The Mycobacterium bovis BCG GroEL1 Contributes to Isoniazid Tolerance in a Dormant-Like State Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Cultures
2.2. Wayne Dormancy Model
2.3. Drug Treatment and Viability Determination
2.4. RNA Extraction, Reverse Transcription, and Real-Time PCR Analysis
2.5. ROS Determination
2.6. ATP Determination
2.7. Statistical Analysis
3. Results and Discussion
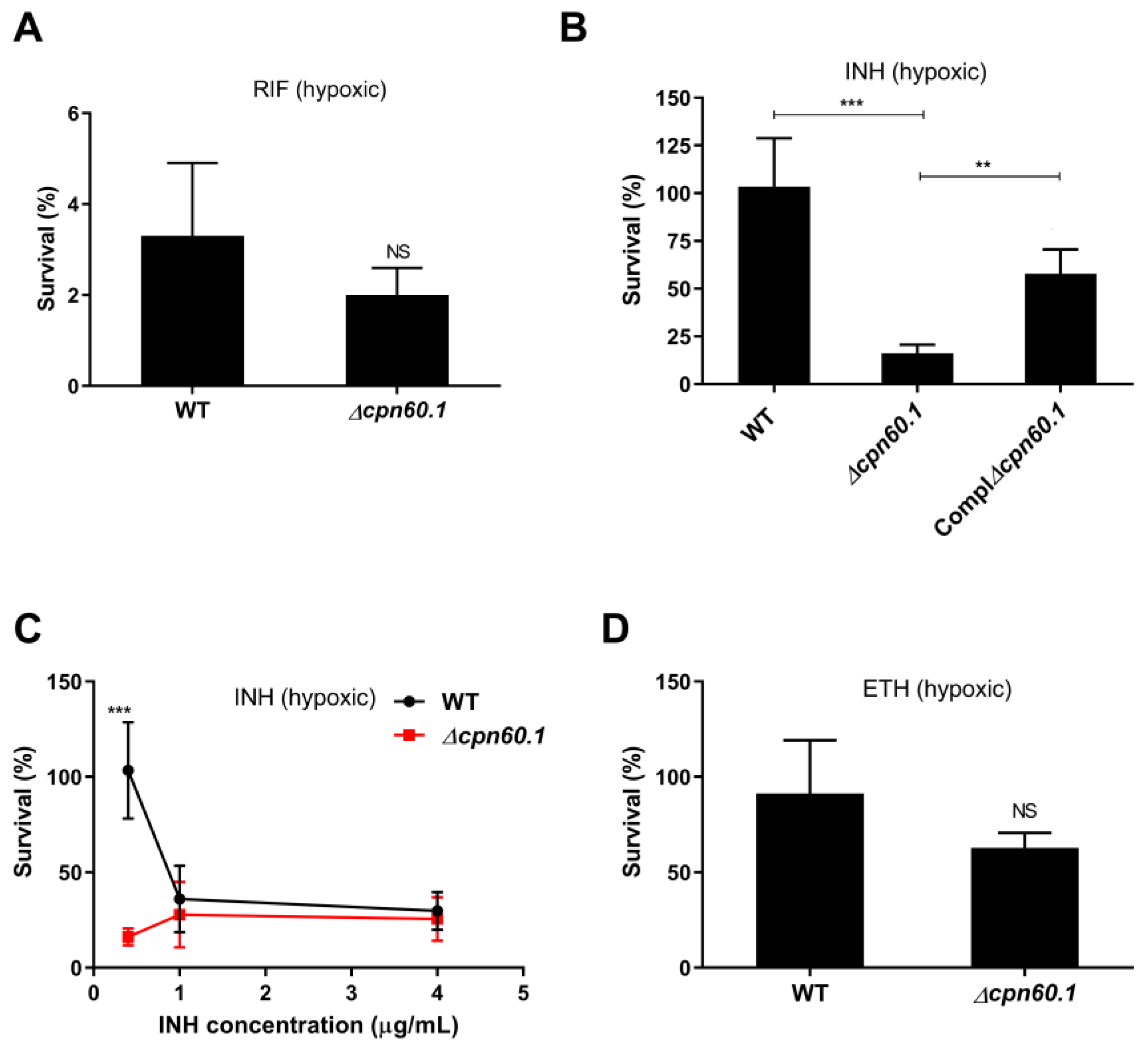
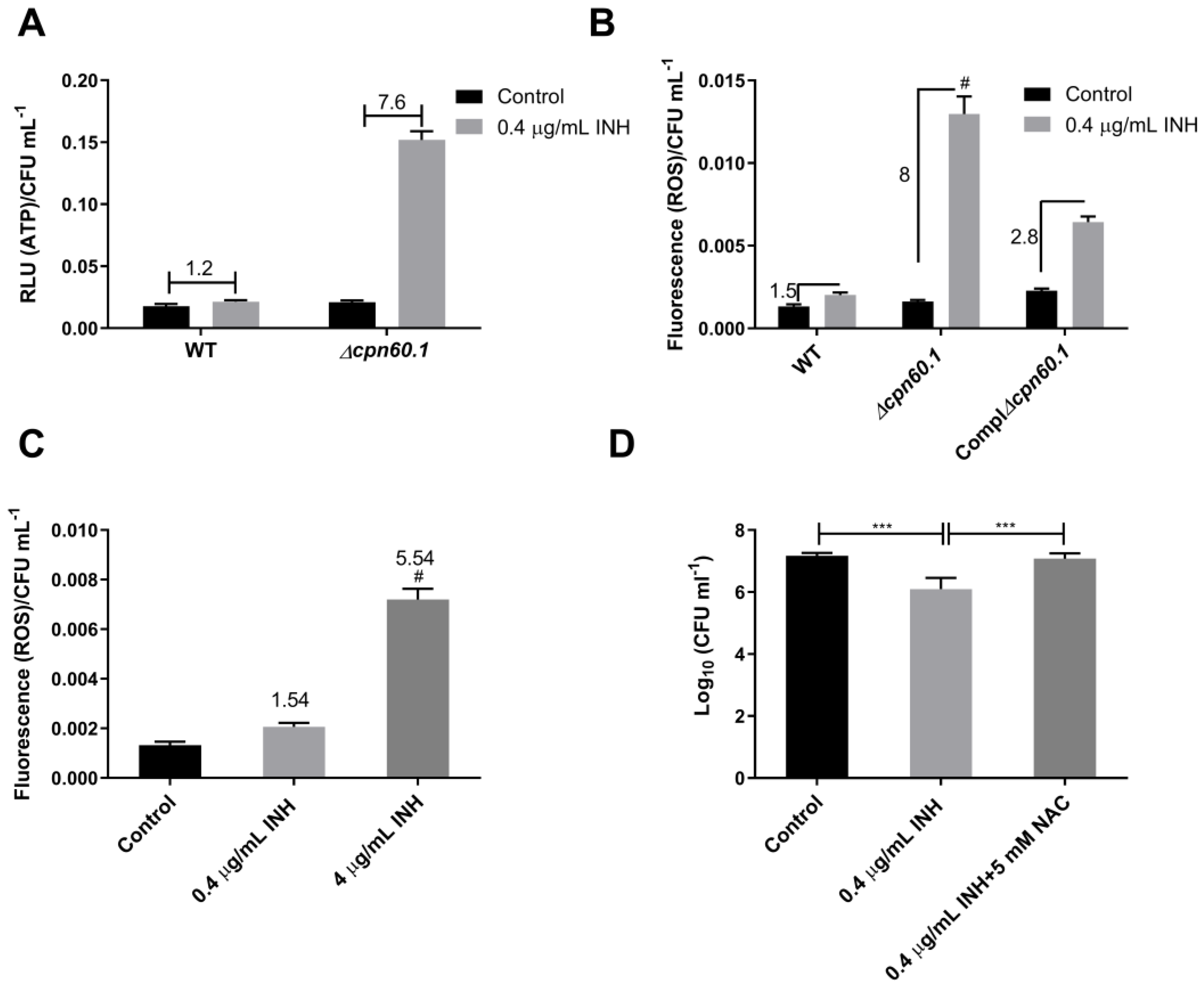
3.1. GroEL1 Contributes to Isoniazid Tolerance in the Wayne Dormant-like State Model
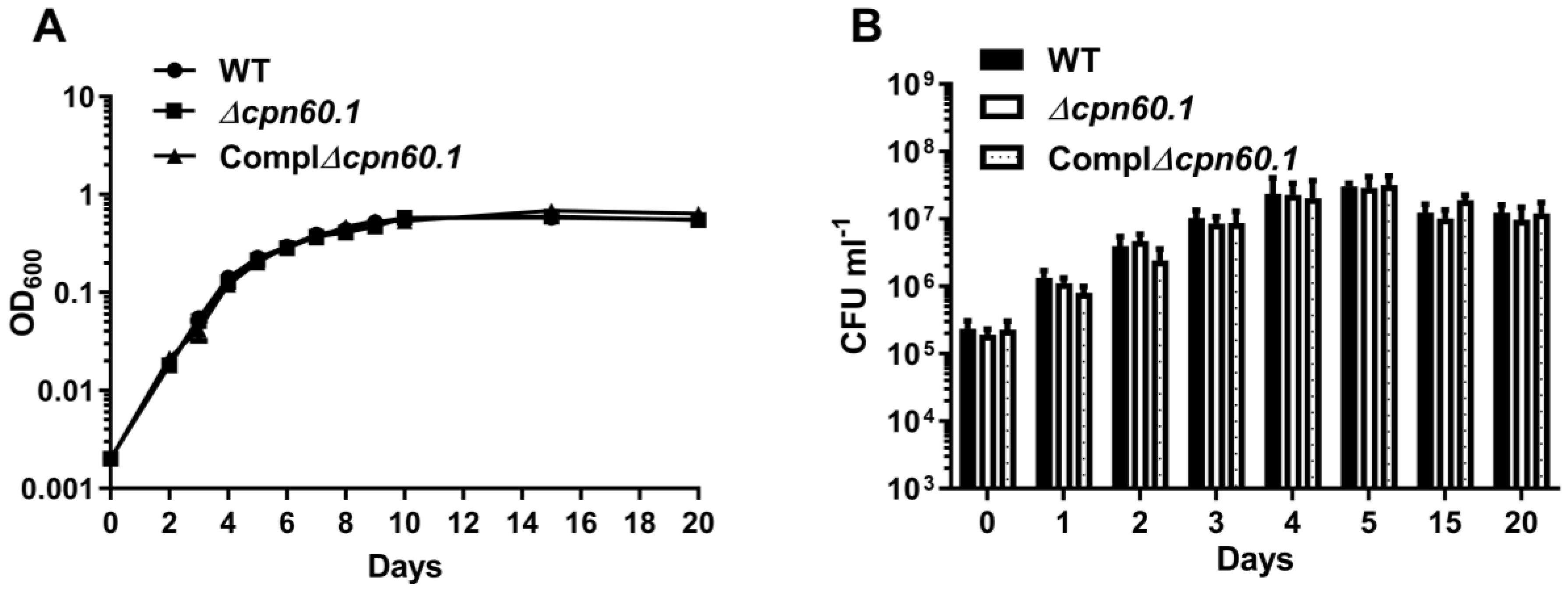
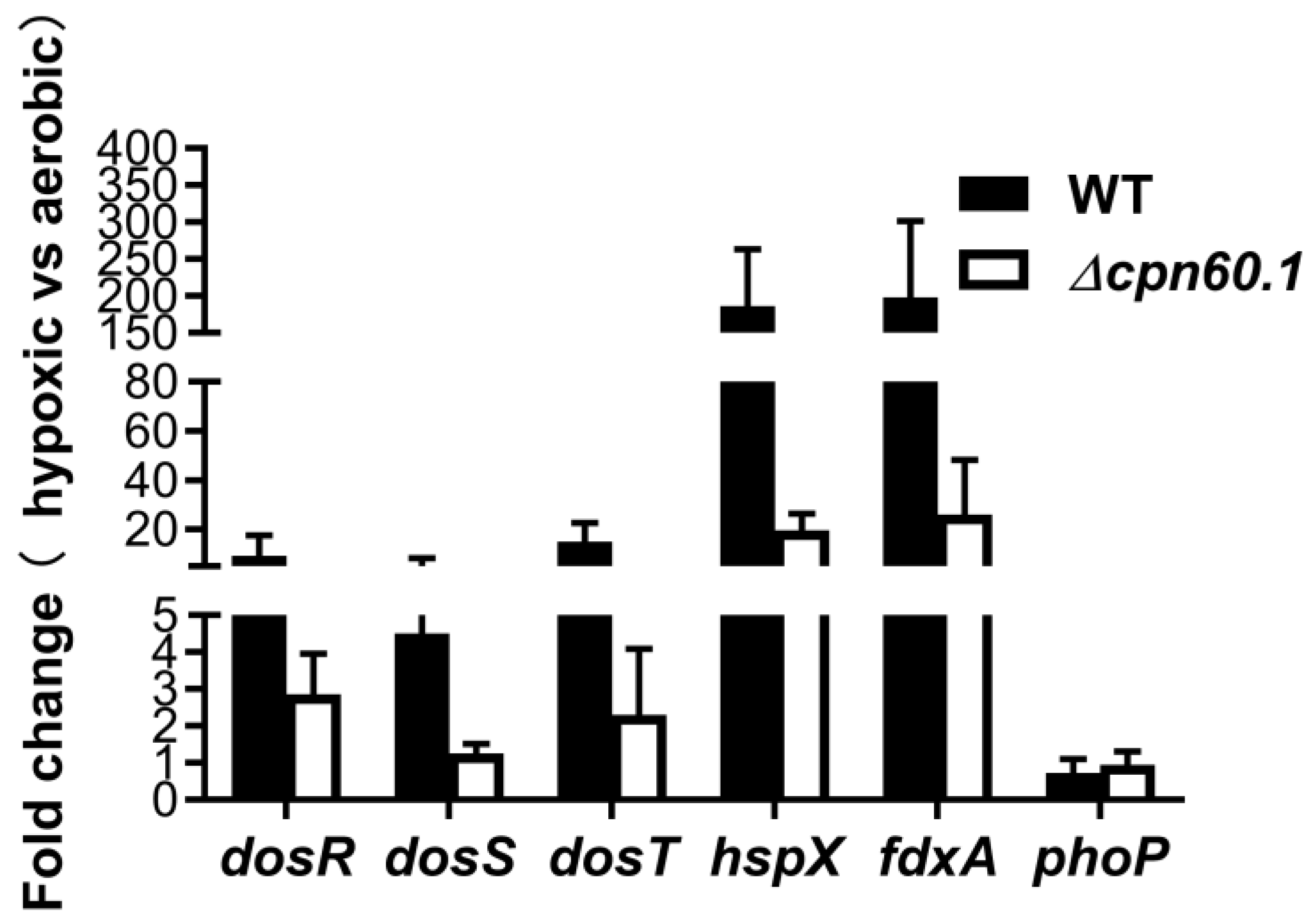
3.2. GroEL1 Is Not Required for Hypoxic Growth Arrest but Contributes to the DosR Regulon Optimal Induction
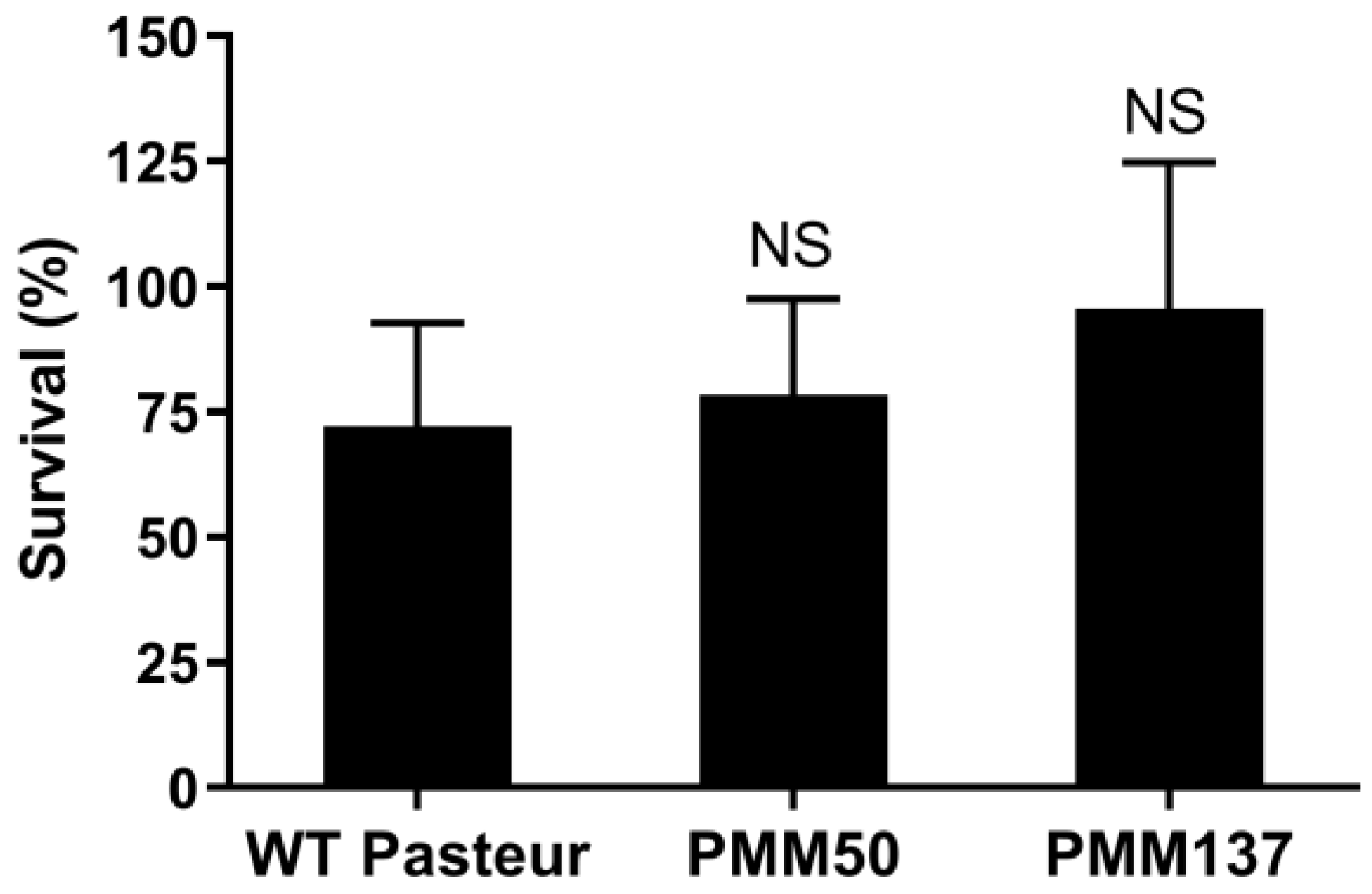
3.3. PDIM Doesn’t Contribute to Isoniazid Tolerance during the Dormant-like State Induced by Hypoxia
3.4. GroEL1 Contributes to the Metabolic Adaptation Required for INH Tolerance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Iacobino, A.; Piccaro, G.; Giannoni, F.; Mustazzolu, A.; Fattorini, L. Activity of drugs against dormant Mycobacterium tuberculosis. Int. J. Mycobacteriol. 2016, 5 (Suppl. 1), S94–S95. [Google Scholar] [CrossRef] [PubMed]
- Betts, J.C.; Lukey, P.T.; Robb, L.C.; McAdam, R.A.; Duncan, K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002, 43, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Wayne, L.G.; Hayes, L.G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996, 64, 2062–2069. [Google Scholar] [CrossRef] [PubMed]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Jones, S.E.; Lennon, J.T. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. USA 2010, 107, 5881–5886. [Google Scholar] [CrossRef] [PubMed]
