Long Noncoding RNAs in Plant Viroids and Viruses: A Review
Abstract
1. Introduction: lncRNAs
2. Viroids
3. Replicative lncRNAs: Defective and Defective-Interfering RNAs
4. Replicative RNAs: Satellite RNAs
5. Nonreplicative Lnc RNAs of Plant Viruses
6. Host lncRNAs during Plant-Virus Interactions
7. Final Remarks
Supplementary Materials
Funding
Conflicts of Interest
References
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef] [PubMed]
- DiStefano, J.K. The emerging role of long noncoding RNAs in human disease. In Disease Gene Identification; Springer: New York, NY, USA, 2018; pp. 91–110. [Google Scholar]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-L.V.; Chekanova, J.A. Long noncoding RNAs in plants. In Long Non Coding RNA Biology; Springer: Singapore, 2017; pp. 133–154. [Google Scholar]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef]
- Jiang, D.; Kong, N.C.; Gu, X.; Li, Z.; He, Y. Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 2011, 7, e1001330. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Matzke, M.A.; Kanno, T.; Matzke, A.J.M. RNA-directed DNA methylation: The evolution of a complex epigenetic pathway in flowering Plants. Annu. Rev. Plant. Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef]
- Seo, J.S.; Sun, H.X.; Park, B.S.; Huang, C.H.; Yeh, S.D.; Jung, C.; Chua, N.H. ELF18-INDUCED LONG-NONCODING RNA associates with mediator to enhance expression of innate immune response genes in arabidopsis. Plant. Cell 2017, 29, 1024–1038. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Ding, B. The biology of viroid-host interactions. Annu. Rev. Phytopathol. 2009, 47, 105–131. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Hernández, C.; Alba, A.E.M.D.; Daròs, J.A.; Serio, F.D. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 2005, 43, 117–139. [Google Scholar] [CrossRef] [PubMed]
- Shimura, H.; Masuta, C. Plant subviral RNAs as a long noncoding RNA (lncRNA): Analogy with animal lncRNAs in host–virus interactions. Virus Res. 2016, 212, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Adkar-Purushothama, C.R.; Perreault, J.P. Current overview on viroid–host interactions. Wiley Interdiscip. Rev. RNA 2020, 11, e1570. [Google Scholar] [CrossRef]
- Flores, R.; Delgado, S.; Gas, M.-E.; Carbonell, A.; Molina, D.; Gago, S.; De la Pena, M. Viroids: The minimal non-coding RNAs with autonomous replication. FEBS Lett. 2004, 567, 42–48. [Google Scholar] [CrossRef]
- Jiang, J.; Smith, H.N.; Ren, D.; Mudiyanselage, S.D.D.; Dawe, A.L.; Wang, L.; Wang, Y. Potato spindle tuber viroid modulates its replication through a direct interaction with a splicing regulator. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Dissanayaka Mudiyanselage, S.D.; Qu, J.; Tian, N.; Jiang, J.; Wang, Y. Potato spindle tuber viroid RNA-templated transcription: Factors and regulation. Viruses 2018, 10, 503. [Google Scholar] [CrossRef]
- Dissanayaka Mudiyanselage, S.D.; Wang, Y. Evidence Supporting That RNA polymerase II catalyzes de novo transcription using potato spindle tuber viroid circular RNA templates. Viruses 2020, 12, 371. [Google Scholar] [CrossRef]
- Flores, R.; Gas, M.E.; Molina-Serrano, D.; Nohales, M.Á.; Carbonell, A.; Gago, S.; De la Peña, M.; Daròs, J.A. Viroid replication: Rolling-circles, enzymes and ribozymes. Viruses 2009, 1, 317–334. [Google Scholar] [CrossRef]
- Nohales, M.-Á.; Flores, R.; Daròs, J.-A. Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc. Natl. Acad. Sci. USA 2012, 109, 13805–13810. [Google Scholar] [CrossRef]
- Daròs, J.-A. Viroids: Small noncoding infectious RNAs with the remarkable ability of autonomous replication. In Current Research Topics in Plant Virology; Springer: Berlin/Heidelberg, Germany, 2016; pp. 295–322. [Google Scholar]
- Navarro, J.-A.; Vera, A.; Flores, R. A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology 2000, 268, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Navarro, J.A.; Flores, R. Characterization of the initiation sites of both polarity strands of a viroid RNA reveals a motif conserved in sequence and structure. EMBO J. 2000, 19, 2662–2670. [Google Scholar] [CrossRef] [PubMed]
- Delgado, S.; de Alba, Á.E.M.; Hernández, C.; Flores, R. A short double-stranded RNA motif of peach latent mosaic viroid contains the initiation and the self-cleavage sites of both polarity strands. J. Virol. 2005, 79, 12934–12943. [Google Scholar] [CrossRef] [PubMed]
