Abstract
The taxonomic status of two gram-negative, whitish-pigmented motile bacteria KMM 9576T and KMM 9553 isolated from a sandy sediment sample from the Sea of Japan seashore was defined. Phylogenetic analysis revealed that strains KMM 9576T and KMM 9553 represent a distinct lineage within the family Rhizobiaceae, sharing 100% 16S rRNA sequence similarity and 99.5% average nucleotide identity (ANI) to each other. The strains showed the highest 16S rRNA sequence similarities of 97.4% to Sinorhizobium garamanticum LMG 24692T, 96.9% to Ensifer adhaerens NBRC 100388T, and 96.8% to Pararhizobium giardinii NBRC 107135T. The ANI values between strain KMM 9576T and Ensifer adhaerens NBRC 100388T, Sinorhizobium fredii USDA 205T, Pararhizobium giardinii NBRC 107135T, and Rhizobium leguminosarum NBRC 14778T were 79.9%, 79.6%, 79.4%, and 79.2%, respectively. The highest core-proteome average amino acid identity (cpAAI) values of 82.1% and 83.1% were estimated between strain KMM 9576T and Rhizobium leguminosarum NBRC 14778T and ‘Rhizobium album’ NS-104, respectively. The DNA GC contents were calculated from a genome sequence to be 61.5% (KMM 9576T) and 61.4% (KMM 9553). Both strains contained the major ubiquinone Q-10 and C18:1ω7c as the dominant fatty acid followed by 11-methyl C18:1ω7c and C19:0 cyclo, and polar lipids consisted of phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, an unidentified aminophospholipid, and two unidentified phospholipids. Based on phylogenetic and phylogenomic analyses, and phenotypic characterization, strains KMM 9576T and KMM 9553 are concluded to represent a novel genus and species, for which the name Fererhizobium litorale gen. nov., sp. nov. is proposed. The type strain of the type species is strain KMM 9576T (=NRIC 0957T).
1. Introduction
The genus Rhizobium, the type genus of the family Rhizobiaceae Conn 1938 [1], first described by Frank (1889) [2] and emended by Young et al. (2001) [3], has been the object of numerous taxonomic studies for a long time. It has been reported that the 16S rRNA gene sequencing has limitations in differentiating Rhizobiaceae members, whereas a multilocus sequence analysis (MLSA) of housekeeping genes proves to be more effective in resolving closely related species of the taxonomic position [4,5,6,7,8]. The use of MLSA demonstrated high heterogeneity of the genera Rhizobium and Agrobacterium within the family Rhizobiaceae, which led to the reclassification of the species group “Rhizobium galegae” as a novel genus Neorhizobium [7]. In addition, based on the MLSA results, the genus named Pararhizobium was proposed to accommodate the cluster of Rhizobium giardinii, R. herbae, “R. helanshanense,” R. sphaerophysae, and Blastobacter capsulatus [7,8]. Recently, in the phylogenomic study based on a whole genome sequencing analysis, Kuzmanovic et al. (2022) [9] have proposed a common approach for the genera delineation in the family Rhizobiaceae, applying a core-genome gene phylogeny and a pairwise core-proteome average amino acid identity (cpAAI) value of approximately 86%. The application of the proposed framework resulted in the reclassification of several Rhizobium species, including a proposal for the new genus Xaviernesmea and the retrieval of Ensifer and Sinorhizobium, which had previously been unified [10,11] as separate genera [9]. Currently, the family Rhizobiaceae comprises 19 genera, including Rhizobium and related Ensifer, Sinorhizobium, Agrobacterium, Pararhizobium, Neorhizobium, Shinella, Ciceribacter, etc. [9]. At the time of writing, the genus Rhizobium contains the largest number of validly described species; 88 as listed at http://lpsn.dsmz.de/genus/rhizobia, accessed on 12 September 2023, including Rhizobium leguminosarum as the type species of the genus [2,9,12,13]. Most rhizobia are able to invade plants of the family Leguminosae and incite the production of tumors or/and root nodules where the bacteria act as intracellular nitrogen-fixing symbionts. The type species Ensifer adhaerens had been reported to be a predator of Micrococcus luteus [14]. Members of the family Rhizobiaceae have been mainly originated from the rhizosphere and the roots of leguminous and other agricultural crops, as well as recovered from aquatic and marine ecosystems, including deep-sea and coastal sediments as non-symbiotic microorganisms [15,16,17,18,19,20,21].
The aim of the present study was to determine the taxonomic status of two aerobic, gram-negative, motile, whitish-pigmented bacteria designated KMM 9576T and KMM 9553 isolated from a sandy sediment sample obtained from the Sea of Japan seashore. Based on molecular and phenotypic data obtained, a novel genus and species, Fererhizobium litorale gen. nov., sp. nov. is described.
