Abstract
Legionella are opportunistic intracellular pathogens that are found throughout the environment. The Legionella contamination of water systems represents a serious social problem that can lead to severe diseases, which can manifest as both Pontiac fever and Legionnaires’ disease (LD) infections. Fluorescence in situ hybridization using nucleic acid mimic probes (NAM-FISH) is a powerful and versatile technique for bacterial detection. By optimizing a peptide nucleic acid (PNA) sequence based on fluorescently selective binding to specific bacterial rRNA sequences, we established a new PNA-FISH method that has been successfully designed for the specific detection of the genus Legionella. The LEG22 PNA probe has shown great theoretical performance, presenting 99.9% specificity and 96.9% sensitivity. We also demonstrated that the PNA-FISH approach presents a good signal-to-noise ratio when applied in artificially contaminated water samples directly on filtration membranes or after cells elution. For water samples with higher turbidity (from cooling tower water systems), there is still the need for further method optimization in order to detect cellular contents and to overcome interferents’ autofluorescence, which hinders probe signal visualization. Nevertheless, this work shows that the PNA-FISH approach could be a promising alternative for the rapid (3–4 h) and accurate detection of Legionella.
1. Introduction
Legionella is a Gram-negative bacteria, ubiquitously found in natural freshwater, as well as in man-made water systems, such as cooling towers, hospitals or whirlpool spas [1]. From the 60 Legionella species, more than 20 are known to infect humans, causing serious illnesses, such as Legionnaires’ disease (LD) and Pontiac fever [2]. From these, Legionella pneumophila is responsible for about 97% of all outbreaks, and the other 3% is mainly due to non-L. pneumophila species, such as L. longbeachae, L. micdadei and L. bozemanii [3]. Upon the inhalation of aerosols, Legionella has the ability to colonize and replicate in the human alveolar macrophages, triggering the disease [4]. Closer surveillance and the use of methods for diagnosis in water management are critical for effectively reducing the public risk caused by Legionella. Moreover, according to official authorities, taking a more active role in minimization of Legionella in water supply systems, including the implementation of stronger programs for monitoring and testing, will be vital in decreasing disease outbreaks [5]. Despite the excellent developments in the area of identification, the current technologies do not fully answer the needs of industrial entities for Legionella monitoring.
The use of the International Organization for Standardization (ISO) 11731:2017 is recommended for the isolation and determination of Legionella in water samples (potable, industrial, waste and natural) using the gold standard culture method [6]. However, this method is a laborious and time-consuming procedure (taking more than a week to provide the results), often with low sensitivity, especially in samples of complex matrices, as in biofilms, and does not allow for the detection of viable but non-cultivable cells (VBNC) [7]. Therefore, it is important to develop culture-independent detection methods for microbial monitoring. PCR-based methods (ISO 12869:2019) [8] have been proposed for monitoring Legionella in environmental systems, but the interpretation of results from environmental samples remains difficult, leading to an overestimation of the actual health risk [9,10]. These molecular methods can detect the number of genome units (GU) per liter, but there is still a need for correspondence with the number of colony forming units (CFU), which is usually lower, probably due to VBNC cells’ presence [11]. Research has been conducted in order to develop, optimize and validate new molecular methods, and fluorescence in situ hybridization (FISH) has been shown to be a promising technology [12,13,14,15,16].
FISH is a visual, culture-independent molecular technique that allows for the rapid detection and quantification of microorganisms [17]. This method is typically based on specific binding of fluorescently labeled nucleic acid probes, generally DNA molecules, to particular rRNA sequences of microorganisms [18]. However, the development of synthetic nucleic acid mimics (NAMs), such as peptide nucleic acid (PNA), has been shown to provide an improved hybridization performance compared to DNA probes [19,20]. PNA chemical structure allows for the bases to be positioned at equivalent spatial arrangement as DNA, which allows for the hybridization with DNA/RNA molecules to occur, obeying the Watson–Crick base-pairing rules [19]. Moreover, they present a stronger affinity to the target sequences and subsequently higher accuracy, mainly explained by the fact that these molecules are neutrally charged, reducing the electrostatic repulsion that occurs between negatively charged DNA/DNA or RNA/RNA duplexes [21,22]. PNA-FISH has been used for the rapid detection (approximately 3 h) of several relevant microorganisms, such as bacteria [23,24,25,26,27], yeasts [28,29] and filamentous fungi [13,30,31]. Regarding Legionella, only one Legionella spp. (PLEG200) probe and one L. pneumophila PNA probe were identified [32] in the literature. These probes were based on pre-existing DNA probes targeting a low-binding-affinity site on the 16S rRNA. In this work, we attempted to design a new PNA-FISH method for the specific detection of the genus Legionella, targeting a distinctive conserved region of 16S rRNA. In addition, preliminary assays, using the PNA-FISH method, were tested in artificial water samples to ensure that this method could be successfully applied to environmental samples.
2. Materials and Methods
2.1. Strain and Growth Conditions
The bacterial strains used in this study are indicated in Table 1. A total of 17 strains of different Legionella species and 19 non-Legionella bacteria were tested. The bacteria collection was supplied by Laboratório de Microbiologia do Departamento de Saúde Ambiental Porto do Instituto Nacional de Saúde Doutor Ricardo Jorge (INSA-DSA ASMIP) and by Prof. Manuel Simões (LEPABE) [33]. All Legionella strains were grown on buffered charcoal yeast extract agar (BCYE) at 37 °C for 2 to 4 days. Non-Legionella strains were grown on tryptic soy agar (TSA) (3% (w/v) tryptic soy broth and 1.5% agar) at 37 °C for 24 h, except Pseudomonas fluorescens, which was incubated at 30 °C.