- Latifi, A.; Bernard, C.; Da Silva, L.; Andéol, Y.; Elleuch, A.; Risoul, V.; Vergne, J.; Maurel, M.C. Replication of avocado sunblotch viroid in the cyanobacterium Nostoc sp. PCC 7120. J. Plant. Pathol. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Flores, R.; Daròs, J.-A.; Hernández, C. Avsunviroidae family: Viroids containing hammerhead ribozymes. Adv. Virus Res. 2000, 55, 271. [Google Scholar]
- Hernandez, C.; Flores, R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. USA 1992, 89, 3711–3715. [Google Scholar] [CrossRef]
- Nohales, M.-Á.; Molina-Serrano, D.; Flores, R.; Daròs, J.-A. Involvement of the chloroplastic isoform of tRNA ligase in the replication of viroids belonging to the family Avsunviroidae. J. Virol. 2012, 86, 8269–8276. [Google Scholar] [CrossRef]
- Moreno, M.; Vazquez, L.; Lopez-Carrasco, A.; Martin-Gago, J.; Flores, R.; Briones, C. Direct visualization of the native structure of viroid RNAs at single-molecule resolution by atomic force microscopy. RNA Biol. 2019, 16, 295–308. [Google Scholar] [CrossRef]
- Xu, W.; Bolduc, F.; Hong, N.; Perreault, J.-P. The use of a combination of computer-assisted structure prediction and SHAPE probing to elucidate the secondary structures of five viroids. Mol. Plant. Pathol. 2012, 13, 666–676. [Google Scholar] [CrossRef]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Di Serio, F. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant. J. 2012, 70, 991–1003. [Google Scholar] [CrossRef]
- Flores, R.; Serra, P.; Minoia, S.; Di Serio, F.; Navarro, B. Viroids: From genotype to phenotype just relying on RNA sequence and structural motifs. Front. Microbiol. 2012, 3, 217. [Google Scholar] [CrossRef]
- Kovalskaya, N.; Hammond, R.W. Molecular biology of viroid–host interactions and disease control strategies. Plant. Sci. 2014, 228, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Dadami, E.; Boutla, A.; Vrettos, N.; Tzortzakaki, S.; Karakasilioti, I.; Kalantidis, K. DICER-LIKE 4 but not DICER-LIKE 2 may have a positive effect on potato spindle tuber viroid accumulation in Nicotiana benthamiana. Mol. Plant. 2013, 6, 232–234. [Google Scholar] [CrossRef] [PubMed]
- De Alba, A.E.M.; Sägesser, R.; Tabler, M.; Tsagris, M. A bromodomain-containing protein from tomato specifically binds potato spindle tuber viroid RNA in vitro and in vivo. J. Virol. 2003, 77, 9685–9694. [Google Scholar] [CrossRef] [PubMed]
- Kalantidis, K.; Denti, M.; Tzortzakaki, S.; Marinou, E.; Tabler, M.; Tsagris, M. Virp1 is a host protein with a major role in Potato spindle tuber viroid infection in Nicotiana plants. J. Virol. 2007, 81, 12872–12880. [Google Scholar] [CrossRef]
- Ding, B.; Itaya, A. Viroid: A useful model for studying the basic principles of infection and RNA biology. Mol. Plant Microbe Interact. 2007, 20, 7–20. [Google Scholar] [CrossRef]
- Lisón, P.; Tárraga, S.; López-Gresa, P.; Saurí, A.; Torres, C.; Campos, L.; Bellés, J.M.; Conejero, V.; Rodrigo, I. A noncoding plant pathogen provokes both transcriptional and posttranscriptional alterations in tomato. Proteomics 2013, 13, 833–844. [Google Scholar] [CrossRef]
- Cottilli, P.; Belda-Palazón, B.; Adkar-Purushothama, C.R.; Perreault, J.-P.; Schleiff, E.; Rodrigo, I.; Ferrando, A.; Lisón, P. Citrus exocortis viroid causes ribosomal stress in tomato plants. Nucleic Acids Res. 2019, 47, 8649–8661. [Google Scholar] [CrossRef]
- Kiefer, M.C.; Owens, R.A.; Diener, T. Structural similarities between viroids and transposable genetic elements. Proc. Natl. Acad. Sci. USA 1983, 80, 6234–6238. [Google Scholar] [CrossRef]
- Flores, R.; Gago-Zachert, S.; Serra, P.; Sanjuán, R.; Elena, S.F. Viroids: Survivors from the RNA world? Annu. Rev. Microbiol. 2014, 68, 395–414. [Google Scholar] [CrossRef]
- Ruminski, D.J.; Webb, C.-H.T.; Riccitelli, N.J.; Lupták, A. Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J. Biol. Chem. 2011, 286, 41286–41295. [Google Scholar] [CrossRef] [PubMed]
- Maurel, M.-C.; Leclerc, F.; Vergne, J.; Zaccai, G. RNA back and forth: Looking through ribozyme and viroid motifs. Viruses 2019, 11, 283. [Google Scholar] [CrossRef] [PubMed]
