Antibiotic Treatment in Anopheles coluzzii Affects Carbon and Nitrogen Metabolism
Abstract
:1. Introduction
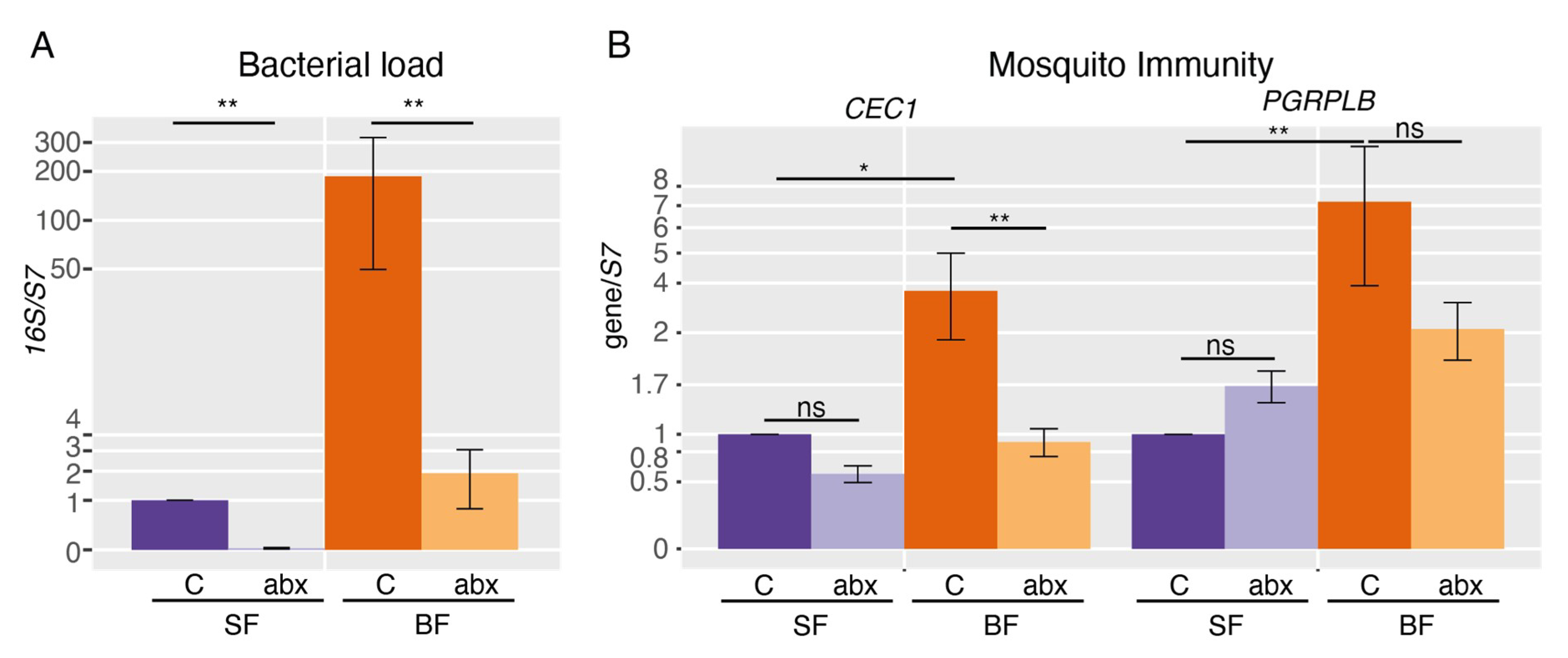
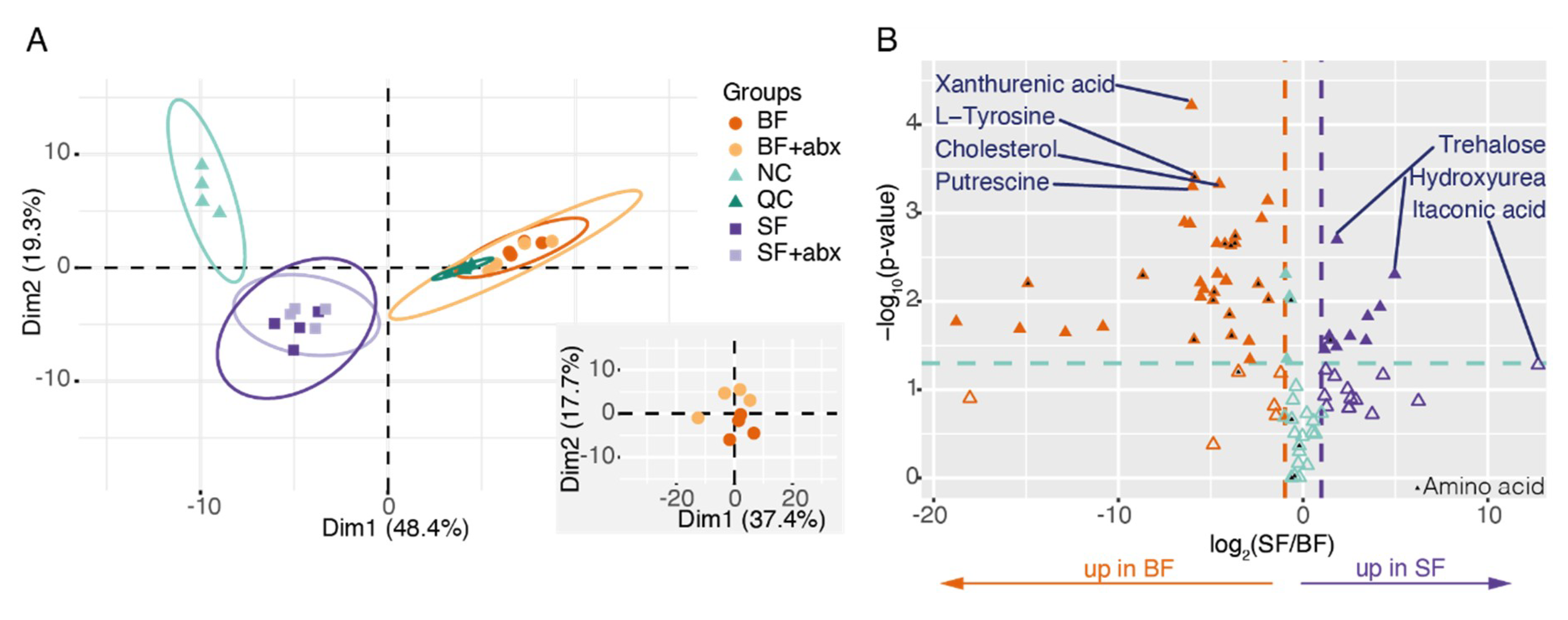