Table 1.
List of strains used in this study, together with the results obtained with the PNA-FISH probe specificity/sensitivity test. ATCC—American Type Culture Collection; WDCM—World Data Centre for Microorganisms.
2.2. In Silico PNA Probe Design for the Specific Detection of Legionella
For the design of a specific Legionella spp. probe, an approach using available alignment programs coupled with the 16S rRNA databases was used according to the methodology described by Teixeira et al. (2021) [34]. From these, a group of 9 target Legionella and 27 non-target sequences were selected from the ARB Silva database (https://www.arb-silva.de/; accessed on May 2020). Only sequences with a length above 1200 bp, high sequence quality (>90%), high alignment quality (>90%) and high pintail quality (>90%) were selected. Regions of interest were identified with MEGA-X and aligned with ClustalW (https://www.megasoftware.net/; last accession on June 2020) (Figure 1). High GC percentage and low number of consecutive self-complementary nucleotides were also criteria to consider in sequence selection.
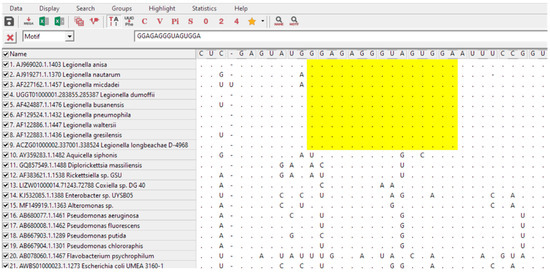
Figure 1.
Partial alignment of 16S rRNA sequences for Legionella probe selection using ClustalW from MEGA-X software. Yellow shadow shows the selected region that matched all Legionella sequences but did not match the nontarget sequences. The sequence shown corresponds with LEG22 complementary reverse sequence.
The theoretical specificity and sensitivity were calculated using the Probe Match tool on the Ribosomal Database Project (RDP-II) (http://rdp.cme.msu.edu; last accession on June 2020) as previously described by Almeida et al. (2010) [35]. Specificity was calculated as nLs/(TnL) × 100, where nLs stands for the number of non-Legionella sequences that did not align with the probe, and TnL is the total of non-Legionella sequences examined. Sensitivity was calculated as Ls/(TLs) × 100, where Ls stands for the number of Legionella sequences detected by the probe, and TLs is the total number of Legionella strains present in the databases. The selected sequence was synthesized (Eurogentec, Seraing, Belgium). The N terminus was labeled with AlexaFluor® 488 via a double 8-amino-3,6-dioxaoctanoic acid (AEEA) linker. Additionally, the theoretical evaluation of a previously published Legionella spp. PNA probe [32] was performed to compare the theoretical accuracy of the selected probe.
2.3. Hybridization Conditions
Hybridization was performed as previously described by the group [13,35,36], with some modifications. As Figure 2A summarizes, suspensions of 2 × 107 cell/mL of L. pneumophila serogroup 1 (WDCM 00107) and L. anisa ATCC 35,292 were dispensed in 8 mm well slides (Marienfeld, Lauda-Königshofen, Germany) and allowed to air dry. For permeabilization and fixation of Legionella spp. cells, 30 µL of 4% (wt/vol) paraformaldehyde followed by 50% (vol/vol) ethanol, for 10 min each, were dispensed in the wells at room temperature and allowed to air dry. The slides were then covered with 20 µL of hybridization solution containing 10% (wt/vol) dextran sulfate, 10 mM NaCl, 30% (vol/vol) formamide, 0.1% (wt/vol) sodium pyrophosphate, 0.2% (wt/vol) polyvinylpyrrolidone, 0.2% (wt/vol) Ficoll, 5 mM disodium EDTA, 0.1% (vol/vol) Triton X-100, 50 mM Tris-HCl (pH 7.5) (all from Sigma-Aldrich, Sintra, Portugal, except disodium EDTA, which was from Pronalab, Lisbon, Portugal) and 200 nM of the PNA probe (Eurogentec, Belgium). The slide wells were covered with coverslips, protected from the light, and incubated for 60 min at different temperatures under study. Following hybridization, the slides were transferred to a coupling jar containing prewarmed washing solution, containing 5 mM Tris base, 15 mM NaCl and 1% (vol/vol) Triton X (pH 10) (all from Sigma-Aldrich, Sintra, Portugal). The samples were allowed to air dry, mounted with a drop of nonfluorescent immersion oil and covered with coverslips.
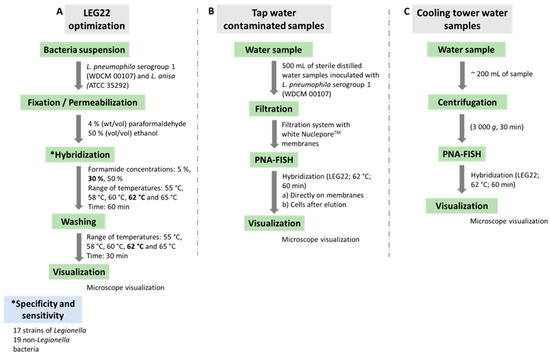
Figure 2.
Representative diagram of the methodology used in the present study. (A) Optimization of LEG22 probe. Numbers in bold correspond to the selected parameters. Specificity and sensitivity were assessed subsequentely; (B,C) Application of PNA-FISH method in tap-water-contaminated samples and in cooling tower water samples, respectively.