- Levin-Reisman, I.; Ronin, I.; Gefen, O.; Braniss, I.; Shoresh, N.; Balaban, N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355, 826–830. [Google Scholar] [CrossRef]
- Rawat, R.; Whitty, A.; Tonge, P.J. The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: Adduct affinity and drug resistance. Proc. Natl. Acad. Sci. USA 2003, 100, 13881–13886. [Google Scholar] [CrossRef]
- Zeng, S.; Soetaert, K.; Ravon, F.; Vandeput, M.; Bald, D.; Kauffmann, J.M.; Mathys, V.; Wattiez, R.; Fontaine, V. Isoniazid bactericidal activity involves electron transport chain perturbation. Antimicrob. Agents Chemother. 2019, 63, e01841-18. [Google Scholar] [CrossRef]
- Zhu, J.H.; Wang, B.W.; Pan, M.; Zeng, Y.N.; Rego, H.; Javid, B. Rifampicin can induce antibiotic tolerance in mycobacteria via paradoxical changes in rpoB transcription. Nat. Commun. 2018, 9, 4218. [Google Scholar] [CrossRef]
- Jones, R.M.; Adams, K.N.; Eldesouky, H.E.; Sherman, D.R. The evolving biology of Mycobacterium tuberculosis drug resistance. Front. Cell. Infect. Microbiol. 2022, 12, 1027394. [Google Scholar] [CrossRef]
- Baek, S.H.; Li, A.H.; Sassetti, C.M. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 2011, 9, e1001065. [Google Scholar] [CrossRef]
- Sirakova, T.D.; Dubey, V.S.; Deb, C.; Daniel, J.; Korotkova, T.A.; Abomoelak, B.; Kolattukudy, P.E. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology 2006, 152, 2717–2725. [Google Scholar] [CrossRef] [PubMed]
- Santucci, P.; Johansen, M.D.; Point, V.; Poncin, I.; Viljoen, A.; Cavalier, J.F.; Kremer, L.; Canaan, S. Nitrogen deprivation induces triacylglycerol accumulation, drug tolerance and hypervirulence in mycobacteria. Sci. Rep. 2019, 9, 8667. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, M.; Nathan, C.; Rhee, K.Y. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat. Commun. 2014, 5, 4306. [Google Scholar] [CrossRef] [PubMed]