2. Materials and Methods
2.1. Isolation and Phenotypic Characterization of Bacteria
A sandy sediment sample was collected from offshore of the Sea of Japan, Russia, (42° 54.8416 N 131° 43.0430 E) at a water depth of 0.3 m on 25 October, 2012. Strains KMM 9576T and KMM 9553 were isolated using the dilution plating technique on tryptic soya agar (TSA) and the seawater medium (SWM), as described in a previous paper [22]. The strains were grown aerobically on Marine Agar 2216 (MA 2216) or Marine Broth 2216 (MB 2216), Tryptic Soya Agar (TSA) or Tryptic Soya Broth (TSB), and R2A agar (all BD Difco), yeast extract-mannitol agar (YMA) or broth (YMB), containing 10 g of mannitol; 0.5 g of KH2PO4; 0.2 g of MgSO4 7H2O; 0.1 g of NaCl; 4 g of CaCO3; 0.4 g of yeast extract; 15 g of agar, distilled water of 1 l, and stored at −70 °C in the MB supplemented with 20% (v/v) glycerol. No growth was observed when they were tested for the anaerobic growth on MA 2216 for a week using an AnaeroPackTM (Mitsubishi Gas Chemical America, Inc., New York, NY, USA). Strains KMM 9553 and KMM 9576T have been deposited to the Collection of Marine Microorganisms (KMM), G.B. Elyakov Pacific Institute of Bioorganic Chemistry, Far Eastern Branch of the Russian Academy of Sciences, Vladivostok, Russia, and the type strain KMM 9576T was deposited to the NODAI Culture Collection Center (NRIC), Tokyo University of Agriculture, Japan, under number of NRIC 0957T. The type strains Rhizobium leguminosarum NBRC 14778T, Ensifer adhaerens NBRC 100388T, Sinorhizobium garamanticum LMG 24692T, Rhizobium rhizogenes NBRC 13257T, Agrobacterium radiobacter NBRC 13532T, Pararhizobium giardinii NBRC 107135T, and Mesorhizobium mediterraneum JCM 21565T were kindly provided by the respective culture collections. All strains used in this study for phenotypic tests and lipid analyses were grown on/in YMA and YMB, if not stated otherwise. Gram-staining, oxidase, catalase, and motility (the hanging drop method) tests were examined according to the standard methods described by Smibert and Krieg (1994) [23]. The morphology of cells negatively stained with a 1% phosphotungstic acid was examined by electronic transmission microscopy Libra 120 (Carl Zeiss, Oberkochen, Germany), provided by the Far Eastern Centre of electronic microscopy, A.V. Zhirmunsky National Scientific Center of Marine Biology, Far Eastern Branch of the Russian Academy of Sciences, using cells grown in diluted (1/10) TSB on carbon-coated 200-mesh copper grids. The tests, including the hydrolysis of starch, gelatin, L-tyrosine, chitin, casein, DNA, and nitrate reduction (sulfanilic acid/α-naphthylamine test), as well as the growth at different salinities (0–10% NaCl), temperatures (5–40 °C), and pH values (4.0–11.5) were conducted as described by Smibert and Krieg (1994) [23]. The medium YMA (or YMB) was used as a basal, while mannitol and CaCO3 were omitted for determination of substrate hydrolysis and pH, respectively. Formation of H2S from thiosulfate was tested in the YMB using a lead acetate paper strip. Biochemical tests for all studied using API 20E, and strains KMM 9576T and KMM 9553 using API ID32 GN (bioMérieux, Marcy-l’Étoile, France), were performed as described by the manufacturer. Carbon source utilization was performed with the API 50 CHB/E tests (bioMérieux, Marcy-l’Étoile, France) according to the manufacturer’s instructions.
Antibiotic susceptibility of strains studied was examined on YMA plates using commercial paper discs (Research Centre of Pharmacotherapy, St. Petersburg, Russia) impregnated with the following antibiotics (µg per disc, unless otherwise indicated): ampicillin (10), benzylpenicillin (10 U), vancomycin (30), gentamicin (10), kanamycin (30), carbenicillin (100), chloramphenicol (30), neomycin (30), oxacillin (10), oleandomycin (15), lincomycin (15), ofloxacin (5), rifampicin (5), polymyxin (300 U), streptomycin (30), cephazolin (30), cephalexin (30), erythromycin (15), nalidixic acid (30), tetracycline (30), and doxocycline (10). For polar lipid and fatty acid analyses, strains KMM 9576T and KMM 9553 and seven related type strains were cultivated on YMA at 28 °C for 48 h. Lipids were extracted using the extraction method of Folch et al. (1957) [24]. Two-dimensional thin layer chromatography of polar lipids was conducted on Silica gel 60 F254 (10 × 10 cm, Merck, Darmstadt, Germany) using chloroform–methanol–water (65:25:4, v/v) for the first direction, and chloroform–methanol–acetic acid–water (80:12:15:4, v/v) for the second [25]. Lipids were detected by spraying with cerium–ammonium molybdate (CAM) containing 5 g of ammonium molybdate and 0.2 g of ceric sulphate in 100 mL of 10% sulphuric acid, followed by heating at 110 °C. Amino-containing lipids were determined with ninhydrin, phospholipids with molybdate reagent, glycolipids with alpha–naphthol, and choline-containing lipids with Dragendorff’s reagent. Respiratory lipoquinones were analyzed by the reversed-phase high-performance thin-layer chromatography as described by Mitchell and Fallon (1990) [26]. Fatty acid methyl esters (FAMEs) were prepared according to the procedure of the Microbial Identification System (MIDI) [27]. The analysis of FAMEs was performed using the GC–17A chromatograph (Shimadzu, Kyoto, Japan) equipped with a capillary column (30 m × 0.25 mm I.D.) coated with Supecowax-10 and SPB-5 phases (Supelco, Bellefonte, PA, USA). Identification of FAMEs was accomplished by equivalent chain-length values and comparing the retention times of the samples to those of standards. In addition, FAMEs were analyzed using a GLC–MS Shimadzu GC–MS model QP5050 (Column MDM–5S, the temperature program from 140 °C to 250 °C, at a rate of 2 °C/min).
2.2. 16S rRNA Gene Sequence and Phylogenetic Analysis
Genomic DNAs of strains KMM 9576T and KMM 9553 extracted according to the method of Saito and Miura [28] were used to determine 16S rRNA gene sequences as described by Shida et al. (1997) [29]. The 16S rRNA gene sequences were compared with those of the closest relatives using EzBioCloud service [30]. Phylogenies were performed on the GGDC web server (http://ggdc.dsmz.de/, accessed on 12 September 2023) [31] using the DSMZ phylogenomics pipeline [32] adapted to single genes. Maximum likelihood (ML) and maximum parsimony (MP) trees were inferred from the alignment with RAxML [33] and TNT [34], respectively. The robustness of phylogenetic trees was estimated by the bootstrap analysis of 1000 replicates.
2.3. Whole-Genome Sequencing, Phylogenomic, and Comparative Analyses
The genomic DNAs were obtained from the strains KMM 9576T and KMM 9553 using the High Pure PCR Template Preparation Kit (Roche, Basel, Switzerland). The quantity and quality of the genomic DNAs were measured using DNA gel electrophoresis and the Qubit 4.0 Fluorometer (Thermo Fisher Scientific, Singapore, Singapore). Preparation of the DNAs sequencing libraries was conducted using Nextera DNA Flex kits (Illumina, San Diego, CA, USA) and whole-genome sequencing were performed subsequently using paired-end runs on an Illumina MiSeq platform with a 150-bp read length. The reads were trimmed using Trimmomatic version 0.39 [35], and their quality was assessed using FastQC version 0.11.8 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed on 21 August 2021). Filtered reads were assembled into contigs with SPAdes version 3.15.3 [36], and genome metrics were calculated with the help of QUAST version 5.0.2 [37]. The genome completeness and contamination were estimated by CheckM version 1.1.3 based on the taxonomic-specific workflow (family Rhizobiaceae) [38].