Several temperatures (55 °C, 58 °C, 60 °C, 62 °C and 65 °C) and formamide concentrations (5%, 30% and 50%) (at temperatures 55 and 60 °C) were studied for signal-to-noise ratio assessment. Slides were stored in the dark for a maximum of 24 h before microscopy visualization. After optimization of the hybridization conditions, the probe was applied for the other Legionella and non-Legionella strains described in Table 1 to assess the specificity and sensitivity of the probe.
2.4. PNA-FISH Method Testing in Water Samples
The first phase of Legionella spp. detection methodology comprises a concentration of the water samples, usually by filtration or centrifugation [6,11]. To evaluate the PNA-FISH detection ability by filtration, 500 mL of sterile distilled water samples was inoculated with 1 mL of different concentrations of L. pneumophila serogroup 1 (WDCM 00107) (corresponding to a final concentration from 105 to 10−1 CFU/L), using a standard filtration system (PALL, Show Low, AZ, USA) with white NucleporeTM (WhatmanTM, Maidstone, UK) membranes with a diameter of 47 mm and a pore size of 0.22 µm. These concentrations correspond to a concentration range of approximately 108 to 102 CFU in the filtration membrane.
To optimize the PNA-FISH method after filtration, 2 protocols were used: direct detection and elution. For direct detection, after filtering the sample, the membranes were allowed to air dry and overlaid with 4% (wt/vol) paraformaldehyde followed by 50% (vol/vol) ethanol, for 10 min each, at room temperature. The membrane was then placed on a glass slide and allowed to air dry. For the hybridization, 60 µL of hybridization solution (pH 7.5) containing 200 nM of the PNA probe was spread within the membrane with the help of a coverslip and incubated with light refraining. Subsequently, the filters were removed gently from the slide, transferred into a Petri dish, also protected from light, and filled with a prewarmed washing solution (pH 10) and incubated for 30 min. The samples were then allowed to dry, mounted with a drop of nonfluorescent immersion oil and covered with coverslips. Slides were stored in the dark for a maximum of 24 h before microscopy visualization (Figure 2B).
For the elution test, in order to resuspend adherent cells from the membrane, after filtering the sample, the membranes were agitated (270 rpm) in 5 mL of sterile distilled water in a Falcon tube for 20 min at room temperature. Then, the entire volume of the sample was centrifuged at 15,000× g for 10 min, and the supernatant was removed. The sample was resuspended in 100 µL of sterile distilled water and placed on a microscopic slide and allowed to air dry. The PNA-FISH protocol was carried out as described above. To determine the number of CFU per sample in all the experiments, 100 µL of the appropriate dilutions was plated onto BCYE plates at 37 °C for 4 days.
To test the PNA-FISH implementation in real water samples, preliminary assays using 10 cooling tower water samples supplied by INSA-DSA ASMIP were also performed (Figure 2C). These samples were firstly tested by INSA-DSA ASMIP, using the culturing method based on the ISO 11731:2017 [6], and all were <103 CFU/L, one of them negative. As these samples have more interferents, we performed a preliminary centrifugation step before applying the hybridization procedure, based on the ISO 11731:2017 [6]. Briefly, 200 mL of each sample was centrifuged at 3000× g for 30 min; then, the supernatant was carefully removed, leaving only 1 mL. Subsequently, the hybridization step was performed as described above.
2.5. Microscopy Visualization
Microscopy visualization was performed using a Nikon Eclipse 80i (Japan) epifluorescence microscope with a camera NikonDS-Fi1 (Izasa, Japan), equipped with a filter sensitive to the Alexa Fluor 488 molecule attached to the PNA probe (excitation, 482/35 nm; emission, 536/40 nm). Visualization was also assessed with the other filters present in the microscope that are not sensitive to the probe fluorescence signal. For every experiment, a negative control was performed simultaneously, for which all the steps described above were carried out, but no probe was added during the hybridization procedure. All the images were acquired using NIS-Elements B.R. 3.2 (Izasa, Japan) software with a magnification of ×100.
2.6. Statistical Analysis
The statistical validity of specificity and sensitivity parameters and respective 95% confidence intervals (CIs) were determined using the VassarStats: Website for Statistical Computation (http://vassarstats.net; assessed on June 2020 and June 2022).
3. Results
3.1. Probe Design
To make a first selection of possible regions for probe design, 16S rRNA gene sequences of Legionella were aligned with the closest relatives’ strains and with other bacteria that might be present in water. The selection was based on regions that showed differences between the Legionella sequences to the non-target strains. Hence, the selected probe, with the best compromise between the number of targets and non-targets detection, was N terminal-TCC ACT ACC CTC TCC-C terminal.
The sequence targeted the 16S rRNA between positions 634 and 648 of the Legionella pneumophila subsp. pneumophila JCM 7571 (Accession number AB594755; SILVA database). The probe was named LEG22, regarding the numbering of sequences tested in 16S rRNA regions analyzed.
The theoretical specificity and sensitivity of the probe were further evaluated using the Probe Match tool (RDP-II) program. The search confirmed that LEG22 had 99.9% (95% CI, 99.8–99.9) specificity and 96.9% (95% CI, 96.5–97.2) sensitivity. In order to compare the probe developed in this study with other Legionella spp. PNA probes found in the literature, the theoretical specificity and sensitivity of the probe PLEG200 [32] were also evaluated with the same software program (Table 2). The search showed that PLEG200 presented an acceptable level of specificity (100%; 95% CI, 99.8–99.9) but lower sensitivity (76.4%; 95% CI, 74.0–78.8).

Table 2.