- Kitzenberg, D.A.; Lee, J.S.; Mills, K.B.; Kim, J.S.; Liu, L.; Vazquez-Torres, A.; Colgan, S.P.; Kao, D.J. Adenosine awakens metabolism to enhance growth-independent killing of tolerant and persister bacteria across multiple classes of antibiotics. MBio 2022, 13, e0048022. [Google Scholar] [CrossRef]
- Vilcheze, C.; Hartman, T.; Weinrick, B.; Jain, P.; Weisbrod, T.R.; Leung, L.W.; Freundlich, J.S.; Jacobs, W.R., Jr. Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2017, 114, 4495–4500. [Google Scholar] [CrossRef]
- Boon, C.; Dick, T. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 2002, 184, 6760–6767. [Google Scholar] [CrossRef]
- Onwueme, K.C.; Vos, C.J.; Zurita, J.; Ferreras, J.A.; Quadri, L.E. The dimycocerosate ester polyketide virulence factors of mycobacteria. Prog. Lipid Res. 2005, 44, 259–302. [Google Scholar] [CrossRef]
- Cox, J.S.; Chen, B.; McNeil, M.; Jacobs, W.R., Jr. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 1999, 402, 79–83. [Google Scholar] [CrossRef]
- Yu, J.; Tran, V.; Li, M.; Huang, X.; Niu, C.; Wang, D.; Zhu, J.; Wang, J.; Gao, Q.; Liu, J. Both phthiocerol dimycocerosates and phenolic glycolipids are required for virulence of Mycobacterium marinum. Infect. Immun. 2012, 80, 1381–1389. [Google Scholar] [CrossRef]
- Astarie-Dequeker, C.; Le Guyader, L.; Malaga, W.; Seaphanh, F.K.; Chalut, C.; Lopez, A.; Guilhot, C. Phthiocerol dimycocerosates of M. tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog. 2009, 5, e1000289. [Google Scholar] [CrossRef] [PubMed]
- Quigley, J.; Hughitt, V.K.; Velikovsky, C.A.; Mariuzza, R.A.; El-Sayed, N.M.; Briken, V. The Cell Wall Lipid PDIM Contributes to Phagosomal Escape and Host Cell Exit of Mycobacterium tuberculosis. MBio 2017, 8, e00148-17. [Google Scholar] [CrossRef] [PubMed]
