Rapid, Sensitive, and Selective Quantification of Bacillus cereus Spores Using xMAP Technology
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Reagents
2.2. Microorganism Preparations and Plasmid Miniprep
2.3. Purification of Recombinant BclA Protein
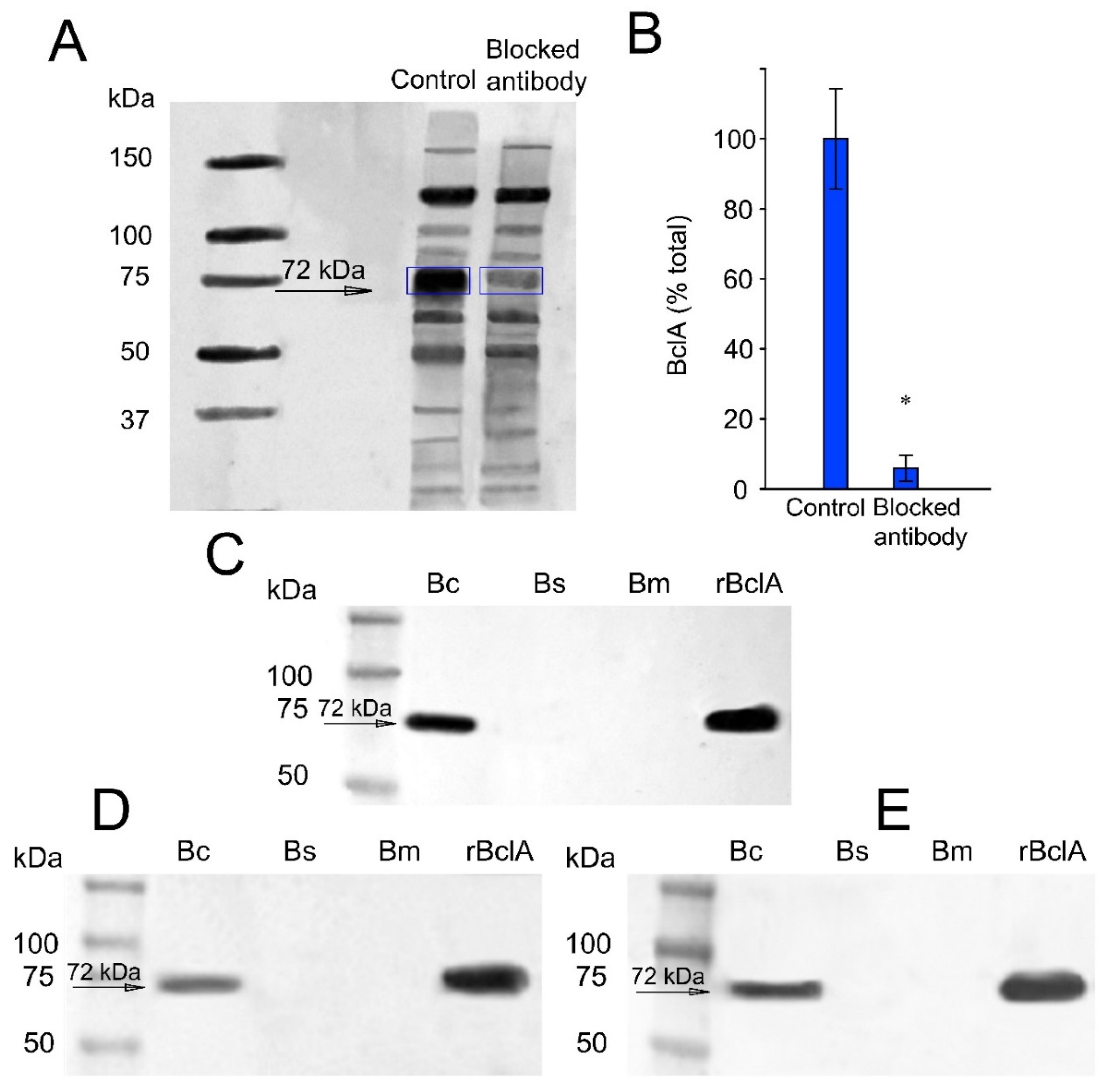
2.4. Immunoblotting
2.5. Paratope Blocking Assay
2.6. Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. xMAP Multiplexing Assay
Magnetic Bead Assembly for Detection by xMAP Technology
2.8. Statistical Data Analysis
3. Results
3.1. Screening of Capture Reagents
3.1.1. Antibody Affinity to B. cereus Spore BclA Protein
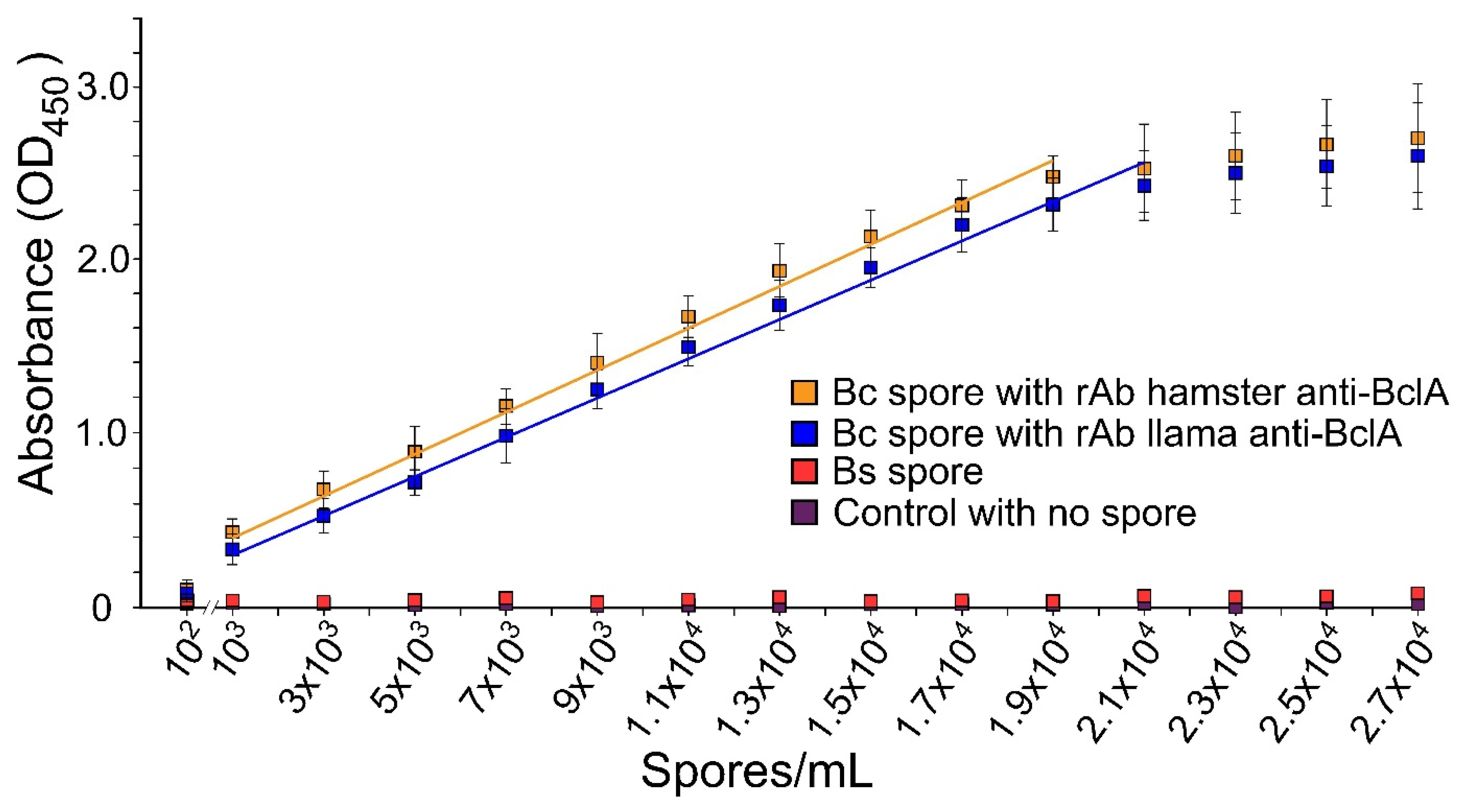
3.1.2. Detection Range of B. cereus Spores with rAbs
3.2. Detection of B. cereus Spores with Aptamers
3.2.1. Selectivity of Detection with ELISA
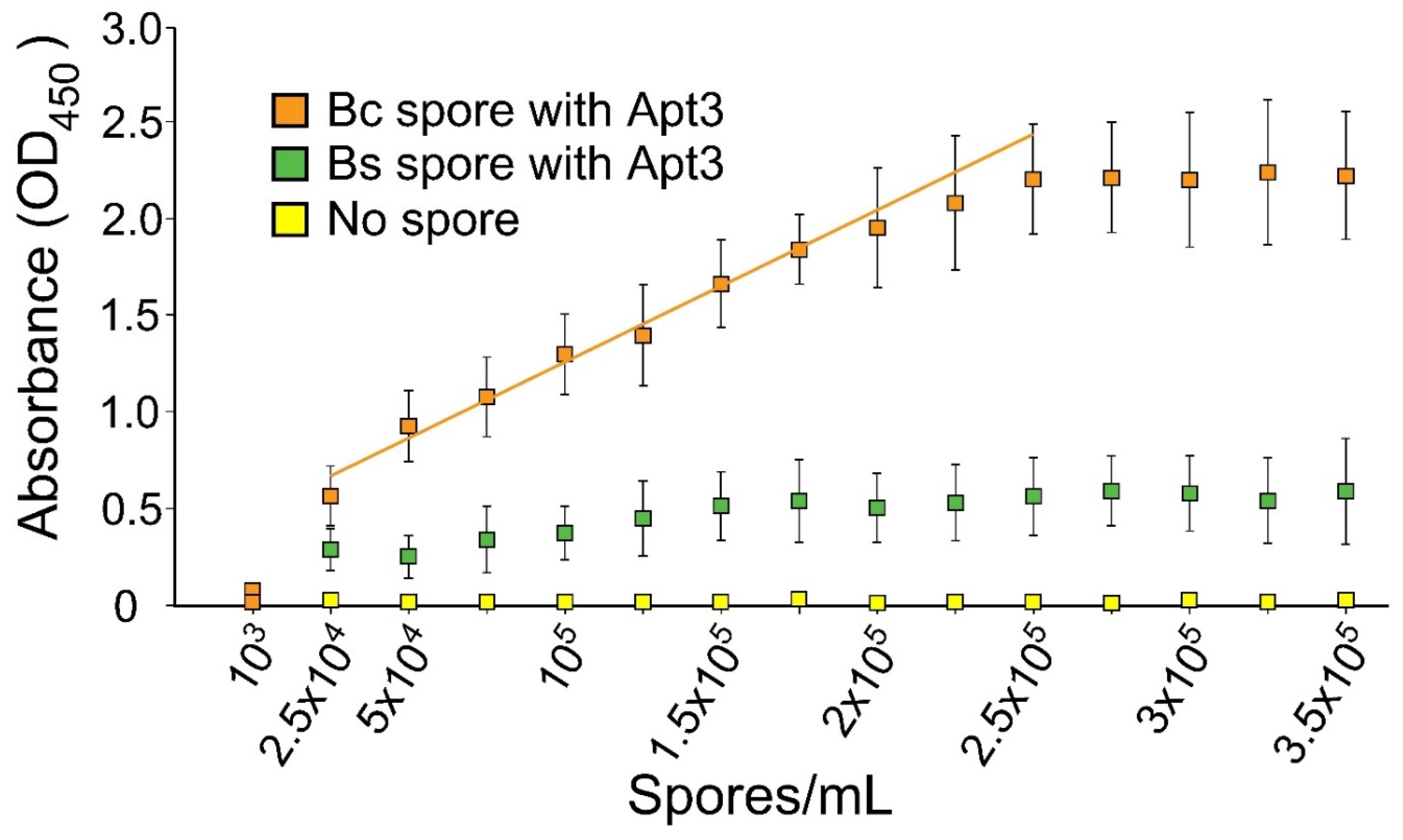
3.2.2. Range of Detection with ELISA
3.3. Detection with xMAP Technology
3.3.1. Antibody-Based Detection of B. cereus Spores
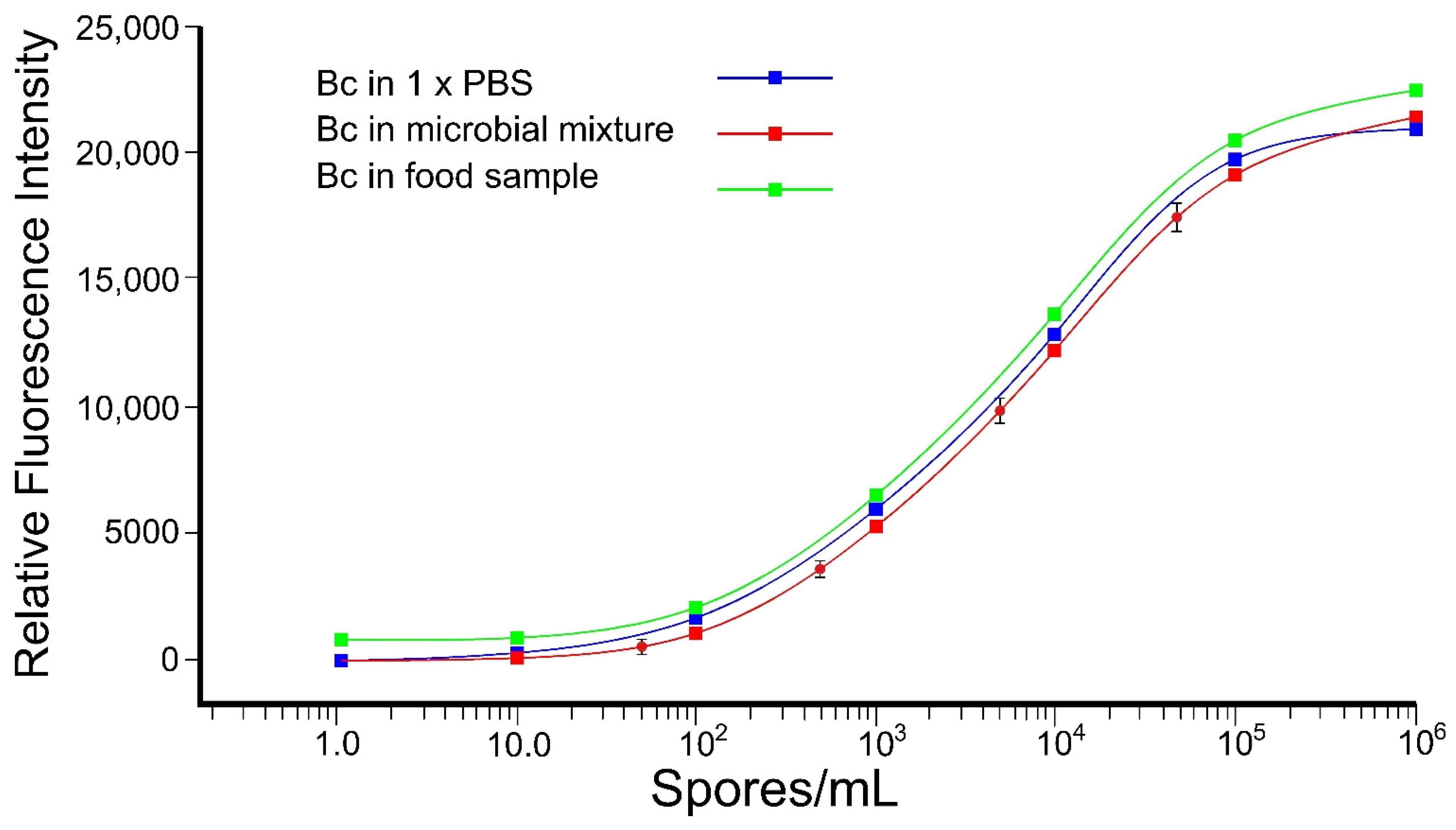
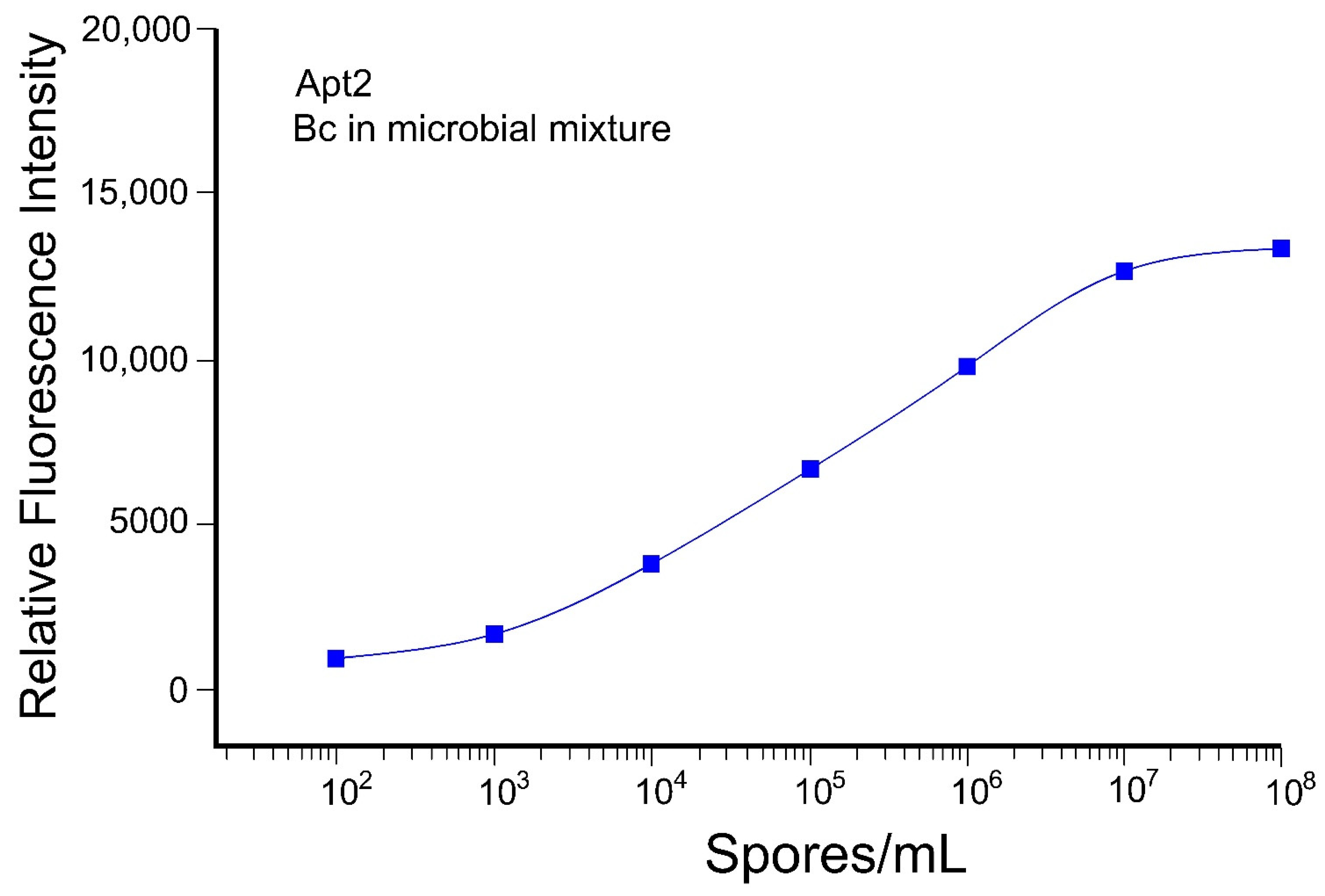
3.3.2. Aptamer-Based Detection of B. cereus Spores
3.3.3. Multi-Analyte Detection of Four Different Microorganisms
4. Discussion
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nicholson, W.L.; Munakata, N.; Horneck, G.; Melosh, H.J.; Setlow, P. Resistance of Bacillus Endospores to Extreme Terrestrial and Extraterrestrial Environments. Microbiol. Mol. Biol. Rev. 2000, 64, 548–572. [Google Scholar] [CrossRef] [PubMed]
- Hayrapetyan, H.; Abee, T.; Groot, M.N. Sporulation dynamics and spore heat resistance in wet and dry biofilms of Bacillus cereus. Food Control 2016, 60, 493–499. [Google Scholar] [CrossRef]
- Tewari, A.; Abdullah, S. Bacillus cereus food poisoning: International and Indian perspective. J. Food Sci. Technol. 2015, 52, 2500–2511. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, P.H.; Granum, P.E.; Bresciani, J.; Torregrossa, M.V.; Eilenberg, J.; Valentino, L. Characterization of Bacillus thuringiensis isolated from infections in burn wounds. FEMS Immunol. Med. Microbiol. 1997, 18, 47–53. [Google Scholar] [CrossRef]
