012, 7, e30058. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.Y.; Lee, I.; Cho, Y.; Lee, Y.M.; Jung, Y.J.; Baek, K.; Nam, S.; Lee, H.K. Sediminicola arcticus sp. nov., a psychrophilic bacterium isolated from deep-sea sediment, and emended description of the genus Sediminicola. Int. J. Syst. Evol. Microbiol. 2015, 65, 1567–1571. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).