- Mohandas, P.; Budell, W.C.; Mueller, E.; Au, A.; Bythrow, G.V.; Quadri, L.E. Pleiotropic consequences of gene knockouts in the phthiocerol dimycocerosate and phenolic glycolipid biosynthetic gene cluster of the opportunistic human pathogen Mycobacterium marinum. FEMS Microbiol. Lett. 2016, 363, fnw016. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Constant, P.; Yang, D.; Baulard, A.; Lefevre, P.; Daffe, M.; Wattiez, R.; Fontaine, V. Cpn60.1 (GroEL1) contributes to mycobacterial Crabtree effect: Implications for biofilm formation. Front. Microbiol. 2019, 10, 1149. [Google Scholar] [CrossRef] [PubMed]
- Soetaert, K.; Rens, C.; Wang, X.M.; De Bruyn, J.; Laneelle, M.A.; Laval, F.; Lemassu, A.; Daffe, M.; Bifani, P.; Fontaine, V.; et al. Increased Vancomycin Susceptibility in Mycobacteria: A new approach to identify synergistic activity against multidrug-resistant mycobacteria. Antimicrob. Agents Chemother. 2015, 59, 5057–5060. [Google Scholar] [CrossRef]
- Wang, X.M.; Lu, C.; Soetaert, K.; S’Heeren, C.; Peirs, P.; Laneelle, M.A.; Lefevre, P.; Bifani, P.; Content, J.; Daffe, M.; et al. Biochemical and immunological characterization of a cpn60.1 knockout mutant of Mycobacterium bovis BCG. Microbiology 2011, 157, 1205–1219. [Google Scholar] [CrossRef]
- Aravindhan, V.; Christy, A.J.; Roy, S.; Ajitkumar, P.; Narayanan, P.R.; Narayanan, S. Mycobacterium tuberculosis groE promoter controls the expression of the bicistronic groESL1 operon and shows differential regulation under stress conditions. FEMS Microbiol. Lett. 2009, 292, 42–49. [Google Scholar] [CrossRef]
- Brosch, R.; Gordon, S.V.; Garnier, T.; Eiglmeier, K.; Frigui, W.; Valenti, P.; Dos Santos, S.; Duthoy, S.; Lacroix, C.; Garcia-Pelayo, C.; et al. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. USA 2007, 104, 5596–5601. [Google Scholar] [CrossRef]