- Catalán, P.; Elena, S.F.; Cuesta, J.A.; Manrubia, S. Parsimonious scenario for the emergence of viroid-like replicons de novo. Viruses 2019, 11, 425. [Google Scholar] [CrossRef] [PubMed]
- Hadidi, A. Next-generation sequencing and CRISPR/Cas13 editing in viroid research and molecular diagnostics. Viruses 2019, 11, 120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qi, S.; Tang, N.; Zhang, X.; Chen, S.; Zhu, P.; Ma, L.; Cheng, J.; Xu, Y.; Lu, M. Discovery of replicating circular RNAs by RNA-seq and computational algorithms. PLoS Pathog. 2014, 10, e1004553. [Google Scholar] [CrossRef]
- Jiang, D.-M.; Wang, M.; Li, S.-F.; Zhang, Z.-X. High-throughput sequencing analysis of small RNAs derived from coleus blumei viroids. Viruses 2019, 11, 619. [Google Scholar] [CrossRef]
- Glouzon, J.-P.S.; Bolduc, F.; Wang, S.; Najmanovich, R.J.; Perreault, J.-P. Deep-sequencing of the peach latent mosaic viroid reveals new aspects of population heterogeneity. PLoS ONE 2014, 9, e87297. [Google Scholar] [CrossRef]
- Wang, Y.; Atta, S.; Wang, X.; Yang, F.; Zhou, C.; Cao, M. Transcriptome sequencing reveals novel Citrus bark cracking viroid (CBCVd) variants from citrus and their molecular characterization. PLoS ONE 2018, 13, e0198022. [Google Scholar] [CrossRef]
- Simon, A.E.; Roossinck, M.J.; Havelda, Z. Plant virus satellite and defective interfering RNAs: New paradigms for a new century. Annu. Rev. Phytopathol. 2004, 42, 415–437. [Google Scholar] [CrossRef]
- Pogany, J.; Romero, J.; Huang, Q.; Sgro, J.-Y.; Shang, H.; Bujarski, J.J. De novo generation of defective interfering-like RNAs in broad bean mottle bromovirus. Virology 1995, 212, 574–586. [Google Scholar] [CrossRef]
- Romero, J.; Huang, Q.; Pogany, J.; Bujarski, J.J. Characterization of defective interfering RNA components that increase symptom severity of broad bean mottle virus infections. Virology 1993, 194, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Collmer, C.W.; Howell, S.H. Role of satellite RNA in the expression of symptoms caused by plant viruses. Annu. Rev. Phytopathol. 1992, 30, 419–442. [Google Scholar] [CrossRef] [PubMed]
- Matthews, R.E.F.; Hull, R. Matthews’ Plant. Virology; Gulf Professional Publishing: Houston, TX, USA, 2002. [Google Scholar]
- Roossinck, M.; Sleat, D.; Palukaitis, P. Satellite RNAs of plant viruses: Structures and biological effects. Microbiol. Mol. Biol. Rev. 1992, 56, 265–279. [Google Scholar] [CrossRef]
- White, K.; Morris, T. Defective and defective interfering RNAs of monopartite plus-strand RNA plant viruses. In Satellites and Defective Viral RNAs; Springer: Berlin/Heidelberg, Germany, 1999; pp. 1–17. [Google Scholar]
- Szittya, G.; Molnár, A.; Silhavy, D.; Hornyik, C.; Burgyán, J. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant. Cell 2002, 14, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Graves, M.V.; Pogany, J.; Romero, J. Defective interfering RNAs and defective viruses associated with multipartite RNA viruses of plants. In Seminars in VIROLOGY; Academic Press: Cambridge, MA, USA, 1996. [Google Scholar]
- Graves, M.V.; Roossinck, M.J. Characterization of defective RNAs derived from RNA 3 of the Fny strain of cucumber mosaic cucumovirus. J. Virol. 1995, 69, 4746–4751. [Google Scholar] [CrossRef]
- Kaplan, I.B.; Lee, K.-C.; Canto, T.; Wong, S.-M.; Palukaitis, P. Host-specific encapsidation of a defective RNA 3 of Cucumber mosaic virus. J. Gen. Virol. 2004, 85, 3757–3763. [Google Scholar] [CrossRef]
- Vignuzzi, M.; López, C.B. Defective viral genomes are key drivers of the virus–host interaction. Nat. Microbiol. 2019, 4, 1075–1087. [Google Scholar] [CrossRef]
- Molenkamp, R.; Greve, S.; Spaan, W.J.; Snijder, E.J. Efficient homologous RNA recombination and requirement for an open reading frame during replication of equine arteritis virus defective interfering RNAs. J. Virol. 2000, 74, 9062–9070. [Google Scholar] [CrossRef][Green Version]
- Pathak, K.B.; Nagy, P.D. Defective interfering RNAs: Foes of viruses and friends of virologists. Viruses 2009, 1, 895–919. [Google Scholar] [CrossRef]
- Kim, M.-J.; Kao, C. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: Implications for RNA–RNA recombination. Proc. Natl. Acad. Sci. USA 2001, 98, 4972–4977. [Google Scholar] [CrossRef]
- Cheng, C.-P.; Nagy, P.D. Mechanism of RNA recombination in carmo-and tombusviruses: Evidence for template switching by the RNA-dependent RNA polymerase in vitro. J. Virol. 2003, 77, 12033–12047. [Google Scholar] [CrossRef] [PubMed]