Evaluation of the Legionella spp. PNA probes available.
3.2. PNA-FISH Perfomance
Although the protocol is generally straightforward, some aspects of the hybridization conditions had to be optimized to evaluate the optimal conditions for the LEG22 PNA probe to work. To guarantee that the probe efficiently accesses and hybridizes with the target sequence, different formamide concentrations (5%, 30% and 50%) were evaluated at 55 °C and 60 °C (the range of temperatures that usually presents higher stringency). As the results were better at 30% formamide, we then tested different hybridization temperatures (between 55 °C and 65 °C) to adjust the best hybridization conditions. The best performance in terms of strongest signal-to-noise ratio was achieved at 62 °C with 30% formamide (Figure 3 and Figure 4).
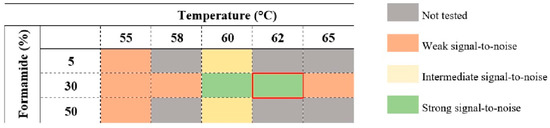
Figure 3.
Temperature and formamide concentration assessments for hybridization conditions optimization. The selected conditions for the best performance are 62 °C and 30% formamide (red bracket).
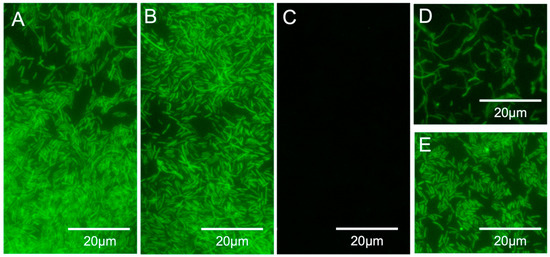
Figure 4.
(A–C) PNA-FISH specificity and sensitivity results for the LEG22 peptide nucleic acid probe. (A) Legionella pneumophila serogroup 1, WDCM 00107; (B) Legionella anisa, ATCC 35292; (C) Escherichia coli (INSA isolate); (D,E) L. pneumophila serogroup 1, WDCM 00107 in filtered artificially contaminated water samples (108 CFU per sample). (D) PNA-FISH on the membranes; (E) PNA-FISH on eluted cells. Images were obtained with equal exposure times.
Once the hybridization method was optimized, the specificity and sensitivity of the LEG22 PNA probe was tested. For this, the optimized protocol was applied to 17 Legionella and to 19 non-Legionella strains. As shown in Table 1 and Figure 4A–C, all Legionella strains were detected, whereas no hybridization was observed for the other species used. Therefore, 100% (95% CI, 77.1–100) specificity and 100% (95% CI, 79.1–100) sensitivity for the probe was obtained.
3.3. Detection in Water Samples
After probe optimization, we tested the method in the water filtration procedure using artificially contaminated tap water (from 108 to 102 CFU per sample) with L. pneumophila. We were able to detect the bacteria after filtration, both directly on the filter and by eluting the cells (Figure 4D,E and Table 3) above 101 CFU/L.

Table 3.
Artificially inoculated tap water results with PNA-FISH (both directly in the membrane and in the eluted cells) and with traditional culture.
Nevertheless, several cooling tower water samples provided by INSA-DSA ASMIP were tested directly with PNA-FISH. All the samples were tested by culture using ISO11731:2017 [6] (<103 CFU/L, one negative). Nonetheless, no positive result was observed with PNA-FISH for all the samples, which demonstrates that there is still a need to further optimize the protocol in order to detect cells in more turbid samples, since signal interference was observed in some of the samples, as they presented more residues.
4. Discussion
The cases of LD reported have been rising worldwide in recent decades. In Portugal, significant LD outbreaks were identified between 2014 and 2020, with more than 550 confirmed cases and 30 deaths [37,38,39]. European guidelines for Legionella identification require the implementation of monitoring and water treatment actions for risk assessment [9]. As water systems are the most significant sources of Legionella infections, it is imperative from a public health perspective to survey these aquatic environments for the presence of these bacteria [7].
Notwithstanding, as stated above, the currently available approaches to routine do not fully comply with the requirements for Legionella testing. FISH can take a step forward as a powerful and rapid tool for identification of microbial populations without the need for time-consuming microorganisms culturing [13,40,41]. Additionally, PNA-FISH overcomes problems related to PCR-based molecular technologies, such as susceptibility to inhibitors, cross-contamination, false positive results and even the requirements for specialized personnel [24,42]. Critically, in addition to technical issues, PCR-based methods can be troublesome and require specific and expensive equipment (for nucleic acid extraction and amplification steps), limiting their use to centralized laboratories. PNA-FISH is a rapid method, as it takes approximately 3 h to provide the result, with a simple workflow, as it does not consist of a technically demanding protocol, and apart from the visualization equipment, it does not require any overpriced equipment. This work intends to give insights about the development of a PNA-FISH method for the detection of Legionella spp. in water.