- Simeone, R.; Leger, M.; Constant, P.; Malaga, W.; Marrakchi, H.; Daffe, M.; Guilhot, C.; Chalut, C. Delineation of the roles of FadD22, FadD26 and FadD29 in the biosynthesis of phthiocerol dimycocerosates and related compounds in Mycobacterium tuberculosis. FEBS J. 2010, 277, 2715–2725. [Google Scholar] [CrossRef]
- Howell Wescott, H.A.; Roberts, D.M.; Allebach, C.L.; Kokoczka, R.; Parish, T. Imidazoles induce reactive oxygen species in Mycobacterium tuberculosis which is not associated with cell death. ACS Omega 2017, 2, 41–51. [Google Scholar] [CrossRef]
- Kumar, A.; Toledo, J.C.; Patel, R.P.; Lancaster, J.R., Jr.; Steyn, A.J. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. USA 2007, 104, 11568–11573. [Google Scholar] [CrossRef] [PubMed]
- Honaker, R.W.; Dhiman, R.K.; Narayanasamy, P.; Crick, D.C.; Voskuil, M.I. DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. J. Bacteriol. 2010, 192, 6447–6455. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo-Asensio, J.; Mostowy, S.; Harders-Westerveen, J.; Huygen, K.; Hernandez-Pando, R.; Thole, J.; Behr, M.; Gicquel, B.; Martin, C. PhoP: A missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS ONE 2008, 3, e3496. [Google Scholar] [CrossRef]
- Lee, J.J.; Lee, S.K.; Song, N.; Nathan, T.O.; Swarts, B.M.; Eum, S.Y.; Ehrt, S.; Cho, S.N.; Eoh, H. Transient drug-tolerance and permanent drug-resistance rely on the trehalose-catalytic shift in Mycobacterium tuberculosis. Nat. Commun. 2019, 10, 2928. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.P.; Cai, W.; Zill, N.A.; Zhang, W.; Ojha, A.K. Adaptation of Mycobacterium tuberculosis to biofilm growth is genetically linked to drug tolerance. Antimicrob. Agents Chemother. 2019, 63, e01213-19. [Google Scholar] [CrossRef]