The draft genome assemblies were annotated using NCBI Prokaryotic Genome Annotation Pipeline (PGAP) and Rapid Annotation using Subsystem Technology (RAST) [39,40]. Comparisons of the Average Nucleotide Identity (ANI), Average Amino Acid Identity (AAI), and digital DNA–DNA hybridization (dDDH) values of the strains KMM 9576T and KMM 9553 and their closest neighbors were performed with the online server ANI/AAI–Matrix [41] and TYGS platform [31], respectively. Core-proteome average amino acid identity (cpAAI) values between the strains and representatives of the family Rhizobiaceae were computed using a pipeline proposed by Kuzmanović et al. (https://github.com/flass/cpAAI_Rhizobiaceae, accessed on 10 January 2023) [9]. The core-proteome phylogeny based on 170 non-recombining core protein markers, including various members of Rhizobium genus, was inferred using IQ–TREE software under the LG + F + I + I + R8 model with bootstrapping using 100 replicates [42].
Annotation of secondary metabolite biosynthetic gene clusters was conducted using antiSMASH server version 6.1.1 (https://antismash.secondarymetabolites.org/#!/start, accessed on 13 March 2023). To identify carbohydrate-active enzymes (CAZymes), the dbCAN3 meta server version 11 was used with default settings (http://cys.bios.niu.edu/dbCAN3, accessed on 27 March 2023) [43]. Predictions by two of the three algorithms integrated within the server were considered sufficient for CAZy family assignments. Genome-wide analysis of orthologous clusters was performed using OrthoVenn2 and OrthoVenn3 (https://orthovenn3.bioinfotoolkits.net/home, accessed on 31 June 2023) [44,45]. The unique genes of strains KMM 9576T and KMM 9553 were functionally annotated using eggNOG-mapper v2 server (http://eggnog-mapper.embl.de/, accessed on 12 January 2023) [46].
3. Results and Discussion
3.1. Phylogenetic and Phylogenomic Analyses
The 16S rRNA gene sequences of strains KMM 9576T and KMM 9553 submitted to the GenBank under accession numbers LC126306 and LC126307 were 100% identical to the 16S rRNA gene sequences ON040664 and ON040663, respectively, and retracted from their genomic sequences. Phylogenetic analysis based on 16S rRNA gene sequences revealed that strains KMM 9576T and KMM 9553 are close to each other (100% identity), representing a distinct lineage within the family Rhizobiaceae (Figure 1). In the phylogenetic trees, Hoeflea anabaenae WH2KT was a sister clade. However, the strains shared the highest sequence similarity to S. garamanticum LMG 24692T (97.7%), followed by E. adhaerens NBRC 100388T (96.9%) and P. giardinii NBRC 107135T (96.8%). The similarity to other Rhizobiaceae members did not exceed 96.8%.
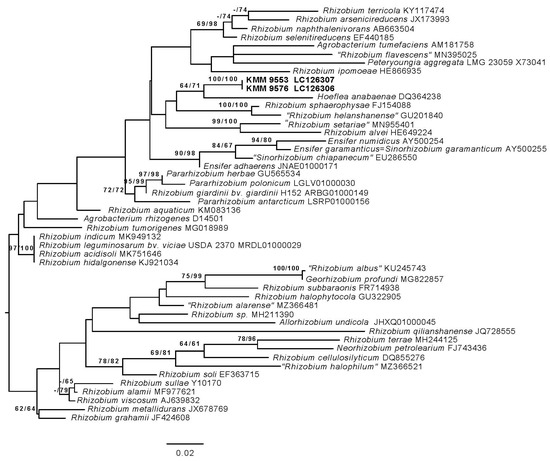
Figure 1.
ML tree based on 16S rRNA gene sequences available from the GenBank database showing relationships of the new strains KMM 9576T, KMM 9553 (in bold), and related members of the family Rhizobiaceae. The tree was inferred under the GTR + GAMMA model and rooted by midpoint rooting. The branches are scaled in terms of the expected number of substitutions per site. The numbers above the branches are support values when larger than 60% from ML (left) and MP (right) bootstrapping (1000 replicates).
The ANI values between strain KMM 9576T and E. adhaerens NBRC 100388T, S. fredii USDA 205T, P. giardinii NBRC 107135T, and R. leguminosarum NBRC 14778T were 79.9%, 79.6%, 79.4%, and 79.2%, respectively. The AAI values between strains KMM 9576T and KMM 9553, and members of related genera, varied from 66.7 to 69.8% (Table S2). The AAI values fell into the range of 60–80% [47] and did not exceed a cut-off value of 75% accepted for the delineation of Rhizobiaceae genera [48]. Genome sequences of strains KMM 9576T and KMM 9553 exhibited ANI/AAI values of the 99.5%/99.7%, and the dDDH values of 99.6%, confirming their non-clonal origin (Table S1).
In addition, the highest cpAAI values of 82.1% and 83. 1% were estimated between strain KMM 9576T and R. leguminosarum NBRC 14778T and “Rhizobium album” NS-104T [49], respectively, which is below a threshold value of 86% proposed by Kuzmanovic et al. (2022) [9] for the genera discrimination in the family Rhizobiaceae (Figure 2, Table S2). The core-proteome phylogeny provided by the cpAAI pipeline based on 97 strains of Rhizobiaceae genera (github.com/flass/cpAAI_Rhizobiaceae) showed that the strains KMM 9576T and KMM 9553 were more closely related to “R. album” NS-104T and “leguminosarum–rhizogene” clades (Figure 3). The position of strain “R. album” NS-104 on the phylogenetic tree (Figure 3), as well as its pairwise cpAAI values (Table S2), which indicate that it was incorrectly named and should be designated as a new Rhizobiaceae genus. Phylogenomic analysis confirmed that the novel strains form a distinct lineage adjacent to R. leguminosarum NBRC 14778T, which indicated that they may represent a novel genus of the family Rhizobiaceae.