Traditionally, probes target the 16S/23S rRNA sequence of the bacteria, since these structures are abundant and highly conserved in cells (e.g., 104–105 for Escherichia coli) [43,44]. PNA, as shorter and neutrally charged molecules, can confer extra efficiency to the reaction, as they can present improved affinity to the target, hence increasing specificity [19]. From the sequences analyzed in different regions of the 16S rRNA, LEG22 was the probe selected for having the best theoretical performance, presenting 99.9% specificity and 96.9% sensitivity. In order to compare the probe developed in this study with other potential probes developed earlier, we conducted an extensive literature search. To the best of our knowledge, only one Legionella spp. PNA probe previously published was found in the literature, PLEG200 [32]. Nevertheless, PLEG200 was based on an existing DNA probe sequence (LEG226), which targets a low-binding-affinity site in the 16S rRNA [15,32]. In that work, ribosomal databases analysis indicated that this probe only bound to the Legionella spp. sequences deposited. Here, and 15 years later, we evaluated this probe again, together with LEG22, with the Probe Match tool (RDP-II) (Table 2) (last accessed in October 2021). The search showed that both probes presented acceptable levels of specificity. However, regarding sensitivity, PLEG200 showed to be less sensitive (76.4%). This may be explained, as these databases are constantly being updated, new sequences being deposited over time, and these changes can affect the accuracy of the probes, since these parameters will be related to the quality and quantity of sequences available in the analysis period [19]. Nevertheless, in practical terms, the criterion for the probe’s choice will fall on the aim of the study. If the criterion is focused on physiology studies or on the inoculation of a specific bacteria in a biofilm system, the sensitivity is not so relevant, as long as the strain of interest is detected. Conversely, for the development of a diagnostic method for online monitoring and risk assessment, it is important to have very high sensitivity. Nonetheless, in the particular case of Legionella, as almost 100% of the bacteria that causes outbreaks is L. pneumophila, as long as the probes can detect this species, the problem should be less relevant. Nevertheless, both probes, PLEG200 [32] and the newly designed LEG22, were able to detect all the Legionella strains tested.
We carefully designed the LEG22 probe to target a conserved region in Legionella strains. In fact, no false negative or false positive results were found when we tested the LEG22 probe against Legionella spp. and non-Legionella spp. strains (Table 1), which corroborates the performance of this probe, providing a suitable alternative for studies in pure cultures but also in biofilms. The main advantage of using PNA rRNA probes is related with the visualization of the whole cells and spatial distribution and metabolic activity of the cells [45], allowing a better knowledge of the organization and functional development of biofilms.
In a FISH protocol, the variables temperature and formamide concentration must be well adjusted in order to optimize the hybridization performance [46]. Formamide destabilizes double-stranded molecules by interfering with hydrogen bonds [47,48], and therefore, reducing the hybridization temperature. It is expected that for each 1% (vol/vol) formamide added, the temperature decreases about 1 °C. It is common to test formamide concentrations ranging from 5% to 70% (vol/vol), depending on the type of microorganism, and to use a range of temperatures near 50–60 °C [46]. Here, we tested 55 and 60 °C with 5, 30 and 50% (vol/vol) formamide. Although the best condition was shown to be 30% formamide and 60 °C, we further tested different temperatures, and 62 °C was chosen as the final temperature, as it provided a better signal-to-noise result.
To assess if the method could be applied in environmental samples, tap water samples artificially contaminated with different L. pneumophila serogroup 1 (WDCM 00107) CFUs were used to adapt a filtration protocol based on ISO 11731:2017 [6]. For both protocols used (PNA-FISH directly on the membrane or after cells elution from the filter), the signal was suitable up to 101 CFU/L with a good signal-to-noise ratio. For the less concentrated samples, detection with LEG22 was not possible, as the number of cells could have been below the detection limit of FISH using a microscope equipment. It was previously defined that for a trustworthy analysis, each microscopic field should contain at least five cells, which may not be the case in the 100 and 10−1 CFU/L samples [15,49], even though this technique appears to be appropriate for the rapid detection of Legionella in contaminated water samples.
For samples with higher turbidity, further optimization of the method will be carried out to improve the detection. The cooling tower water samples were analyzed with an initial centrifugation protocol based on ISO 11731:2017 [6] due to the high presence of residues. The samples, although testing positive for culture (except for one sample) (<103 CFU/L), were negative with PNA-FISH. One of the issues observed was the presence of autofluorescent water interferents that may hinder the detection of the cells’ fluorescent signal. In order to overcome the interference, the water samples pre-treatment will be a future concern. The implementation of a tangential flow filtration system to isolate and concentrate bacterial cells can effectively solve the problem [50]. However, compared to culture, which takes several days to provide results, PNA-FISH can be considered a rapid and reliable method for Legionella detection, as it takes only 3–4 h to perform the whole protocol.
FISH is a very versatile tool, and allied with microfluidic platforms, this method can, in the future, be implemented in routine monitoring practices in industrial settings.
5. Conclusions
This work describes the development of a new detection method for Legionella spp. in water, based on PNA-FISH. This method, using a LEG22 PNA probe, proved to be a very sensitive and specific method for Legionella spp. detection in pure cultures and in contaminated water samples. It was observed that for water samples with higher turbidity, coming from cooling towers, an improved filtration protocol is still needed. Nevertheless, the results showed that the PNA-FISH method remains a good alternative for rapid and accurate detection of Legionella.
6. Patents
A Portuguese patent was submitted, resulting from the work reported in this manuscript.
Author Contributions
Conceptualization, L.C., N.F.A. and C.A.; Methodology, L.C., M.N.-V., A.S.A. and G.N.A.; Investigation, M.N.-V., I.A. and A.B.; Validation, M.N.-V. and A.B.; Resources, C.P. and G.N.A.; Writing, M.N.-V., A.B. and L.C.; Review/Editing, L.C., A.S.A., C.A. and N.F.A.; Supervision, C.P., N.F.A., C.A. and L.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by: LA/P/0045/2020 (ALiCE), UIDB/00511/2020 and UIDP/00511/2020 (LEPABE), funded by national funds through the FCT/MCTES (PIDDAC); Projects POCI-01-0145-FEDER-029961, POCI-01-0145-FEDER-031011 and POCI-01-0145-FEDER-030431 funded by FEDER funds through COMPETE2020—Programa Operacional Competitividade e Internacionalização (POCI) and by national funds (PIDDAC) through FCT/MCTES. Montserrat Nácher-Vázquez and Laura Cerqueira were also financed by Project POCI-01-0145-FEDER-029961.