- Zheng, H.; Colvin, C.J.; Johnson, B.K.; Kirchhoff, P.D.; Wilson, M.; Jorgensen-Muga, K.; Larsen, S.D.; Abramovitch, R.B. Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence. Nat. Chem. Biol. 2017, 13, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Cho, H.J.; Kim, Y.M.; Oh, J.I.; Kang, B.S. Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J. Biol. Chem. 2009, 284, 13057–13067. [Google Scholar] [CrossRef]
- Sharma, A.; Rustad, T.; Mahajan, G.; Kumar, A.; Rao, K.V.; Banerjee, S.; Sherman, D.R.; Mande, S.C. Towards understanding the biological function of the unusual chaperonin Cpn60.1 (GroEL1) of Mycobacterium tuberculosis. Tuberculosis 2016, 97, 137–146. [Google Scholar] [CrossRef]
- Matta, S.K.; Kumar, D. Hypoxia and classical activation limits Mycobacterium tuberculosis survival by Akt-dependent glycolytic shift in macrophages. Cell Death Discov. 2016, 2, 16022. [Google Scholar] [CrossRef]
- Sherman, D.R.; Mdluli, K.; Hickey, M.J.; Arain, T.M.; Morris, S.L.; Barry, C.E., 3rd; Stover, C.K. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 1996, 272, 1641–1643. [Google Scholar] [CrossRef]
- Koshkin, A.; Zhou, X.T.; Kraus, C.N.; Brenner, J.M.; Bandyopadhyay, P.; Kuntz, I.D.; Barry, C.E., 3rd; Ortiz de Montellano, P.R. Inhibition of Mycobacterium tuberculosis AhpD, an element of the peroxiredoxin defense against oxidative stress. Antimicrob. Agents Chemother. 2004, 48, 2424–2430. [Google Scholar] [CrossRef]
- Bryk, R.; Lima, C.D.; Erdjument-Bromage, H.; Tempst, P.; Nathan, C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 2002, 295, 1073–1077. [Google Scholar] [CrossRef]