- Jensen, G.B.; Hansen, B.M.; Eilenberg, J.; Mahillon, J. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 2003, 5, 631–640. [Google Scholar] [CrossRef]
- Kumari, S.; Sarkar, P.K. Bacillus cereus hazard and control in industrial dairy processing environment. Food Control 2016, 69, 20–29. [Google Scholar] [CrossRef]
- Schreiber, N.; Hackl, G.; Reisinger, A.C.; Zollner-Schwetz, I.; Eller, K.; Schlagenhaufen, C.; Pietzka, A.; Czerwenka, C.; Stark, T.D.; Kranzler, M.; et al. Acute liver failure after ingestion of fried rice balls: A case series of Bacillus cereus food poisonings. Toxins 2022, 14, 12. [Google Scholar] [CrossRef]
- Beecher, D.J.; Schoeni, J.L.; Wong, A.C. Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect. Immun. 1995, 63, 4423–4428. [Google Scholar] [CrossRef]
- Lund, T.; Granum, P.E. Characterisation of a non-haemolytic enterotoxin complex from Bacillus cereus isolated after a foodborne outbreak. FEMS Microbiol. Lett. 1996, 141, 151–156. [Google Scholar] [CrossRef]
- Gaviria Rivera, A.M.; Granum, P.E.; Priest, F.G. Common occurrence of enterotoxin genes and enterotoxicity in Bacillus thuringiensis. FEMS Microbiol. Lett. 2000, 190, 151–155. [Google Scholar] [CrossRef]
- Lund, T.; De Buyser, M.L.; Granum, P.E. A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 2000, 38, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Tran, S.L.; Guillemet, E.; Gohar, M.; Lereclus, D.; Ramarao, N. CwpFM (EntFM) is a Bacillus cereus potential cell wall peptidase implicated in adhesion, biofilm formation, and virulence. J. Bacteriol. 2010, 192, 2638–2642. [Google Scholar] [CrossRef] [PubMed]
- Berthold-Pluta, A.; Pluta, A.; Garbowska, M.; Stefańska, I. Prevalence and Toxicity Characterization of Bacillus cereus in Food Products from Poland. Foods 2019, 8, 269. [Google Scholar] [CrossRef] [PubMed]
- Ehling-Schulz, M.; Frenzel, E.; Gohar, M. Food-bacteria interplay: Pathometabolism of emetic Bacillus cereus. Front. Microbiol. 2015, 6, 704. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, R.; Jessberger, N.; Ehling-Schulz, M.; Märtlbauer, E.; Granum, P.E. The Food Poisoning Toxins of Bacillus cereus. Toxins 2021, 13, 98. [Google Scholar] [CrossRef] [PubMed]