Acknowledgments
We would like to thank Manuel Simões and INSA-DSA ASMIP for providing the strains and water samples used in this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chauhan, D.; Shames, S.R. Pathogenicity and Virulence of Legionella: Intracellular Replication and Host Response. Virulence 2021, 12, 1122–1144. [Google Scholar] [CrossRef]
- Hamilton, K.A.; Prussin, A.J.; Ahmed, W.; Haas, C.N. Outbreaks of Legionnaires’ Disease and Pontiac Fever 2006–2017. Curr. Environ. Health Rep. 2018, 5, 263–271. [Google Scholar] [CrossRef]
- Cross, K.E.; Mercante, J.W.; Benitez, A.J.; Brown, E.W.; Diaz, M.H.; Winchell, J.M. Simultaneous Detection of Legionella Species and L. Anisa, L. Bozemanii, L. Longbeachae and L. Micdadei Using Conserved Primers and Multiple Probes in a Multiplex Real-Time PCR Assay. Diagn. Microbiol. Infect. Dis. 2016, 85, 295–301. [Google Scholar] [CrossRef][Green Version]
- Ziltener, P.; Reinheckel, T.; Oxenius, A. Neutrophil and Alveolar Macrophage-Mediated Innate Immune Control of Legionella Pneumophila Lung Infection via TNF and ROS. PLoS Pathog. 2016, 12, e1005591. [Google Scholar] [CrossRef]
- Mercante, J.W.; Winchell, J.M. Current and Emerging Legionella Diagnostics for Laboratory and Outbreak Investigations. Clin. Microbiol. Rev. 2015, 28, 95–133. [Google Scholar] [CrossRef]
- International Organization for Standardization. ISO 11731:2017 Water Quality—Enumeration of Legionella; International Organization for Standardization: Geneva, Switzerland, 2017; Volume 2017. [Google Scholar]
- Sciuto, E.L.; Laganà, P.; Filice, S.; Scalese, S.; Libertino, S.; Corso, D.; Faro, G.; Coniglio, M.A. Environmental Management of Legionella in Domestic Water Systems: Consolidated and Innovative Approaches for Disinfection Methods and Risk Assessment. Microorganisms 2021, 9, 577. [Google Scholar] [CrossRef]
- ISO. ISO/TS 12869 Water Quality—Detection and Quantification of Legionella Spp. and/or Legionella Pneumophila by Concentration and Genic Amplification by Quantitative Polymerase Chain Reaction (QPCR); ISO: Geneva, Switzerland, 2019. [Google Scholar]
- European Working Group for Legionella Infections (EWGLI). European Technical Guidelines for the Prevention, Control and Investigation, of Infections Caused by Legionella Species; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2017; p. 126.
- Young, C.; Smith, D.; Wafer, T.; Crook, B. Rapid Testing and Interventions to Control Legionella Proliferation Following a Legionnaires’ Disease Outbreak Associated with Cooling Towers. Microorganisms 2021, 9, 615. [Google Scholar] [CrossRef]
- Walker, J.T.; McDermott, P.J. Confirming the Presence of Legionella Pneumophila in Your Water System: A Review of Current Legionella Testing Methods. J. AOAC Int. 2021, 104, 1135–1147. [Google Scholar] [CrossRef]
- Azevedo, A.S.; Sousa, I.M.; Fernandes, R.M.; Azevedo, N.F.; Almeida, C. Optimizing Locked Nucleic Acid/2′-O-Methyl-RNA Fluorescence In Situ Hybridization (LNA/2′OMe-FISH) Procedure for Bacterial Detection. PLoS ONE 2019, 14, e0217689. [Google Scholar] [CrossRef]
- Cerqueira, L.; Moura, S.; Almeida, C.; Vieira, M.J.; Azevedo, N.F. Establishment of a New PNA-FISH Method for Aspergillus Fumigatus Identification: First Insights for Future Use in Pulmonary Samples. Microorganisms 2020, 8, 1950. [Google Scholar] [CrossRef]
- Kuo, J.-T.; Chang, L.-L.; Yen, C.-Y.; Tsai, T.-H.; Chang, Y.-C.; Huang, Y.-T.; Chung, Y.-C. Development of Fluorescence In Situ Hybridization as a Rapid, Accurate Method for Detecting Coliforms in Water Samples. Biosensors 2021, 11, 8. [Google Scholar] [CrossRef]
- Manz, W.; Amann, R.; Szewzyk, R.; Szewzyk, U.; Stenström, T.A.; Hutzler, P.; Schleifer, K.H. In Situ Identification of Legionellaceae Using 16S RRNA-Targeted Oligonucleotide Probes and Confocal Laser Scanning Microscopy. Microbiology 1995, 141, 29–39. [Google Scholar] [CrossRef]
- Rocha, R.; Santos, R.S.; Madureira, P.; Almeida, C.; Azevedo, N.F. Optimization of Peptide Nucleic Acid Fluorescence In Situ Hybridization (PNA-FISH) for the Detection of Bacteria: The Effect of PH, Dextran Sulfate and Probe Concentration. J. Biotechnol. 2016, 226, 1–7. [Google Scholar] [CrossRef]
- Emerson, J.B.; Adams, R.I.; Román, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s Microbes: Tools for Distinguishing the Living from the Dead in Microbial Ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef]