- Nambi, S.; Long, J.E.; Mishra, B.B.; Baker, R.; Murphy, K.C.; Olive, A.J.; Nguyen, H.P.; Shaffer, S.A.; Sassetti, C.M. The oxidative stress network of Mycobacterium tuberculosis reveals coordination between radical detoxification systems. Cell Host Microbe 2015, 17, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Lamprecht, D.A.; Finin, P.M.; Rahman, M.A.; Cumming, B.M.; Russell, S.L.; Jonnala, S.R.; Adamson, J.H.; Steyn, A.J. Turning the respiratory flexibility of Mycobacterium tuberculosis against itself. Nat. Commun. 2016, 7, 12393. [Google Scholar] [CrossRef] [PubMed]
- Shetty, A.; Dick, T. Mycobacterial cell wall synthesis inhibitors cause lethal ATP burst. Front. Microbiol. 2018, 9, 1898. [Google Scholar] [CrossRef] [PubMed]
- Heikal, A.; Hards, K.; Cheung, C.Y.; Menorca, A.; Timmer, M.S.; Stocker, B.L.; Cook, G.M. Activation of type II NADH dehydrogenase by quinolinequinones mediates antitubercular cell death. J. Antimicrob. Chemother. 2016, 71, 2840–2847. [Google Scholar] [CrossRef]
- Yano, T.; Kassovska-Bratinova, S.; Teh, J.S.; Winkler, J.; Sullivan, K.; Isaacs, A.; Schechter, N.M.; Rubin, H. Reduction of clofazimine by mycobacterial type 2 NADH:quinone oxidoreductase: A pathway for the generation of bactericidal levels of reactive oxygen species. J. Biol. Chem. 2011, 286, 10276–10287. [Google Scholar] [CrossRef]
- Van Acker, H.; Coenye, T. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 2017, 25, 456–466. [Google Scholar] [CrossRef]
- Singh, N.; Sharma, N.; Singh, P.; Pandey, M.; Ilyas, M.; Sisodiya, L.; Choudhury, T.; Gosain, T.P.; Singh, R.; Atmakuri, K. HupB, a nucleoid-associated protein, is critical for survival of Mycobacterium tuberculosis under host-mediated stresses and for enhanced tolerance to key first-line antibiotics. Front. Microbiol. 2022, 13, 937970. [Google Scholar] [CrossRef]
- Basu, D.; Khare, G.; Singh, S.; Tyagi, A.; Khosla, S.; Mande, S.C. A novel nucleoid-associated protein of Mycobacterium tuberculosis is a sequence homolog of GroEL. Nucleic Acids Res. 2009, 37, 4944–4954. [Google Scholar] [CrossRef]