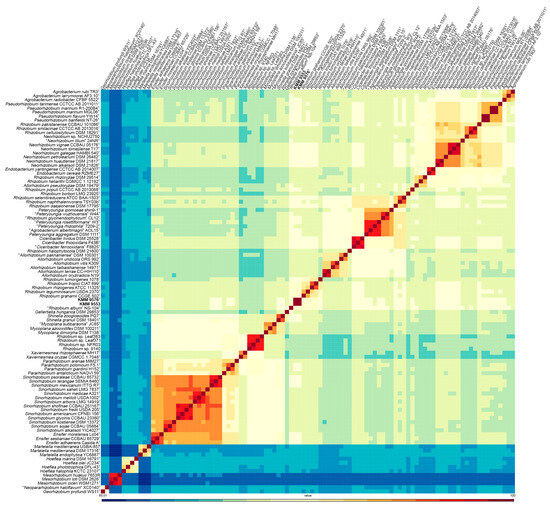
Figure 2.
Heatmap of pairwise cpAAI values between the strains KMM 9576T and KMM 9553 and the members of Rhizobiaceae clades. The Mesorhizobium spp. strains and Georhizobium profindi WS11 were included as outgroups. The KMM 9576T and KMM 9553 are marked in bold.
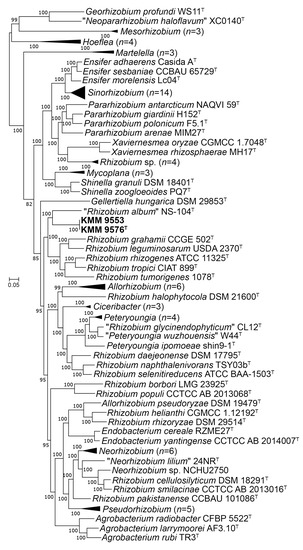
Figure 3.
ML tree based on concatenated 170 protein sequences showing a phylogenetic position of the novel strains KMM 9576T, KMM 9553, and related members of the family Rhizobiaceae. The tree was inferred using IQ–TREE software under the LG + F + I + I + R8 model with bootstrapping of 100 replicates. The Mesorhizobium spp. strains and Georhizobium profindi WS11 were used as outgroups. Bar, 0.05 substitutions per amino acid position. Collapsed branches are shown in black triangles.
Thus, these phylogenetic and phylogenomic data suggested that strains KMM 9576T and KMM 9553 represent a novel species and a novel genus within the family Rhizobiaceae.
3.2. Genomic Characteristics
According to an evaluation by the CheckM tool [38], the genome sequences of strains KMM 9576T and KMM 9553 were obtained with high completeness (98.2% and 100%) at low values of contaminations (0.04% and 0.22%), respectively. The draft genomes of KMM 9576T and KMM 9553 were de novo assembled into 67 and 94 contigs, with N50 values of 350,002 and 211,989 bp, and L50 values of 5 and 8, respectively. The genome sizes were estimated to be 5,252,096 and 5,494,465 bp in length with a coverage of 61× and 41×, respectively. The genome sequences were in accordance with the proposed minimal standards for bacterial taxonomy [50]. The comparison of genome features with genomes of type strains of closely and distantly related Rhizobiaceae genera is presented in Table 1. The genome sequences contain from 4981 (KMM 9576T) to 7623 (R. leguminosarum USDA 2370T) genes, from 45 (KMM 9576T) to 64 (E. adhaerens ATCC 33212T) tRNAs, and from 1 up to 5 ribosomal RNA operon copies (E. adhaerens ATCC 33212T).

Table 1.
Genomic features of strains KMM 9576T, KMM 9553, and type strains of closely related Rhizobiaceae genera.
According to the RAST annotation, about 1400 genes for both strains KMM 9576T and KMM 9553 are in subsystems, among which the largest number of genes was assigned to “Amino Acids and Derivatives,” “Carbohydrates,” and “Cofactors, Vitamins, Prosthetic Groups, Pigments.” According to the KEGG annotation, both strains have genes encoding the Embden–Meyerhof pathway (except for phosphofructokinase and fructose–bisphosphatase ones, but diphosphate-dependent phosphofructokinase is present), the pentose phosphate pathway, the Krebs cycle, and the glyoxylate cycle. In addition, there is a complete pathway for glycogen biosynthesis and degradation. According to the dbCAN3 server, KMM 9576T and KMM 9553 encode a total of 109 and 113 CAZymes, respectively, with glycosyltransferases (GT) being more abundant in the GT2 and GT14 families.
To identify Fererhizobium specific genes, orthologous groups were analyzed using the translated proteomes of KMM 9576T and KMM 9553 in comparison with those of the three closely related genera “R. album”, R. leguminosarum, and R. rhizogenes (Figure 3). A total of 6487 orthologous clusters comprising 31,339 proteins were found. The core genome shared by the five compared strains was represented by 2547 orthologous clusters (Figure 4a). Among them, 1781 were single-copy gene clusters.
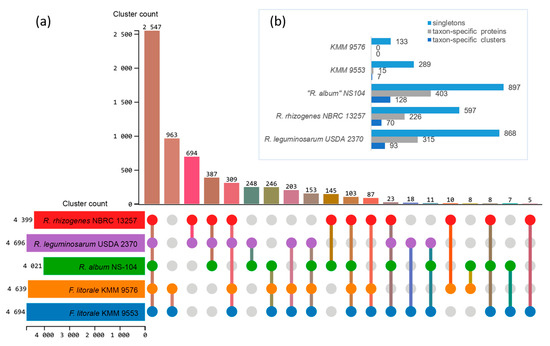
Figure 4.
Proteome comparison of Fererhizobium strains and type strains of closely related genera “R. album” NS-104T, R. leguminosarum NBRC 14778T, and R. rhizogenes NBRC 13257T using OrthoVenn3. UpSet–JS table displays common (a) and unique (b) orthologous clusters, and singletons (b). Horizontal bar chart on the left shows the number of orthologous clusters per strain. Right vertical bar chart indicates the number of orthologous clusters shared among the strains. The lines represent intersecting sets. Upper horizontal bar chart (b) shows the number of taxon-specific clusters including their proteins and singletons per strain.