- Wierzchoslawski, R.; Bujarski, J.J. Efficient in vitro system of homologous recombination in brome mosaic bromovirus. J. Virol. 2006, 80, 6182–6187. [Google Scholar] [CrossRef] [PubMed]
- Wierzchoslawski, R.; Dzianott, A.; Kunimalayan, S.; Bujarski, J.J. A transcriptionally active subgenomic promoter supports homologous crossovers in a plus-strand RNA virus. J. Virol. 2003, 77, 6769–6776. [Google Scholar] [CrossRef] [PubMed]
- Pogany, J.; Nagy, P.D. Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J. Virol. 2008, 82, 5967–5980. [Google Scholar] [CrossRef] [PubMed]
- Eliasco, E.; Livieratos, I.; Müller, G.; Guzman, M.; Salazar, L.; Coutts, R. Sequences of defective RNAs associated with potato yellow vein virus. Arch. Virol. 2006, 151, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Havelda, Z.; Szittya, G.; Burgyán, J. Characterization of the molecular mechanism of defective interfering RNA-mediated symptom attenuation in tombusvirus-infected plants. J. Virol. 1998, 72, 6251–6256. [Google Scholar] [CrossRef]
- Llamas, S.; Sandoval, C.; Babin, M.; Pogany, J.; Bujarski, J.J.; Romero, J. Effect of the host and temperature on the formation of defective RNAs associated with broad bean mottle virus infection. Phytopathology 2004, 94, 69–75. [Google Scholar] [CrossRef]
- Nagy, P.D.; Dzianott, A.; Ahlquist, P.; Bujarski, J.J. Mutations in the helicase-like domain of protein 1a alter the sites of RNA-RNA recombination in brome mosaic virus. J. Virol. 1995, 69, 2547–2556. [Google Scholar] [CrossRef]
- Poirier, E.Z.; Mounce, B.C.; Rozen-Gagnon, K.; Hooikaas, P.J.; Stapleford, K.A.; Moratorio, G.; Vignuzzi, M. Low-fidelity polymerases of alphaviruses recombine at higher rates to overproduce defective interfering particles. J. Virol. 2016, 90, 2446–2454. [Google Scholar] [CrossRef]
- Fodor, E.; Crow, M.; Mingay, L.J.; Deng, T.; Sharps, J.; Fechter, P.; Brownlee, G.G. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 2002, 76, 8989–9001. [Google Scholar] [CrossRef]
- Nagy, P.D. Mutations in the RNA-binding domains of tombusvirus replicase proteins affect RNA recombination in vivo. Virology 2003, 317, 359–372. [Google Scholar]
- Vasilijevic, J.; Zamarreño, N.; Oliveros, J.C.; Rodriguez-Frandsen, A.; Gómez, G.; Rodriguez, G.; Pérez-Ruiz, M.; Rey, S.; Barba, I.; Pozo, F. Reduced accumulation of defective viral genomes contributes to severe outcome in influenza virus infected patients. PLoS Pathog. 2017, 13, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Odagiri, T.; Tobita, K. Mutation in NS2, a nonstructural protein of influenza A virus, extragenically causes aberrant replication and expression of the PA gene and leads to generation of defective interfering particles. Proc. Natl. Acad. Sci. USA 1990, 87, 5988–5992. [Google Scholar] [CrossRef]
- Pfaller, C.K.; Mastorakos, G.M.; Matchett, W.E.; Ma, X.; Samuel, C.E.; Cattaneo, R. Measles virus defective interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA-1-like hypermutations. J. Virol. 2015, 89, 7735–7747. [Google Scholar] [CrossRef] [PubMed]
- Lukhovitskaya, N.I.; Thaduri, S.; Garushyants, S.K.; Torrance, L.; Savenkov, E.I. Deciphering the mechanism of defective interfering RNA (DI RNA) biogenesis reveals that a viral protein and the DI RNA act antagonistically in virus infection. J. Virol. 2013, 87, 6091–6103. [Google Scholar] [CrossRef]
- Nagy, P.D.; Bujarski, J.J. Homologous RNA recombination in brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J. Virol. 1996, 70, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D.; Bujarski, J.J. Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J. Virol. 1997, 71, 3799–3810. [Google Scholar] [CrossRef]
- Sztuba-Solińska, J.; Fanning, S.W.; Horn, J.R.; Bujarski, J.J. Mutations in the coat protein-binding cis-acting RNA motifs debilitate RNA recombination of Brome mosaic virus. Virus Res. 2012, 170, 138–149. [Google Scholar] [CrossRef]
- Jaag, H.M.; Nagy, P.D. The combined effect of environmental and host factors on the emergence of viral RNA recombinants. PLoS Pathog. 2010, 6, e1001156. [Google Scholar] [CrossRef]
- Cattaneo, R.; Schmid, A.; Eschle, D.; Baczko, K.; ter Meulen, V.; Billeter, M.A. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 1988, 55, 255–265. [Google Scholar] [CrossRef]