- Bottone, E.J. Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 2010, 23, 382–398. [Google Scholar] [CrossRef] [PubMed]
- Rishi, E.; Rishi, P.; Sengupta, S.; Jambulingam, M.; Madhavan, H.N.; Gopal, L.; Therese, K.L. Acute postoperative Bacillus cereus endophthalmitis mimicking toxic anterior segment syndrome. Ophthalmology 2013, 120, 181–185. [Google Scholar] [CrossRef]
- Ikeda, M.; Yagihara, Y.; Tatsuno, K.; Okazaki, M.; Okugawa, S.; Moriya, K. Clinical characteristics and antimicrobial susceptibility of Bacillus cereus blood stream infections. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 43. [Google Scholar] [CrossRef]
- Doménech-Sánchez, A.; Laso, E.; Pérez, M.J.; Berrocal, C.I. Emetic disease caused by Bacillus cereus after consumption of tuna fish in a beach club. Foodborne Pathog. Dis. 2011, 8, 835–837. [Google Scholar] [CrossRef]
- Sloan-Gardner, T.S.; Glynn-Robinson, A.J.; Roberts-Witteveen, A.; Krsteski, R.; Rogers, K.; Kaye, A.; Moffatt, C.R.M. An outbreak of gastroenteritis linked to a buffet lunch served at a Canberra restaurant. Commun. Dis. Intell. Q. Rep. 2014, 38, E273–E278. [Google Scholar]
- Delbrassinne, L.; Botteldoorn, N.; Andjelkovic, M.; Dierick, K.; Denayer, S. An emetic Bacillus cereus outbreak in a kindergarten: Detection and quantification of critical levels of cereulide toxin. Foodborne Pathog. Dis. 2015, 12, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Schmid, D.; Rademacher, C.; Kanitz, E.E.; Frenzel, E.; Simons, E.; Allerberger, F.; Ehling-Schulz, M. Elucidation of enterotoxigenic Bacillus cereus outbreaks in Austria by complementary epidemiological and microbiological investigations, 2013. Int. J. Food Microbiol. 2016, 232, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Glasset, B.; Herbin, S.; Guillier, L.; Cadel-Six, S.; Vignaud, M.L.; Grout, J.; Pairaud, S.; Michel, V.; Hennekinne, J.A.; Ramarao, N.; et al. Bacillus cereus-induced food-borne outbreaks in France, 2007 to 2014: Epidemiology and genetic characterisation. Eurosurveillance 2016, 21, 30413. [Google Scholar] [CrossRef] [PubMed]
- Thein, C.C.; Trinidad, R.M.; Pavlin, B.I. A large foodborne outbreak on a small Pacific island. Pac. Health Dialog 2010, 16, 75–80. [Google Scholar] [PubMed]