The distinctive feature of the Fererhizobium core genome was an excessive number of genes in the clusters of L-arabinose transport ATP-binding protein (GO:0015407; 11–12 genes versus 1–3), and putative adenylate cyclase 3 (GO: 0035556; 9–10 genes versus one). In pairwise comparison, the largest number of orthologous clusters of 963 was revealed for the pair KMM 9576T/KMM 9553, following R. rhizogenes/R. leguminosarum (694), “R. album”/R. leguminosarum (248), and “R. album”/R. rhizogenes (145). Both Fererhizobium strains shared the most of clusters with “R. album” NS-104T (246), followed by R. leguminosarum NBRC 13257T (203) and R. rhizogenes NBRC 13257T (87) (Figure 4a). Among 963 Fererhizobium-specific orthologous clusters, functions of the most were associated with biological processes, such as transcription (GO:0006351) and regulation of transcription (GO:0006355), transmembrane transport (GO:0055085), and sodium ion transport (GO:0006814). A number of specific clusters (Figure 4b) for every strain were 128 for “R. album” NS-104T, 93 for R. leguminosarum NBRC 14778T, 70 for R. rhizogenes NBRC 13257T, and only 7 for KMM 9553. According to the RAST and eggNOG-mapper annotations, more than 50% of singletons were not annotated. Singletons for both strains KMM 9576T and KMM 9553 were mostly enriched in genes encoding mobile or selfish elements, restriction–modification systems, transport systems, and regulatory elements. Both strains do not have key nitrogen-fixation (nifHDK) and nodulation (nodABC) genes in their genomes. In addition, genes that are involved in the biosynthesis of capsular polysaccharide, exopolysaccharide, and lipopolysaccharide were revealed in KMM 9576T and KMM 9553. According to antiSMASH, the annotation of secondary metabolite biosynthetic gene clusters uncovered two homoserine lactone biosynthetic clusters, one redox-cofactor, such as PQQ (NC_021985:1458906-1494876), one thioamitide RiPP, as found in JOBF01000011, one other unspecified ribosomally synthesized and post-translationally modified peptide product (RiPP), one N-acyl amino acid, and one terpene biosynthetic cluster.
3.3. Phenotypic Characterization and Chemotaxonomy
Bacteria KMM 9576T and KMM 9553 were small rod- or ovoid-shaped motile by one or two subpolar flagella, and cells containing more than two flagella were also observed (Figure 5a,c). Novel bacteria could grow in salinities from 0 to 4% and grew well on/in SWM, MA 2216, MB 2216, R2A agar, TSB, TSA, YMA, and YMB. On carbohydrate-containing media, novel strains produced copious extracellular material, particularly strain KMM 9553. Individual cells of strain KMM 9553 could be observed as non-motile probably due to the large amount of mucus the strain tends to produce (Figure 5b).
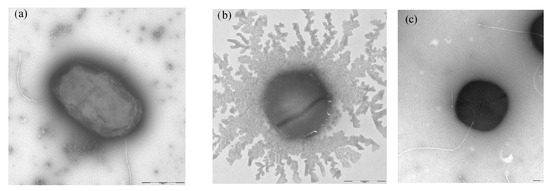
Figure 5.
Transmission electron micrographs of strains (a) KMM 9576T and (b,c) KMM 9553. Bar, 500 nm.
The novel strains were not identical in their phenotypic traits. Strain KMM 9576T, unlike KMM 9553, demonstrated a weak reaction of DNA hydrolysis, which tested negative for the oxidation of L-arabinose in API 20E and proved to be resistant to cephazolin and oleandomycin (Table 2) and utilized different carbon sources in API 50 CHB/E tests (Table S3). Strains KMM 9576T and KMM 9553 were similar to A. radiobacter NBRC 13532T in being able to grow in 0–4% of NaCl, and to hydrolyze DNA. Unlike other relatives, strains KMM 9576T and KMM 9553 and R. leguminosarum NBRC 14778T were able to produce H2S and hydrolyze tyrosine (Table 2). Strains KMM 9576T and KMM 9553 differed from E. adhaerens NBRC 100388T and P. giardinii NBRC 107135T in not being able to grow in 2–4% of NaCl and in their carbohydrate utilization patterns (Table 2 and Table S3).

Table 2.
Differential characteristics of strains KMM 9576T and KMM 9553 and type strains of related genera.
Both strains KMM 9576T and KMM 9553 contained Q-10 as the major ubiquinone; the predominant fatty acid was detected to be C18:1ω7c (52.47 and 46.16%), followed by 11-Methyl C18:1ω7c (14.91 and 14.11%), C19:0cyclo (13.01 and 15.27%), C18:0 (10.98 and 7.09%), and C14:0 3-OH (3.86 and 12.77%) in both strains, respectively (Table 3). The fatty acid profiles of strains KMM 9576T and KMM 9553 were found to be similar to that of R. leguminosarum NBRC 14778T. Strains E. adhaerens, NBRC 100388T, and S. garamanticum LMG 24692T differed from the novel strains in the presence of small amounts of C16:0 3-OH, C18:0 3-OH, and C17:0 cyclo (in E. adhaerens NBRC 100388T), while P. giardinii NBRC 107135T differed in the presence of C18:0 3-OH and C17:0 cyclo, as shown in Table 3. Strains M. mediterraneum JCM 21565T did not contain C14:0 3-OH and revealed a distinction in the presence of iso-C17:0 as compared with other related strains (Table 3). R. rhizogenes NBRC 13257T was significantly distinguished from all bacteria studied by the content of C15:0 3-OH and C19:0. These findings corroborate the results reported by Tighe et al. [51]. Unlike related type strains, R. rhizogenes NBRC 13257T and A. radiobacter NBRC 13532T contained high proportions of C14:0 3-OH and C16:0 3-OH, whereas novel strains contained a high amount of 11-Methyl C18:1ω7c (Table 3).

Table 3.
Cellular fatty acid composition (%) of strains KMM 9576T and KMM 9553 and type strains of related Rhizobiaceae members.
The polar lipid compositions of strains KMM 9576T, KMM 9553, and related bacteria tested did not significantly differ and included phosphatidylcholine (PC), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), an unidentified aminophospholipid (APL), and two unidentified phospholipids (PL and PL1) (Figure S1). The type strains R. rhizogenes NBRC 13257T, S. garamanticum LMG 24692T, E. adhaerens NBRC 100388T, and A. radiobacter NBRC 13532T differed from novel strains by the presence of additional unidentified phospholipids (PL2) in their lipid compositions. R. rhizogenes NBRC 13257T and S. garamanticum LMG 24692T contained additional unidentified lipids (L and/or L1). M. mediterraneum JCM 21565T contained a minor amount of PE compared to those of novel strains and included an unidentified lipid (L2) (Figure S1).