- Ward, S.V.; Sternsdorf, T.; Woods, N.-B. Targeting expression of the leukemogenic PML-RARα fusion protein by lentiviral vector-mediated small interfering RNA results in leukemic cell differentiation and apoptosis. Hum. Gene Ther. 2011, 22, 1593–1598. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Kim, E.J.; Felt, S.A.; Taylor, L.J.; Agarwal, D.; Grant, G.R.; Lopez, C.B. Correction: A specific sequence in the genome of respiratory syncytial virus regulates the generation of copy-back defective viral genomes. PLoS Pathog. 2019, 15, e1008099. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Minicka, J.; Zarzyńska-Nowak, A.; Budzyńska, D.; Elena, S.F. Defective RNA particles derived from Tomato black ring virus genome interfere with the replication of parental virus. Virus Res. 2018, 250, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Pospieszny, H.; Hasiów-Jaroszewska, B.; Borodynko-Filas, N.; Elena, S.F. Effect of defective interfering RNA on the vertical transmission of Tomato black ring virus. Plant. Prot. Sci. 2020. [Google Scholar] [CrossRef]
- Chang, Y.; Borja, M.; Scholthof, H.; Jackson, A.; Morris, T. Host effects and sequences essential for accumulation of defective interfering RNAs of cucumber necrosis and tomato bushy stunt tombusviruses. Virology 1995, 210, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Hornyik, C.; Havelda, Z.; Burgyán, J. Identification of sequence elements of tombusvirus-associated defective interfering RNAs required for symptom modulation. Arch. Virol. 2006, 151, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-C.; Hsu, Y.-H.; Lin, N.-S. Satellite RNAs and satellite viruses of plants. Viruses 2009, 1, 1325–1350. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-B.; Bian, X.-Y.; Wu, L.-M.; Liu, L.-X.; Smith, N.A.; Isenegger, D.; Wu, R.-M.; Masuta, C.; Vance, V.B.; Watson, J.M. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA 2004, 101, 3275–3280. [Google Scholar] [CrossRef]
- Smith, N.A.; Eamens, A.L.; Wang, M.-B. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 2011, 7, e1002022. [Google Scholar] [CrossRef]
- KAPER, J.M.; Waterworth, H.E. Cucumber Mosaic Virus Associated RNA 5: Causal Agent for Tomato Necrosis. Science 1977, 196, 429–431. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Kalantidis, K.; Rao, A.L.N. A bromodomain-containing host protein mediates the nuclear importation of a satellite RNA of cucumber mosaic virus. J. Virol. 2014, 88, 1890–1896. [Google Scholar] [CrossRef] [PubMed]
- Palani, P.V.; Kasiviswanathan, V.; Chen, J.C.-F.; Chen, W.; Hsu, Y.-H.; Lin, N.-S. The Arginine-Rich Motif of Bamboo mosaic virus Satellite RNA-Encoded P20 Mediates Self-Interaction, Intracellular Targeting, and Cell-to-Cell Movement. Mol. Plant. Microbe Interact. 2006, 19, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Roossinck, M.J. Chapter 60-Small Linear Satellite RNAs. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 649–658. [Google Scholar]
- Palukaitis, P.; García-Arenal, F. Cucumoviruses. Adv. Virus. Res. 2003, 62, 241–323. [Google Scholar] [PubMed]
- Kouadio, K.T.; De Clerck, C.; Agneroh, T.A.; Parisi, O.; Lepoivre, P.; Jijakli, H. Role of satellite RNAs in cucumber mosaic virus-host plant interactions. A review. Biotechnol. Agron. Société Environ. 2013, 17, 644–650. [Google Scholar]
- Sleat, D.E.; Zhang, L.; Palukaitis, P. Mapping determinants within cucumber mosaic virus and its satellite RNA for the induction of necrosis in tomato plants. Mol. Plant Microbe Interact. 1994, 7, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Irian, S.; Dai, X.; Zhao, P.X.; Roossinck, M.J. Regulation of a virus-induced lethal disease in tomato revealed by LongSAGE analysis. Mol. Plant. Microbe Interact. 2007, 20, 1477–1488. [Google Scholar] [CrossRef]
- Shen, W.X.; Au, P.C.K.; Shi, B.J.; Smith, N.A.; Dennis, E.S.; Guo, H.S.; Wang, M.B.; Zhou, C.Y. Satellite RNAs interfere with the function of viral RNA silencing suppressors. Front. Plant. Sci. 2015, 6, 281. [Google Scholar] [CrossRef]
- Shimura, H.; Pantaleo, V.; Ishihara, T.; Myojo, N.; Inaba, J.-i.; Sueda, K.; Burgyán, J.; Masuta, C.A. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 2011, 7, e1002021. [Google Scholar] [CrossRef]