- Cerqueira, L.; Fernandes, R.M.; Ferreira, R.M.; Oleastro, M.; Carneiro, F.; Brandão, C.; Pimentel-Nunes, P.; Dinis-Ribeiro, M.; Figueiredo, C.; Keevil, C.W.; et al. Validation of a Fluorescence In Situ Hybridization Method Using Peptide Nucleic Acid Probes for Detection of Helicobacter Pylori Clarithromycin Resistance in Gastric Biopsy Specimens. J. Clin. Microbiol. 2013, 51, 1887–1893. [Google Scholar] [CrossRef]
- Cerqueira, L.; Azevedo, N.F.; Almeida, C.; Jardim, T.; Keevil, C.W.; Vieira, M.J. DNA Mimics for the Rapid Identification of Microorganisms by Fluorescence In Situ Hybridization (FISH). Int. J. Mol. Sci. 2008, 9, 1944–1960. [Google Scholar] [CrossRef]
- Stender, H.; Fiandaca, M.; Hyldig-Nielsen, J.J.; Coull, J. PNA for Rapid Microbiology. J. Microbiol. Methods 2002, 48, 1–17. [Google Scholar] [CrossRef]
- Chan, J.H.; And, S.L.; Wong, W.F. Antisense Oligonucleotides: From Design To Therapeutic Application. Clin. Exp. Pharmacol. Physiol. 2006, 33, 480–481. [Google Scholar] [CrossRef]
- Schoch, K.M.; Miller, T.M. Antisense Oligonucleotides: Translation from Mouse Models to Human Neurodegenerative Diseases Kathleen. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Cerqueira, L.; Fernandes, R.M.; Ferreira, R.M.; Carneiro, F.; Dinis-Ribeiro, M.; Figueiredo, C.; Keevil, C.W.; Azevedo, N.F.; Vieira, M.J. PNA-FISH as a New Diagnostic Method for the Determination of Clarithromycin Resistance of Helicobacter Pylori. BMC Microbiol. 2011, 11, 101. [Google Scholar] [CrossRef]
- Rocha, R.; Sousa, J.M.; Cerqueira, L.; Vieira, M.J.; Almeida, C.; Azevedo, N.F. Development and Application of Peptide Nucleic Acid Fluorescence In Situ Hybridization for the Specific Detection of Listeria Monocytogenes. Food Microbiol. 2019, 80, 1–8. [Google Scholar] [CrossRef]
- Lehtola, M.J.; Torvinen, E.; Miettinen, I.T.; Keevil, C.W. Fluorescence In Situ Hybridization Using Peptide Nucleic Acid Probes for Rapid Detection of Mycobacterium Avium Subsp. Avium and Mycobacterium Avium Subsp. Paratuberculosis in Potable-Water Biofilms. Appl. Environ. Microbiol. 2006, 72, 848–853. [Google Scholar] [CrossRef]
- Lehtola, M.J.; Loades, C.J.; Keevil, C.W. Advantages of Peptide Nucleic Acid Oligonucleotides for Sensitive Site Directed 16S RRNA Fluorescence In Situ Hybridization (FISH) Detection of Campylobacter Jejuni, Campylobacter Coli and Campylobacter Lari. J. Microbiol. Methods 2005, 62, 211–219. [Google Scholar] [CrossRef]
- Bragança, S.M.; Azevedo, N.F.; Simöes, L.C.; Keevil, C.W.; Vieira, M.J. Use of Fluorescent In Situ Hybridisation for the Visualisation of Helicobacter Pylori in Real Drinking Water Biofilms. Water Sci. Technol. 2007, 55, 387–393. [Google Scholar] [CrossRef]
- Hall, L.; Le Febre, K.M.; Deml, S.M.; Wohlfiel, S.L.; Wengenack, N.L. Evaluation of the Yeast Traffic Light PNA FISH Probes for Identification of Candida Species from Positive Blood Cultures. J. Clin. Microbiol. 2012, 50, 1446–1448. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.S.; Lima, C.C.; Carvalho, D.; Meireles, F.; Guimarães, N.; Azevedo, N.F. Response Surface Methodology to Optimize Peptide Nucleic Acid Fluorescence In Situ Hybridization (PNA-FISH) in Saccharomyces Cerevisiae. LWT Food Sci. Technol. 2017, 80, 27–31. [Google Scholar] [CrossRef]
- Shinozaki, M.; Okubo, Y.; Sasai, D.; Nakayama, H.; Murayama, S.Y.; Ide, T.; Wakayama, M.; Hiruta, N.; Shibuya, K. Identification of Fusarium Species in Formalin-Fixed and Paraffin-Embedded Sections by In Situ Hybridization Using Peptide Nucleic Acid Probes. J. Clin. Microbiol. 2011, 49, 808–813. [Google Scholar] [CrossRef]
- Teertstra, W.R.; Lugones, L.G.; Wösten, H.A.B. In Situ Hybridisation in Filamentous Fungi Using Peptide Nucleic Acid Probes. Fungal Genet. Biol. 2004, 41, 1099–1103. [Google Scholar] [CrossRef]
- Wilks, S.A.; Keevil, C.W. Targeting Species-Specific Low-Affinity 16S RRNA Binding Sites by Using Peptide Nucleic Acids for Detection of Legionellae in Biofilms. Appl. Environ. Microbiol. 2006, 72, 5453–5462. [Google Scholar] [CrossRef]
- Simões, L.C.; Simões, M.; Oliveira, R.; Vieira, M.J. Potential of the Adhesion of Bacteria Isolated from Drinking Water to Materials. J. Basic Microbiol. 2007, 47, 174–183. [Google Scholar] [CrossRef]