The DNA GC contents of 61.4% and 61.5% were calculated from a genome sequence of strains KMM 9576T and KMM 9553, respectively, which is close to the values of 49–68 mol% reported for the members of the family Rhizobiaceae [1]. The phylogenetic distinctness of strains KMM 9576T and KMM 9553 was supported by phenotypic differences in the temperature and salinity ranges that provided their growth, substrate hydrolysis ability, and carbohydrate utilization patterns. Differential phenotypic and physiological characteristics are indicated in Table 2 and Table S2. Based on the combination of phylogenomic analyses and phenotypic characteristics, it is proposed to classify strains KMM 9576T and KMM 9553 as a novel genus and species, Fererhizobium litorale.
4. Conclusions
Description of Fererhizobium gen. nov.
Fererhizobium gen. nov. (Fe.re.rhi.zo’bi.um. L. adv. fere, nearly, almost; N.L. neut. n. Rhizobium a bacterial generic name; N.L. neut. n. Fererhizobium, a genus adjacent to Rhizobium).
Gram-stain negative, aerobic non-spore-forming rod-shaped or ovoid cells. Oxidase- and catalase-positive. The major respiratory quinone is ubiquinone-10. The major fatty acid is C18:1ω7c, followed by 11-Methyl C18:1ω7c and C19:0cyclo. The polar lipids include phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, an unidentified aminophospholipid, and two unidentified phospholipids. Phylogenetically related to members of the genera Rhizobium, Sinorhizobium, and Pararhizobium within the family Rhizobiaceae, class Alphaproteobacteria. The type species is Fererhizobium litorale sp. nov.
Description of Fererhizobium litorale sp. nov.
Fererhizobium litorale (li.to.ra’le. L. neut. adj. litorale of or belonging to the seashore).
In addition to the properties described for the genus, the species is characterized as follows. Whitish-pigmented, rod- or ovoid-shaped cells, 0.5–0.7 μm in diameter and 0.8–1.4 μm in length, encapsulated, motile by one or two subpolar flagella. Cells may be observed as non-motile, with growth on/in YMA, YMB, TSA, TSB, MA 2216, MB 2216, and SWM. On carbohydrate-containing media, extracellular material is produced abundantly. Growth occurs in 0–4% of NaCl (optimal is 0.5–1%), and at 5–38 °C (optimal is 28–30 °C); there is weak growth at 38 °C and no growth at 40 °C. The pH range for growth is 6.0–10.0, with an optimum of 7.0–8.0. It is negative for hydrolysis of gelatin, casein, starch, chitin, Tween 80, and nitrate reduction, and positive for DNA hydrolysis and H2S production in conventional tests. On the L-tyrosine-containing medium, it forms a transparent zone but does not produce black–brown pigments.
According to the ID32 GN, it is positive for the assimilation of L-rhamnose, N-acetylglucosamine, D-sucrose, inositol, D-maltose, lactic acid, L-alanine, D-mannitol, D-glucose, salicin, L-fucose, D-sorbitol, L-arabinose, propionic acid, L-histidine, potassium 2-ketogluconate, and L-proline; weakly positive for the assimilation of D-ribose and 3-hydroxybutyric acid; and negative for the assimilation of itaconic acid, suberic acid, sodium malonate, sodium acetate, potassium 5-ketogluconate, glycogen, 3-hydroxybenzoic acid, L-serine, D-melibiose, capric acid, valeric acid, trisodium citrate, and 4-hydroxybutyric acid.
According to the API 20E, tests were positive for citrate utilization, PNPG test, urease production under anaerobic conditions, and oxidation of D-sucrose (weak reaction), while they were negative for gelatin hydrolysis, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, H2S production under anaerobic conditions and tryptophane deaminase, indole production, acetoin production (Voges–Proskauer reaction), and an oxidation of D-glucose, D-mannitol, inositol, D-sorbitol, L-rhamnose, D-melibiose, and amygdalin; oxidation of L-arabinose is strain-dependent (reaction of the type strain is negative).
According to the API 50CH/E tests, it was positive for the utilization of aesculin ferric citrate, D-fucose, D-arabinose, D-ribose, and L-rhamnose; negative for the utilization of glycerol, erythritol, D-xylose, D-adonitol, L-xylose, methyl-βD-xylopyranoside, D-galactose, D-glucose, D-fructose, L-sorbose, dulcitol, inositol, D-mannitol, methyl-αD-mannopyranoside, methyl-αD-glucopyranoside, N-acetylglucosamine, amygdalin, arbutin, salicin, D-cellobiose, D-maltose, D-lactose, D-melibiose, D-sucrose, D-tregalose, inulin, D-melezitose, D-raffinose, amidon, glycogen, xylitol, gentiobiose, D-turanose, D-lyxose, D-tagatose, D-arabitol, L-arabitol, potassium gluconate, potassium 2-ketogluconate, and potassium 5-ketogluconate.
The utilization of L-arabinose, D-sorbitol, D-mannose, and L-fucose is strain-dependent (negative reaction of the type strain for L-arabinose, D-sorbitol, D-mannose utilization, and position for L-fucose utilization). Susceptible to (content per disc): gentamicin (10 µg), kanamycin (30 µg), neomycin (30 µg), ofloxacin (5 µg), rifampicin (5 µg), streptomycin (30 µg), and cephalexin (30 µg); and resistant to ampicillin (10 µg), benzylpenicillin (10 U), vancomycin (30 µg), carbenicillin (100 µg), lincomycin (15 µg), chloramphenicol (30 µg), nalidixic acid (30 µg), oxacillin (10 µg), polymyxin B (300 U), tetracycline (30 µg), doxocycline (10 µg), and erythromycin (15 µg); susceptibility to cephazolin (30 µg) and oleandomycin (15 µg) is strain-dependent (the type strain is resistant). The DNA GC content of 61.4–61.5% is calculated from the genome sequence. The major isoprenoid quinone is ubiquinone Q-10. The major fatty acid is C18:1ω7c, followed by 11-Methyl C18:1ω7c and C19:0cyclo. The polar lipids included phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, an unidentified aminophospholipid, and two unidentified phospholipids.