- Obrępalska-Stęplowska, A.; Zmienko, A.; Wrzesińska, B.; Goralski, M.; Figlerowicz, M.; Zyprych-Walczak, J.; Siatkowski, I.; Pospieszny, H. The defense response of Nicotiana benthamiana to peanut stunt virus infection in the presence of symptom exacerbating satellite RNA. Viruses 2018, 10, 449. [Google Scholar] [CrossRef]
- Zhu, H.; Duan, C.-G.; Hou, W.-N.; Du, Q.-S.; Lv, D.-Q.; Fang, R.-X.; Guo, H.-S. Satellite RNA-Derived Small Interfering RNA satsiR-12 Targeting the 3′ Untranslated Region of Cucumber Mosaic Virus Triggers Viral RNAs for Degradation. J. Virol. 2011, 85, 13384–13397. [Google Scholar] [CrossRef]
- Zahid, K.; Zhao, J.-H.; Smith, N.A.; Schumann, U.; Fang, Y.-Y.; Dennis, E.S.; Zhang, R.; Guo, H.-S.; Wang, M.-B. Nicotiana small RNA sequences support a host genome origin of cucumber mosaic virus satellite RNA. PLoS Genet. 2015, 11, e1004906. [Google Scholar] [CrossRef] [PubMed]
- Hajimorad, M.R.; Ghabrial, S.A.; Roossinck, M.J. De novo emergence of a novel satellite RNA of cucumber mosaic virus following serial passages of the virus derived from RNA transcripts. Arch. Virol. 2009, 154, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Pyle, A.M. Looking at LncRNAs with the ribozyme toolkit. Mol. Cell 2014, 56, 13–17. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gellatly, D.; Mirhadi, K.; Venkataraman, S.; AbouHaidar, M.G. Structural and sequence integrity are essential for the replication of the viroid-like satellite RNA of lucerne transient streak virus. J. Gen. Virol. 2011, 92, 1475–1481. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P. Satellite RNAs and satellite viruses. Mol. Plant. Microbe Interact. 2016, 29, 181–186. [Google Scholar] [CrossRef]
- Nelson, J.A.; Shepotinovskaya, I.; Uhlenbeck, O.C. Hammerheads derived from sTRSV show enhanced cleavage and ligation rate constants. Biochemistry 2005, 44, 14577–14585. [Google Scholar] [CrossRef]
- AbouHaidar, M.G.; Venkataraman, S.; Golshani, A.; Liu, B.; Ahmad, T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc. Natl. Acad. Sci. USA 2014, 111, 14542–14547. [Google Scholar] [CrossRef]
- Liao, Q.; Zhu, L.; Du, Z.; Zeng, R.; Feng, J.; Chen, J. Satellite RNA-mediated reduction of cucumber mosaic virus genomic RNAs Accumulation in Nicotiana tabacum. Acta Biochimica et Biophysica Sinica 2007, 39, 217–223. [Google Scholar] [CrossRef]
- Zhang, F.; Simon, A.E. Enhanced viral pathogenesis associated with a virulent mutant virus or a virulent satellite RNA correlates with reduced virion accumulation and abundance of free coat protein. Virology 2003, 312, 8–13. [Google Scholar] [CrossRef]
- Sayama, H.; Kominato, M.; Ubukata, M.; Sato, T. Three-year risk assessment of the practical application of cross-protection in processing tomato fields using an attenuated cucumber mosaic virus (CMV) strain containing an ameliorative satellite RNA. Acta Horticulturae 2001, 542, 47–53. [Google Scholar] [CrossRef]
- Cillo, F.; Finetti-Sialer, M.M.; Papanice, M.A.; Gallitelli, D. Analysis of mechanisms involved in the Cucumber mosaic virus satellite RNA-mediated transgenic resistance in tomato plants. Mol. Plant. Microbe Interact. 2004, 17, 98–108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cao, X.; Liu, S.; Yu, C.; Li, X.; Yuan, X. A new strategy of using satellite RNA to control viral plant diseases: Post-inoculation with satellite RNA attenuates symptoms derived from pre-infection with its helper virus. Plant. Biotechnol. J. 2019, 17, 1856–1858. [Google Scholar] [CrossRef] [PubMed]
- Gnanasekaran, P.; Chakraborty, S. Biology of viral satellites and their role in pathogenesis. Curr. Opin. Virol. 2018, 33, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solińska, J.; Stollar, V.; Bujarski, J.J. Subgenomic messenger RNAs: Mastering regulation of (+)-strand RNA virus life cycle. Virology 2011, 412, 245–255. [Google Scholar] [CrossRef]
- Wierzchoslawski, R.; Urbanowicz, A.; Dzianott, A.; Figlerowicz, M.; Bujarski, J.J. characterization of a novel 5′ subgenomic RNA3a derived from RNA3 of brome mosaic bromovirus. J. Virol. 2006, 80, 12357–12366. [Google Scholar] [CrossRef]
- Roby, J.A.; Pijlman, G.P.; Wilusz, J.; Khromykh, A.A. Noncoding subgenomic flavivirus RNA: Multiple functions in West Nile Virus pathogenesis and modulation of host responses. Viruses 2014, 6, 404–427. [Google Scholar] [CrossRef]