- May, F.J.; Polkinghorne, B.G.; Fearnley, E.J. Epidemiology of bacterial toxin-mediated foodborne gastroenteritis outbreaks in Australia, 2001 to 2013. Commun. Dis. Intell. Q. Rep. 2016, 40, E460–E469. [Google Scholar]
- Ramarao, N.; Tran, S.L.; Marin, M.; Vidic, J. Advanced methods for detection of Bacillus cereus and its pathogenic factors. Sensors 2020, 20, 2667. [Google Scholar] [CrossRef]
- Vidic, J.; Chaix, C.; Manzano, M.; Heyndrickx, M. Food sensing: Detection of Bacillus cereus Spores in Dairy Products. Biosensors 2020, 10, 15. [Google Scholar] [CrossRef]
- White, P.L.; Archer, A.E.; Barnes, R.A. Comparison of non-culture-based methods for detection of systemic fungal infections, with an emphasis on invasive Candida infections. J. Clin. Microbiol. 2005, 43, 2181–2187. [Google Scholar] [CrossRef]
- López-Campos, G.; Martínez-Suárez, J.V.; Aguado-Urda, M.; López-Alonso, V. Microarray Detection and Characterization of Bacterial Foodborne Pathogens; Springer: New York, NY, USA, 2012; ISBN 9781461432500. [Google Scholar]
- Olsen, J.E. DNA-based methods for detection of food-borne bacterial pathogens. Food Res. Int. 2000, 33, 257–266. [Google Scholar] [CrossRef]
- Bursle, E.; Robson, J. Non-culture methods for detecting infection. Aust. Prescr. 2016, 39, 171–175. [Google Scholar] [CrossRef]
- Reslova, N.; Michna, V.; Kasny, M.; Mikel, P.; Kralik, P. xMAP technology: Applications in detection of pathogens. Front. Microbiol. 2017, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Koyuncu, S.; Andersson, M.G.; Häggblom, P. Accuracy and sensitivity of commercial PCR-based methods for detection of Salmonella enterica in feed. Appl. Environ. Microbiol. 2010, 76, 2815–2822. [Google Scholar] [CrossRef] [PubMed]
- Setlow, P. Spores of Bacillus subtilis: Their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Straub, T.M.; Dockendorff, B.P.; Quiñonez-Díaz, M.D.; Valdez, C.O.; Shutthanandan, J.I.; Tarasevich, B.J.; Grate, J.W.; Bruckner-Lea, C.J. Automated methods for multiplexed pathogen detection. J. Microbiol. Methods 2005, 62, 303–316. [Google Scholar] [CrossRef]