The GenBank accession number for the whole-genome shotgun sequence of strain KMM 9576T is JALDYZ010000000.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11102385/s1, Figure S1: Two-dimensional thin-layer chromatograms of polar lipids of strains: (a) KMM 9576T; (b) KMM 9553; (c) Rhizobium leguminosarum NBRC 14778T; (d) Rhizobium radiobacter NBRC 13532T; (e) Rhizobium rhizogenes NBRC 13257T; (f) Ensifer garamanticus LMG 24692T; (g) Mesorhizobium mediterraneum JCM 21565T; (h) Ensifer adhaerens NBRC 100388T; (i) Pararhizobium giardinii NBRC 107135T. Abbreviations: PC, phosphatidylcholine, PE, phosphatidylethanoamine; PG, phosphatidylglycerol; APL, an unknown aminophospholipid; PL, PL1, PL2, unknown phospholipids; L, L1, L2, unknown lipids; Table S1: Average nucleotide identity (ANI), average amino acid identity (AAI), and digital DNA–DNA hybridization (dDDH) values for studied strains KMM 9576T and KMM 9553 and selected taxa from the family Rhizobiaceae; Table S2: Pairwise core-proteome average amino acid identity (cpAAI) values between the strains KMM 9576T and KMM 9553 and members of the family Rhizobiaceae; Table S3: Differential physiological characteristics of strains KMM 9576T and KMM 9553 and the most closely related bacteria. Strains: 1, KMM 9576T; 2, KMM 9553; 3, Rhizobium leguminosarum NBRC 14778T; 4, Ensifer adhaerens NBRC 100388T; 5, Sinorhizobium garamanticum LMG 24692T; 6, Pararhizobium giardinii NBRC 107135T (data were obtained from the present study).
Author Contributions
Investigation, L.R., N.O., N.T., V.K., V.S. and M.I.; Methodology, L.R., N.O., N.T., V.K. and V.S.; Project administration, V.M. and M.I.; Resources, V.M., L.T. and M.I.; Software, N.O. and M.I.; Writing—original draft, L.R., N.O. and M.I.; Writing—review and editing, L.R., V.M. and M.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant from the Ministry of Science and Higher Education, Russian Federation 15.BRK.21.0004 (Contract No. 075-15-2021-1052).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The type strain of the species is strain KMM 9576T (=NRIC 0957T), which was isolated from a sandy sediment sampled from the Sea of Japan seashore, Russia. The DDBJ/ENA/GenBank accession numbers for the 16S rRNA gene and the whole-genome shotgun sequences of strains KMM 9576T and KMM 9553 are LC126306 (ON040664) and LC126307 (ON040663), as well as JALDYZ010000000 and JALDYY010000000, respectively.
Acknowledgments
We would like to thank Moriya Ohkuma, Japan Collection of Microorganisms, JCM, Japan, for providing type strains for comparative analyses. We sincerely appreciate to two unknown reviewers for all valuable comments and suggestions, which helped us to improve the quality of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hördt, A.; Lopez, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Frank, B. Ueber die pilzsymbiose der Leguminosen. Ber. Dtsch. Bot. Ges. 1989, 7, 332–346. [Google Scholar]
- Young, J.M.; Kuykendall, L.D.; Martinez-Romero, E.; Kerr, A.; Sawada, H. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int. J Syst. Evol. Microbiol. 2001, 51, 89–103. [Google Scholar] [PubMed]
- Gaunt, M.W.; Turner, S.L.; Rigottier-Gois, L.; Lloyd-Macgilp, S.A.; Young, J.P. Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int. J. Syst. Evol. Microbiol. 2001, 51, 2037–2048. [Google Scholar] [CrossRef]
- Martens, M.; Dawyndt, P.; Coopman, R.; Gillis, M.; De Vos, P.; Willem, A. Advantages of multilocus sequence analysis for taxonomic studies: A case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int. J. Syst. Evol. Microbiol. 2008, 58, 200–214. [Google Scholar] [CrossRef]
- Merabet, C.; Martens, M.; Mahdhi, M.; Zakhia, F.; Sy, A.; Le Roux, C.; Domergue, O.; Coopman, R.; Bekki, A.; Mars, M.; et al. Multilocus sequence analysis of root nodule isolates from Lotus arabicus (Senegal), Lotus creticus, Argyrolobium uniflorum and Medicago sativa (Tunisia) and description of Ensifer numidicus sp. nov. and Ensifer garamanticus sp. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 664–674. [Google Scholar] [CrossRef]
- Mousavi, S.A.; Österman, J.; Wahlberg, N.; Nesme, X.; Lavire, C.; Vial, L.; Paulin, L.; De Lajudie, P.; Lindström, K. Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst. Appl. Microbiol. 2014, 37, 208–215. [Google Scholar] [CrossRef]
- Mousavi, S.A.; Willems, A.; Nesme, X.; De Lajudie, P.; Lindström, K. Revised phylogeny of Rhizobiaceae: Proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst. Appl. Microbiol. 2015, 38, 84–90. [Google Scholar] [CrossRef]
- Kuzmanovic, N.; Fagorzi, C.; Mengoni, A.; Lassalle, F.; diCenzo, G. Taxonomy of Rhizobiaceae revisited: Proposal of a new framework for genus delimitation. Int. J. Syst. Evol. Microbiol. 2022, 72, 005243. [Google Scholar] [CrossRef]
- Willems, A.; Fernández-López, M.; Muñoz-Adelantado, E.; Goris, J.; De Vos, P.; Martínez-Romero, E.; Toro, N.; Gillis, M. Description of new Ensifer strains from nodules and proposal to transfer Ensifer adhaerens Casida 1982 to Sinorhizobium as Sinorhizobium adhaerens comb. nov. Request for an opinion. Int. J. Syst. Evol. Microbiol. 2003, 43, 1207–1217. [Google Scholar] [CrossRef]
- Young, J.M. The genus name Ensifer Casida 1982 takes priority over Sinorhizobium Chen et al. 1988, and Sinorhizobium morelense Wang et al. 2002 is a later synonym of Ensifer adhaerens Casida 1982. Is the combination ‘Sinorhizobium adhaerens’ (Casida 1982) Willems et al. 2003 legitimate? Request for an Opinion. Int. J. Syst. Evol. Microbiol. 2003, 53, 2107–2110. [Google Scholar] [PubMed]