- Teixeira, H.; Sousa, A.L.; Azevedo, A.S. Bioinformatic Tools and Guidelines for the Design of Fluorescence In Situ Hybridization Probes. In Fluorescence In Situ Hybridization (FISH) for Microbial Cells—Methods and Concepts; Humana: New York, NY, USA, 2021; Volume 2246, pp. 35–50. [Google Scholar]
- Almeida, C.; Azevedo, N.F.; Fernandes, R.M.; Keevil, C.W.; Vieira, M.J. Fluorescence In Situ Hybridization Method Using a Peptide Nucleic Acid Probe for Identification of Salmonella Spp. in a Broad Spectrum of Samples. Appl. Environ. Microbiol. 2010, 76, 4476–4485. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.; Azevedo, A.S.; Mendes, L. Application of Nucleic Acid Mimics in Fluorescence In Situ Hybridization. In Fluorescence In Situ Hybridization (FISH) for Microbial Cells—Methods and Concepts; Humana: New York, NY, USA, 2021; Volume 2246, pp. 69–86. [Google Scholar]
- Ameida, D.Q.; Silva, T.; Rodrigues, V.; Ladeira, R.; Sousa, F.; Capucho, R.; Duarte, G. Surto de Doença Dos Legionários Na Costa Norte de Portugal Durante a Pandemia Da Covid-19. Acta Med. Port. 2021, 34, 2021. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. ELDSNET Operating Procedures for the Surveillance of Travel-Associated Legionnaires’ Disease in the EU/EEA; ECDC: Stockholm, Sweden, 2017; Volume 22.
- George, F.; Shivaji, T.; Pinto, C.S.; Serra, L.A.O.; Valente, J.; Albuquerque, M.J.; Vicêncio, P.C.O.; San-Bento, A.; Diegues, P.; Nogueira, P.J.; et al. A Large Outbreak of Legionnaires’ Disease in an Industrial Town in Portugal. Rev. Port. Saúde Pública 2016, 34, 199–208. [Google Scholar] [CrossRef][Green Version]
- Frickmann, H.; Zautner, A.E.; Moter, A.; Kikhney, J.; Hagen, R.M.; Stender, H.; Poppert, S. Fluorescence In Situ Hybridization (FISH) in the Microbiological Diagnostic Routine Laboratory: A Review. Crit. Rev. Microbiol. 2017, 43, 263–293. [Google Scholar] [CrossRef] [PubMed]
- Moter, A.; Göbel, U.B. Fluorescence In Situ Hybridization (FISH) for Direct Visualization of Microorganisms. J. Microbiol. Methods 2000, 41, 85–112. [Google Scholar] [CrossRef]
- Almeida, C.; Cerqueira, L.; Azevedo, N.F.; Vieira, M.J. Detection of Salmonella Enterica Serovar Enteritidis Using Real Time PCR, Immunocapture Assay, PNA FISH and Standard Culture Methods in Different Types of Food Samples. Int. J. Food Microbiol. 2013, 161, 16–22. [Google Scholar] [CrossRef]
- Amann, R.; Fuchs, B.M. Single-Cell Identification in Microbial Communities by Improved Fluorescence In Situ Hybridization Techniques. Nat. Rev. Microbiol. 2008, 6, 339–348. [Google Scholar] [CrossRef]
- Yilmaz, L.Ş.; Ökten, H.E.; Noguera, D.R. Making All Parts of the 16S RRNA of Escherichia Coli Accessible In Situ to Single DNA Oligonucleotides. Appl. Environ. Microbiol. 2006, 72, 733–744. [Google Scholar] [CrossRef]
- Dar, D.; Dar, N.; Cai, L.; Newman, D.K. In Situ Single-Cell Activities of Microbial Populations Revealed by Spatial Transcriptomics. bioRxiv 2021. [Google Scholar] [CrossRef]
- Almeida, C.; Azevedo, N.F. An Introduction to Fluorescence In Situ Hybridization in Microorganisms. In Fluorescence In Situ Hybridization (FISH) for Microbial Cells—Methods and Concepts; Humana: New York, NY, USA, 2021; Volume 2246, pp. 1–15. [Google Scholar]
- Farrell, R.E. RNA Methodologies A Laboratory Guide for Isolation and Characterization; Academic Press: New York, NY, USA, 2010. [Google Scholar]
- Santos, R.S.; Guimarães, N.; Madureira, P.; Azevedo, N.F. Optimization of a Peptide Nucleic Acid Fluorescence In Situ Hybridization (PNA-FISH) Method for the Detection of Bacteria and Disclosure of a Formamide Effect. J. Biotechnol. 2014, 187, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.; Azevedo, N.F.; Iversen, C.; Fanning, S.; Keevil, C.W.; Vieira, M.J. Development and Application of a Novel Peptide Nucleic Acid Probe for the Specific Detection of Cronobacter Genomospecies (Enterobacter Sakazakii) in Powdered Infant Formula. Appl. Environ. Microbiol. 2009, 75, 2925–2930. [Google Scholar] [CrossRef] [PubMed]
- McEgan, R.; Rodrigues, C.A.P.; Sbodio, A.; Suslow, T.V.; Goodridge, L.D.; Danyluk, M.D. Detection of Salmonella Spp. from Large Volumes of Water by Modified Moore Swabs and Tangential Flow Filtration. Lett. Appl. Microbiol. 2013, 56, 88–94. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).