- Poritz, M.A.; Blaschke, A.J.; Byington, C.L.; Meyers, L.; Nilsson, K.; Jones, D.E.; Thatcher, S.A.; Robbins, T.; Lingenfelter, B.; Amiott, E.; et al. Correction: FilmArray, an Automated Nested Multiplex PCR System for Multi-Pathogen Detection: Development and Application to Respiratory Tract Infection. PLoS ONE 2011, 6, e26047. [Google Scholar] [CrossRef]
- Ikanovic, M.; Rudzinski, W.E.; Bruno, J.G.; Allman, A.; Carrillo, M.P.; Dwarakanath, S.; Bhahdigadi, S.; Rao, P.; Kiel, J.L.; Andrews, C.J. Fluorescence assay based on aptamer-quantum dot binding to bacillus thuringiensis spores. J. Fluoresc. 2007, 17, 193–199. [Google Scholar] [CrossRef]
- Fan, M.; McBurnett, S.R.; Andrews, C.J.; Allman, A.M.; Bruno, J.G.; Kiel, J.L. Aptamer selection express: A novel method for rapid single-step selection and sensing of aptamers. J. Biomol. Tech. 2008, 19, 311. [Google Scholar]
- Bruno, J.G.; Carrillo, M.P. Development of aptamer beacons for rapid presumptive detection of Bacillus spores. J. Fluoresc. 2012, 22, 915–924. [Google Scholar] [CrossRef]
- Moteshareie, H.; Hajikarimlou, M.; Indrayanti, A.M.; Burnside, D.; Dias, A.P.; Lettl, C.; Ahmed, D.; Omidi, K.; Kazmirchuk, T.; Puchacz, N.; et al. Heavy metal sensitivities of gene deletion strains for ITT1 and RPS1A connect their activities to the expression of URE2, a key gene involved in metal detoxification in yeast. PLoS ONE 2018, 13, e0198704. [Google Scholar] [CrossRef]
- Lin, D.Q.; Yao, S.J.; Mei, L.H.; Zhu, Z.Q. Collection and purification of parasporal crystals from Bacillus thuringiensis by aqueous two-phase extraction. Sep. Sci. Technol. 2003, 38, 1665–1680. [Google Scholar] [CrossRef]
- Moteshareie, H.; Hassen, W.M.; Vermette, J.; Dubowski, J.J.; Tayabali, A.F. Strategies for capturing Bacillus thuringiensis spores on surfaces of (001) GaAs-based biosensors. Talanta 2022, 236, 122813. [Google Scholar] [CrossRef] [PubMed]
- Mounsef, J.R.; Salameh, D.; kallassy Awad, M.; Chamy, L.; Brandam, C.; Lteif, R. A simple method for the separation of Bacillus thuringiensis spores and crystals. J. Microbiol. Methods 2014, 107, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Maes, E.; Krzewinski, F.; Garenaux, E.; Lequette, Y.; Coddeville, B.; Trivelli, X.; Ronse, A.; Faille, C.; Guerardel, Y. Glycosylation of BclA glycoprotein from Bacillus cereus and Bacillus anthracis exosporium is domain-specific. J. Biol. Chem. 2016, 291, 9666–9677. [Google Scholar] [CrossRef] [PubMed]