- Ta, M.; Vrati, S. Mov34 protein from mouse brain interacts with the 3′ noncoding region of japanese encephalitis virus. J. Virol. 2000, 74, 5108–5115. [Google Scholar] [CrossRef]
- Lei, Y.; Huang, Y.; Zhang, H.; Yu, L.; Zhang, M.; Dayton, A. Functional interaction between cellular p100 and the dengue virus 3′ UTR. J. Gen. Virol. 2011, 92, 796–806. [Google Scholar] [CrossRef]
- Iwakawa, H.-O.; Mizumoto, H.; Nagano, H.; Imoto, Y.; Takigawa, K.; Sarawaneeyaruk, S.; Kaido, M.; Mise, K.; Okuno, T. A viral noncoding RNA generated by Cis-element-mediated protection against 5′→3′ RNA decay represses both cap-independent and cap-dependent translation. J. Virol. 2008, 82, 10162–10174. [Google Scholar] [CrossRef]
- Peltier, C.; Klein, E.; Hleibieh, K.; D’Alonzo, M.; Hammann, P.; Bouzoubaa, S.; Ratti, C.; Gilmer, D. Beet necrotic yellow vein virus subgenomic RNA3 is a cleavage product leading to stable non-coding RNA required for long-distance movement. J. Gen. Virol. 2012, 93, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-H.; Sun, Y.-D.; Atallah, O.O.; Huguet-Tapia, J.C.; Noble, J.D.; Folimonova, S.Y. A Long Non-Coding RNA of Citrus tristeza virus: Role in the virus interplay with the host immunity. Viruses 2019, 11, 436. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Miller, W.A.; Shen, R.; Staplin, W.; Kanodia, P. Noncoding RNAs of plant viruses and viroids: Sponges of host translation and RNA interference machinery. Mol. Plant. Microbe Interact. 2016, 29, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Kapusta, A.; Kronenberg, Z.; Lynch, V.J.; Zhuo, X.; Ramsay, L.; Bourque, G.; Yandell, M.; Feschotte, C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013, 9, e1003470. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.-H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in arabidopsis. Plant. Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [PubMed]
- Biémont, C. A brief history of the status of transposable elements: From junk DNA to major players in evolution. Genetics 2010, 186, 1085–1093. [Google Scholar] [CrossRef]
- Castel, S.E.; Martienssen, R.A. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013, 14, 100–112. [Google Scholar] [CrossRef]
- Mohebbi, A.; Tahamtan, A.; Eskandarian, S.; Askari, F.S.; Shafaei, M.; Lorestani, N. Viruses and long non-coding RNAs: Implicating an evolutionary conserved region. VirusDisease 2018, 29, 478–485. [Google Scholar] [CrossRef]
- Zaynab, M.; Fatima, M.; Abbas, S.; Umair, M.; Sharif, Y.; Raza, M.A. Long non-coding RNAs as molecular players in plant defense against pathogens. Microbial Pathogenesis 2018, 121, 277–282. [Google Scholar] [CrossRef]
- Nejat, N.; Ramalingam, A.; Mantri, N. Advances in Transcriptomics of Plants. In Plant Genetics and Molecular Biology; Varshney, R.K., Pandey, M.K., Chitikineni, A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 161–185. [Google Scholar]
- Wang, J.; Yu, W.; Yang, Y.; Li, X.; Chen, T.; Liu, T.; Ma, N.; Yang, X.; Liu, R.; Zhang, B. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 2015, 5, 16946. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Y.; Jin, L.; Ling, X.; Liu, T.; Chen, T.; Ji, Y.; Yu, W.; Zhang, B. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant. Biol. 2018, 18, 104. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Chu, S.; Jiao, Y. Present scenario of circular RNAs (circRNAs) in plants. Front. Plant. Sci. 2019, 10, 379. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Bazin, J.; Webb, S.; Crespi, M.; Zubieta, C.; Conn, S.J. CircRNAs in plants. In Circular RNAs; Springer: Singapore, 2018; pp. 329–343. [Google Scholar]
- Zhang, P.; Li, S.; Chen, M. Characterization and function of circular RNAs in plants. Front. Mol. Biosci. 2020, 7, 91. [Google Scholar] [CrossRef]