| Primer Name | Sequence 5′ to 3′ |
|---|---|
| 23S forward primer | GAAGAATGAGCCTGCGAGTC |
| 23S reverse primer | GGTCCAGAACACGCCACTAT |
| sigA forward primer | CTCGGTTCGCGCCTACCTCA |
| sigA reverse primer | GCGCTCGCTAAGCTCGGTCA |
| dosR forward primer | CGGTCGCTGCTGGACAATC |
| dosR reverse primer | TTTCGGCTAGGAACATTCG |
| dosS forward primer | ACCGGCAGCATCGGGTATTGC |
| dosS reverse primer | TCGATGAGCAGCCCGATGAC |
| dosT forward primer | CATCGGTTGGATTTCCGCTG |
| dosT reverse primer | CCATCTGCCTTCTCGGTCAA |
| fdxA forward primer | CCTATGTGATCGGTAGTGA |
| fdxA reverse primer | GGGTTGATGTAGAGCATT |
| hspX forward primer | CGCACCGAGCAGAAGGAC |
| hspX reverse primer | CCGCCACCGACACAGTAA |
| phoP forward primer | TGGGTGGTGACGACTATGTG |
| phoP reverse primer | GTGGTTCCTTGTTGCCCTTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, S.; Yang, D.; Rens, C.; Fontaine, V. The Mycobacterium bovis BCG GroEL1 Contributes to Isoniazid Tolerance in a Dormant-Like State Model. Microorganisms 2023, 11, 286. https://doi.org/10.3390/microorganisms11020286
Zeng S, Yang D, Rens C, Fontaine V. The Mycobacterium bovis BCG GroEL1 Contributes to Isoniazid Tolerance in a Dormant-Like State Model. Microorganisms. 2023; 11(2):286. https://doi.org/10.3390/microorganisms11020286
Chicago/Turabian StyleZeng, Sheng, Dong Yang, Céline Rens, and Véronique Fontaine. 2023. "The Mycobacterium bovis BCG GroEL1 Contributes to Isoniazid Tolerance in a Dormant-Like State Model" Microorganisms 11, no. 2: 286. https://doi.org/10.3390/microorganisms11020286
APA StyleZeng, S., Yang, D., Rens, C., & Fontaine, V. (2023). The Mycobacterium bovis BCG GroEL1 Contributes to Isoniazid Tolerance in a Dormant-Like State Model. Microorganisms, 11(2), 286. https://doi.org/10.3390/microorganisms11020286