- Emanuel, P.A.; Dang, J.; Gebhardt, J.S.; Aldrich, J.; Garber, E.A.; Kulaga, H.; Stopa, P.; Valdes, J.J.; Dion-Schultz, A. Recombinant antibodies: A new reagent for biological agent detection. Biosens. Bioelectron. 2000, 14, 751–759. [Google Scholar] [CrossRef]
- Wingren, C.; Sandström, A.; Segersvärd, R.; Carlsson, A.; Andersson, R.; Löhr, M.; Borrebaeck, C.A.K. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Res. 2012, 72, 2481–2490. [Google Scholar] [CrossRef]
- Porschewski, P.; Grättinger, M.A.M.; Klenzke, K.; Erpenbach, A.; Blind, M.R.; Schäfer, F. Using aptamers as capture reagents in bead-based assay systems for diagnostics and hit identification. J. Biomol. Screen. 2006, 11, 773–781. [Google Scholar] [CrossRef][Green Version]
- Bernard, E.D.; Nguyen, K.C.; DeRosa, M.C.; Tayabali, A.F.; Aranda-Rodriguez, R. Development of a bead-based aptamer/antibody detection system for C-reactive protein. Anal. Biochem. 2015, 472, 67–74. [Google Scholar] [CrossRef]
- Bernard, E.D.; Nguyen, K.C.; DeRosa, M.C.; Tayabali, A.F.; Aranda-Rodriguez, R. Incorporating aptamers in the multiple analyte profiling assays (xMAP): Detection of C-reactive protein. Methods Mol. Biol. 2017, 1575, 303–322. [Google Scholar]
- Dunbar, S.A.; Jacobson, J.W. Quantitative, multiplexed detection of Salmonella and other pathogens by Luminex xMAP suspension array. Methods Mol. Biol. 2007, 394, 1–19. [Google Scholar] [CrossRef]
- Charlermroj, R.; Himananto, O.; Seepiban, C.; Kumpoosiri, M.; Warin, N.; Oplatowska, M.; Gajanandana, O.; Grant, I.R.; Karoonuthaisiri, N.; Elliott, C.T. Multiplex Detection of Plant Pathogens Using a Microsphere Immunoassay Technology. PLoS ONE 2013, 8, e62344. [Google Scholar] [CrossRef]