- Ramírez-Bahena, M.H.; García-Fraile, P.; Peix, A.; Valverde, A.; Rivas, R.; Igual, J.M.; Mateos, P.F.; Martínez-Molina, E.; Velázquez, E. Revision of the taxonomic status of the species Rhizobium leguminosarum (Frank 1879) Frank 889AL, Rhizobium phaseoli 1926AL and Rhizobium trifolii Dangeard 1926AL. R. trifolii is a later synonym of R. leguminosarum. Reclassification of the strain R. leguminosarum DSM 30132 (=NCIMB 11478) as Rhizobium pisi sp. nov. Int. J. Syst. Evol. Microbiol. 2008, 58, 2484–2490. [Google Scholar] [PubMed]
- Young, J.P.W.; Moeskjær, S.; Afonin, A.; Rahi, P.; Maluk, M.; James, E.K.; Cavassim, M.I.A.; Rashid, M.H.-o.; Aserse, A.A.; Perry, B.J.; et al. Defining the Rhizobium leguminosarum species complex. Genes 2021, 12, 111. [Google Scholar] [CrossRef] [PubMed]
- Casida, L.E., Jr. Ensifer adhaerens gen. nov., sp. nov.: A bacterial predator of bacteria in soil. Int. J. Syst. Bacteriol. 1982, 32, 339–345. [Google Scholar] [CrossRef]
- Ramana, C.V.; Parag, B.; Girija, K.R.; Ram, B.R.; Venkata Ramana, V.; Sasikala, C. Rhizobium subbaraonis sp. nov., an endolithic bacterium isolated from beach sand. Int. J. Syst. Evol. Microbiol. 2013, 63, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.-Y.; Huang, H.-W.; Young, C.-C.; Chen, W.-M. Rhizobium alvei sp. nov., isolated from a freshwater river. Int. J. Syst. Evol. Microbiol. 2015, 65, 472–478. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, R.-P.; Ren, C.; Lai, Q.-L.; Zeng, R.-Y. Rhizobium marinum sp. nov., a malachite-green tolerant bacterium isolated from the sea water. Int. J. Syst. Evol. Microbiol. 2015, 65, 4449–4454. [Google Scholar] [CrossRef]
- Li, Y.; Lei, X.; Xu, Y.; Zhu, H.; Xu, M.; Fu, L.; Zheng, W.; Zhang, J.; Zheng, T. Rhizobium albus sp. nov., isolated from lake water in Xiamen Fujian Province of China. Curr. Microbiol. 2017, 74, 42–48. [Google Scholar] [CrossRef]
- Mathe, I.; Toth, E.; Mentes, A.; Szabo, A.; Marialigeti, K.; Schumann, P.; Felfoldi, T. A new Rhizobium species isolated from the water of a crater lake, description of Rhizobium aquaticum sp. nov. Antonie Van Leeuwenhoek 2018, 111, 2175–2183. [Google Scholar] [CrossRef]
- Cao, J.; Wei, Y.; Lai, Q.; Wu, Y.; Deng, J.; Li, J.; Liu, R.; Wang, L.; Fang, J. Georhizobium profundi gen. nov., sp. nov., a piezotolerant bacterium isolated from a deep-sea sediment sample of the New Britain Trench. Int. J. Syst. Evol. Microbiol. 2020, 70, 373–379. [Google Scholar] [CrossRef]
- Wang, X.N.; Wang, L.; He, W.; Yang, Q.; Zhang, D.F. Description of Flavimaribacter sediminis gen. nov., sp. nov., a new member of the family Rhizobiaceae isolated from marine sediment. Curr. Microbiol. 2023, 80, 301. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, L.A.; Uchino, M.; Tebo, B.; Tanaka, N.; Frolova, G.M.; Mikhailov, V.V. Pseudomonas marincola sp. nov. isolated from marine environments. Int. J. Syst. Evol. Microbiol. 2008, 58, 706–710. [Google Scholar] [CrossRef]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for general and molecular bacteriology; Gerhardt, P., Murray, R.G.E., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–655. [Google Scholar]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.D.; Shah, H.N. Fatty acid, menaquinone and polar lipid composition of Rothia dentosacariosa. Arch. Microbiol. 1984, 137, 247–249. [Google Scholar] [CrossRef]
- Mitchell, K.; Fallon, R.J. The determination of ubiquinone profiles by reversed-phase high performance thin-layer chromatography as an aid to the speciation of Legionellaceae. J. Gen. Microbiol. 1990, 136, 2035–2041. [Google Scholar] [CrossRef]
- Sasser, M. Microbial Identification by Gas Chromatographic Analysis of Fatty Acid Methyl esters (GC-FAME); Technical Note 101; MIDI: Newark, DE, USA, 1990. [Google Scholar]
- Saito, H.; Miura, K.I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta. 1963, 72, 619–629. [Google Scholar] [CrossRef]
- Shida, O.; Takagi, H.; Kadowaki, K.; Nakamura, L.K.; Komagata, K. Emended description of Paenibacillus amylolyticus and description of Paenibacillus illinoisensis sp. nov. and Paenibacillus chibensis sp. nov. Int. J. Syst. Bacteriol. 1997, 47, 299–306. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 7, D801–D807. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 8, 10. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Lu, F.; Luo, Y.; Bie, L.; Xu, L.; Wang, Y. OrthoVenn3: An integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2013, 1, 13–14. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. BioRxiv 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Rodriguez-R, L.M.; Konstantinidis, K.T. MyTaxa: An advanced taxonomic classifer for genomic and metagenomic sequences. Nucleic Acids Res. 2014, 42, e73. [Google Scholar] [CrossRef]
- Rahi, P.; Khairnar, M.; Hagir, A.; Narayan, A.; Jain, K.R.; Madamwar, D.; Pansare, A.; Shouche, Y. Peteryoungia gen. nov. with four new species combinations and description of Peteryoungia desertarenae sp. nov., and taxonomic revision of the genus Ciceribacter based on phylogenomics of Rhizobiaceae. Arch. Microbiol. 2021, 203, 3591–3604. [Google Scholar] [CrossRef]
- Hang, P.; Zhang, L.; Zhou, X.Y.; Hu, Q.; Jiang, J.D. Rhizobium album sp. nov., isolated from a propanil-contaminated soil. Antonie Van Leeuwenhoek 2019, 112, 319–327. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Tighe, S.W.; de Lajudie, P.; Dipietro, K.; Lindström, K.; Nick, G.; Jarvis, B.D. Analysis of cellular fatty acids and phenotypic relationships of Agrobacterium, Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium species using the Sherlock Microbial Identification System. Int. J. Syst. Evol. Microbiol. 2000, 50, 787–801. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).