- Ghorbani, A.; Izadpanah, K.; Peters, J.R.; Dietzgen, R.G.; Mitter, N. Detection and profiling of circular RNAs in uninfected and maize Iranian mosaic virus-infected maize. Plant. Sci. 2018, 274, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, H.; Fan, M.; He, Y.; Guo, P. Genome-wide identification of long non-coding RNAs and circular RNAs reveal their ceRNA networks in response to cucumber green mottle mosaic virus infection in watermelon. Arch. Virol. 2020, 165, 1177–1190. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Cui, L.; Zhou, Y.; Zhu, C.; Fan, D.; Gong, H.; Zhao, Q.; Zhou, C.; Zhao, Y.; Lu, D.; et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015, 21, 2076–2087. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Li, D.; Li, L.; Zhang, Q.; Wang, S.; Huang, H. Identification of circular RNAs in kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front. Plant. Sci. 2017, 8, 413. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, M.; Wei, S.; Qin, F.; Zhao, H.; Suo, B. Identification of circular RNAs and their targets in leaves of Triticum aestivum L. under dehydration stress. Front. Plant. Sci. 2017, 7. [Google Scholar] [CrossRef]
- Zuo, J.; Wang, Q.; Zhu, B.; Luo, Y.; Gao, L. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem. Biophys. Res. Commun. 2016, 479, 132–138. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
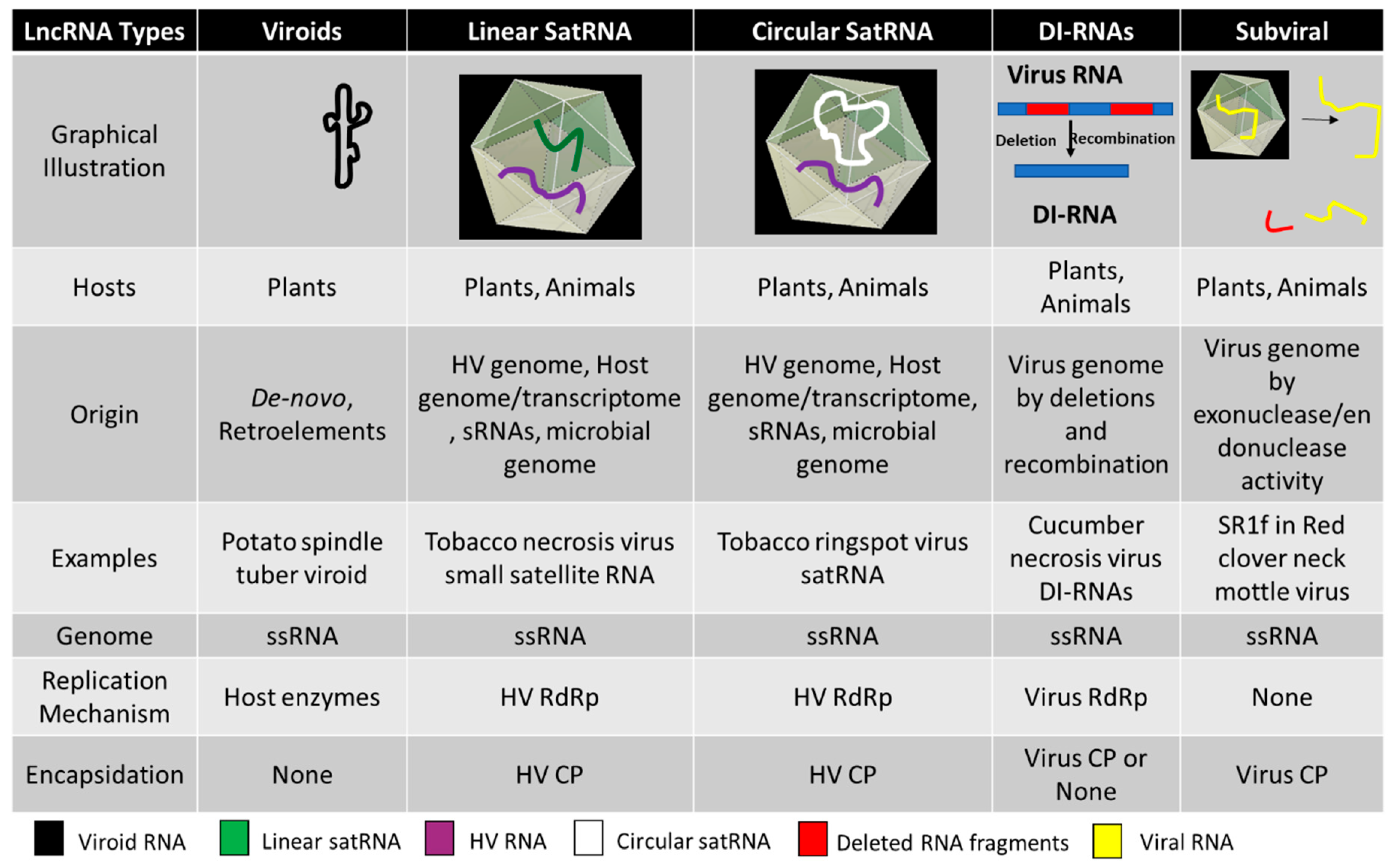

| Viroid Classification | Type Species | Genome and Pathogenesis |
|---|---|---|
| Avsunviroidae | ||
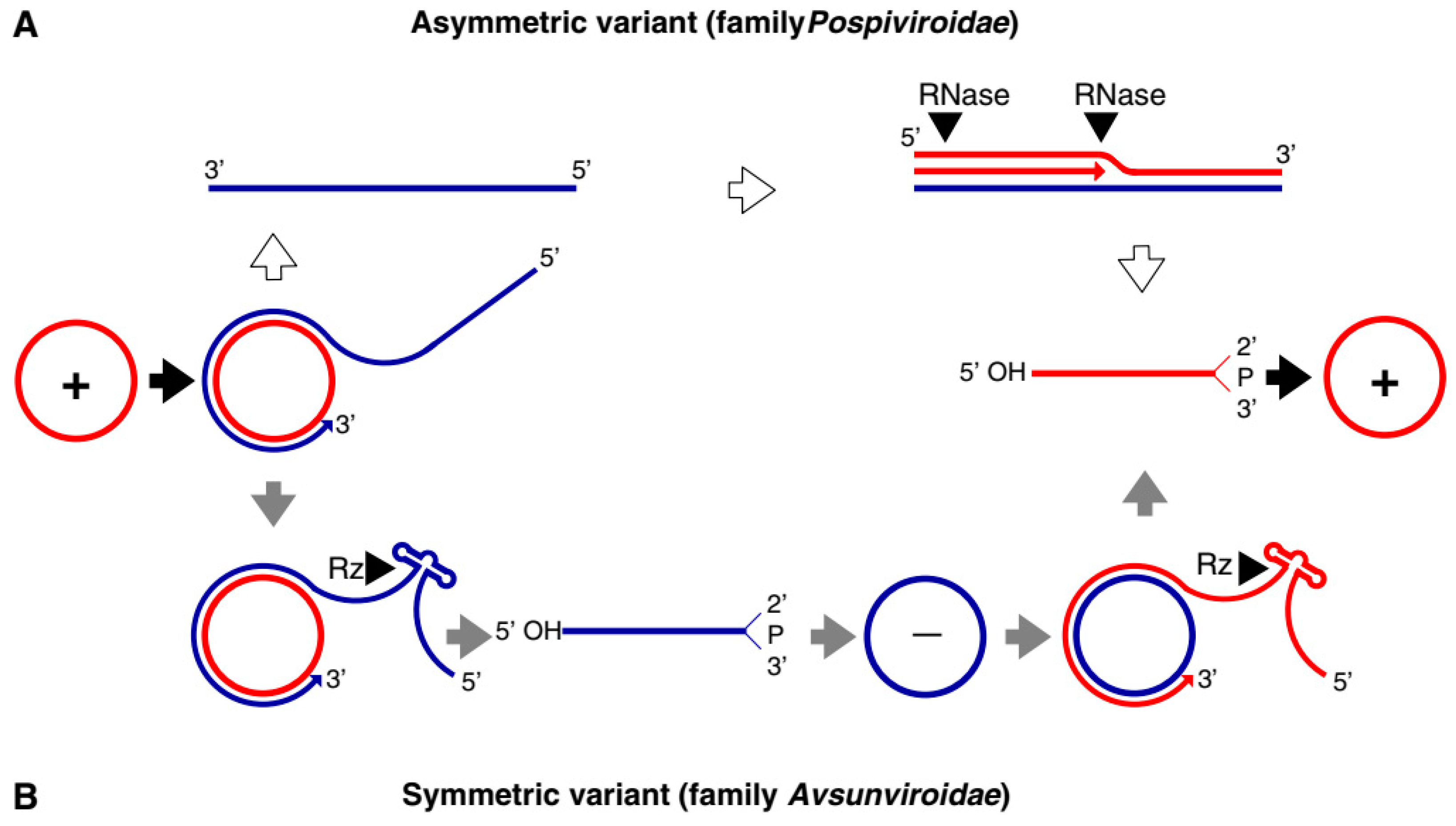
| Avsunviroid | Avocado Sun Blotch Viroid (ASBvd) | The viroids in this family have a circular genome of 247–399 nucleotides. Viroids are characterized by a specific central conserved region (CCR) in the RNA and have hammerhead ribozymes (HHR) required for symmetric rolling circle replication in the chloroplast. Most viroids are symptomatic to the host whereas some can be asymptomatic. |
| Pelamoviroid | Peach latent mosaic viroid (PLMvd) | |
| Elaviroid | Eggplant latent viroid (ELVd) | |
| Pospiviroidae | ||
| Pospiviroid | Potato spindle tuber viroid (PSTVd) | The genome size ranges from 246 to 371 nucleotides. The viroids lack central conserved region (CCR) and ribozyme activity. Replication in the nucleus by asymmetric rolling circle replication is catalyzed completely by host enzymes. It can infect a wide range of hosts including Solanaceae, Asteraceae, Compsitae, and others including various economically important fruit crops like apples, citruses and some ornamental plants. |
| Hostuviroid | Hop stunt viroid (HSV) | |
| Cocadviroid | Coconut cadang-cadang viroid (CCCVd) | |
| Apscaviroid | Apple scar skin viroid (ASSVd) | |
| Coleviroid | Coleus blumei viroid (CBVd) | |
| Family/Genus of Helper Virus | Example | Genome and Pathogenesis |
|---|---|---|
| Linear Small Satellite RNA | ||
| Tombusviridae | Tomato bushy stunt virus (TBSV) satellite RNA | The genome is linear long non-coding RNA, ranging from 339 to 901 nucleotides. Most satRNAs are pathogenic to the helper viruses whereas some, like black beet scorch virus (BBSV) satellite RNA, is known to intensify symptoms and virus accumulation. |
| Bromoviridae | Cucumber mosaic virus (CMV) satellite RNA | |
| Umbravirus | Groundnut rosette virus (GRV) satellite RNA | |
| Circular Sat RNAs | ||
| Secoviridae | Tobacco ringspot virus (TRSV) satellite RNA | Also known as virusoids, they have circular long non-coding RNAs ranging from 220 to 457 nucleotides. The RYMV satRNA is the smallest known pathogenic subviral agent. The RNA secondary structure is conserved with a hammerhead ribozyme. They replicate by rolling circle mechanism using the helper virus machinery and are encapsidated by the helper virus coat protein. Mostly pathogenic to the helper virus, leading to attenuation of the symptoms in host plants. The replication of satRNAs is known to be supported by heterologous helper viruses across different species. |
| Luteoviridae | Barley yellow dwarf virus (BYDV) | |
| Sobemovirus | Rice yellow mottle virus (RYMV) satellite | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shrestha, N.; Bujarski, J.J. Long Noncoding RNAs in Plant Viroids and Viruses: A Review. Pathogens 2020, 9, 765. https://doi.org/10.3390/pathogens9090765
Shrestha N, Bujarski JJ. Long Noncoding RNAs in Plant Viroids and Viruses: A Review. Pathogens. 2020; 9(9):765. https://doi.org/10.3390/pathogens9090765
Chicago/Turabian StyleShrestha, Nipin, and Józef J. Bujarski. 2020. "Long Noncoding RNAs in Plant Viroids and Viruses: A Review" Pathogens 9, no. 9: 765. https://doi.org/10.3390/pathogens9090765
APA StyleShrestha, N., & Bujarski, J. J. (2020). Long Noncoding RNAs in Plant Viroids and Viruses: A Review. Pathogens, 9(9), 765. https://doi.org/10.3390/pathogens9090765