- Hrdy, J.; Vasickova, P.; Nesvadbova, M.; Novotny, J.; Mati, T.; Kralik, P. MOL-PCR and xMAP Technology: A Multiplex System for Fast Detection of Food- and Waterborne Viruses. J. Mol. Diagn. 2021, 23, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Francy, D.S.; Stelzer, E.A.; Brady, A.M.G.; Huitger, C.; Bushon, R.N.; Ip, H.S.; Ware, M.W.; Villegas, E.N.; Gallardo, V.; Lindquist, H.D.A. Comparison of filters for concentrating microbial indicators and pathogens in lake water samples. Appl. Environ. Microbiol. 2013, 79, 1342–1352. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Oh, S.W. Optimization of Bacterial Concentration by Filtration for Rapid Detection of Foodborne Escherichia coli O157:H7 Using Real-Time PCR Without Microbial Culture Enrichment. J. Food Sci. 2019, 84, 3241–3245. [Google Scholar] [CrossRef] [PubMed]
- Cattani, F.; Barth, V.C.; Nasário, J.S.R.; Ferreira, C.A.S.; Oliveira, S.D. Detection and quantification of viable Bacillus cereus group species in milk by propidium monoazide quantitative real-time PCR. J. Dairy Sci. 2016, 99, 2617–2624. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.; Hünniger, T.; Jarck, J.H.; Frohnmeyer, E.; Kallinich, C.; Haase, I.; Hahn, U.; Fischer, M. Food Sensing: Aptamer-Based Trapping of Bacillus cereus Spores with Specific Detection via Real Time PCR in Milk. J. Agric. Food Chem. 2015, 63, 8050–8057. [Google Scholar] [CrossRef]
- Izadi, Z.; Sheikh-Zeinoddin, M.; Ensafi, A.A.; Soleimanian-Zad, S. Fabrication of an electrochemical DNA-based biosensor for Bacillus cereus detection in milk and infant formula. Biosens. Bioelectron. 2016, 80, 582–589. [Google Scholar] [CrossRef]
- Liang, L.; Wang, P.; Qu, T.; Zhao, X.; Ge, Y.; Chen, Y. Detection and quantification of Bacillus cereus and its spores in raw milk by qPCR, and distinguish Bacillus cereus from other bacteria of the genus Bacillus. Food Qual. Saf. 2022, 6, fyab035. [Google Scholar] [CrossRef]

| Number | Capture Reagents | Description |
|---|---|---|
| pAb1 | Polyclonal Antibody: PA1-73114 (Thermo Fisher Scientific, Waltham, MA, USA) | Rabbit anti-B. cereus + B. subtilis spores |
| mAb2 | Monoclonal Antibody: G46D (Thermo Fisher Scientific, Waltham, MA, USA) | Mouse anti-B. anthracis spores |
| rAb3 | Recombinant Antibody: A4D1 (Creative Biolabs, Inc., Shirley, NY, USA) | Hamster anti-BclA protein (hamster ovary cells (CHO)) |
| rAb4 | Recombinant Antibody: A5 (Creative Biolabs, Inc., Shirley, NY, USA) | Llama anti-BclA protein |
| mAb5 | Monoclonal anti-P. aeruginosa antibody: ab35835 (Abcam, Inc., Cambridge, UK) | Mouse anti-P. aeruginosa outer membrane protein (clone B11) |
| mAb6 | Monoclonal anti-S. cerevisiae PGK1 antibody: ab113687 (Abcam, Inc., Cambridge, UK) | Mouse anti-S. cerevisiae PGK1 protein (clone 22C5D8) |
| mAb7 | Monoclonal anti-E. coli antibody: ab137967 (Abcam, Inc., Cambridge, UK) | Rabbit anti-E. coli outer membrane protein |
| Apt1 | DNA aptamer #1 [37] | Developed against Bt spores (61 bases). Modified by 5′ thiolation. |
| Apt2 | DNA aptamer #2 [39] | Developed against B. anthracis spores (72 bases). Modified by 5′ thiolation. |
| Apt3 | DNA aptamer #3 [38] | Developed against B. anthracis spores (80 bases). Modified by 5′ thiolation. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022, Her Majesty the Queen in Right of Canada as represented by Health Canada. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moteshareie, H.; Hassen, W.M.; Dirieh, Y.; Groulx, E.; Dubowski, J.J.; Tayabali, A.F. Rapid, Sensitive, and Selective Quantification of Bacillus cereus Spores Using xMAP Technology. Microorganisms 2022, 10, 1408. https://doi.org/10.3390/microorganisms10071408
Moteshareie H, Hassen WM, Dirieh Y, Groulx E, Dubowski JJ, Tayabali AF. Rapid, Sensitive, and Selective Quantification of Bacillus cereus Spores Using xMAP Technology. Microorganisms. 2022; 10(7):1408. https://doi.org/10.3390/microorganisms10071408
Chicago/Turabian StyleMoteshareie, Houman, Walid M. Hassen, Yasmine Dirieh, Emma Groulx, Jan J. Dubowski, and Azam F. Tayabali. 2022. "Rapid, Sensitive, and Selective Quantification of Bacillus cereus Spores Using xMAP Technology" Microorganisms 10, no. 7: 1408. https://doi.org/10.3390/microorganisms10071408
APA StyleMoteshareie, H., Hassen, W. M., Dirieh, Y., Groulx, E., Dubowski, J. J., & Tayabali, A. F. (2022). Rapid, Sensitive, and Selective Quantification of Bacillus cereus Spores Using xMAP Technology. Microorganisms, 10(7), 1408. https://doi.org/10.3390/microorganisms10071408