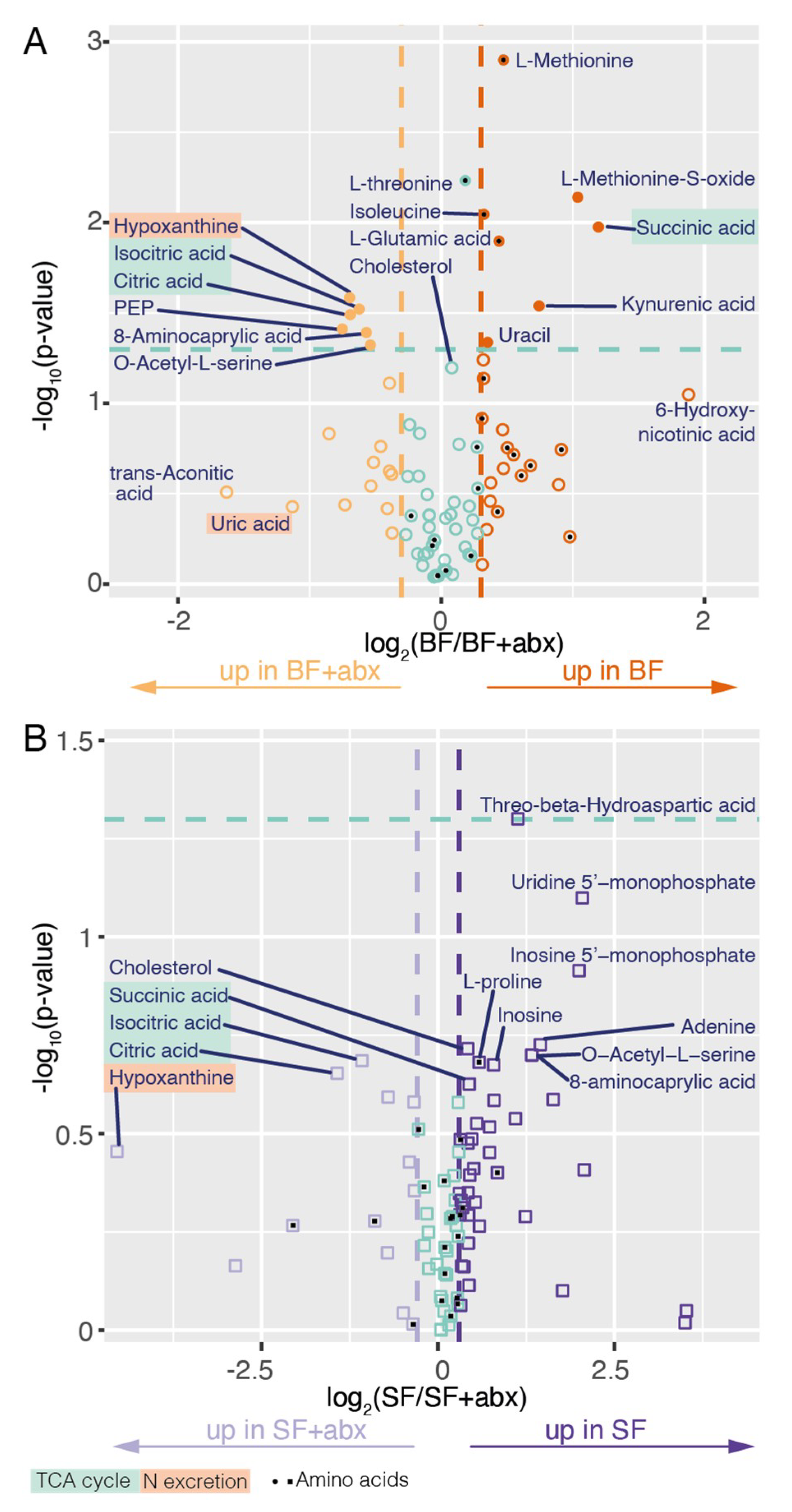
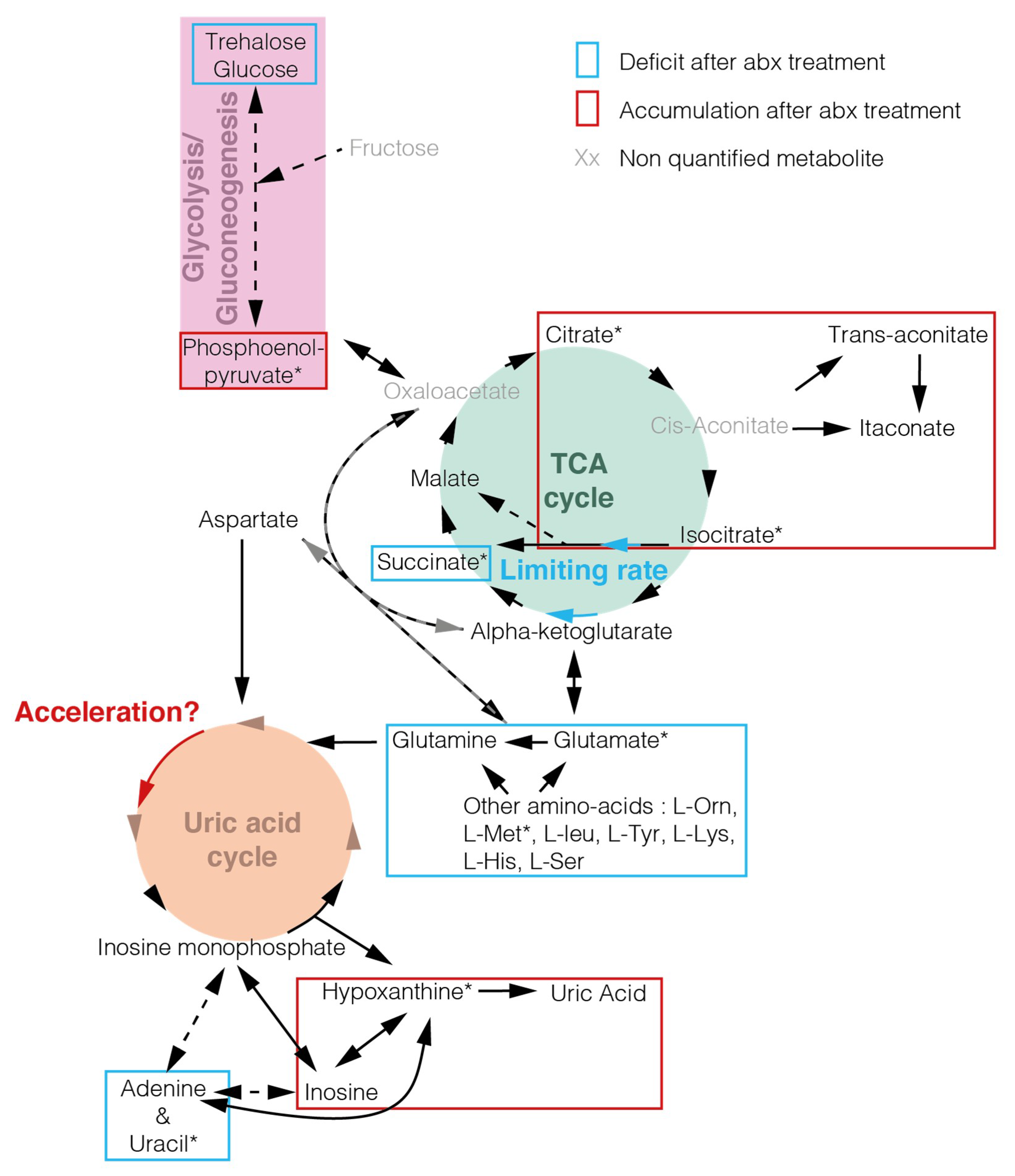
2. Results
3. Discussion
4. Materials and Methods
4.1. Mosquito Rearing
4.2. Blood Feeding
4.3. Antibiotic Treatment
4.4. Midgut Dissection
4.5. RNA Extraction and RT-qPCR
4.6. Metabolomics
4.6.1. Methanol Quenching
4.6.2. Dual Phase Extraction and Sample Preparation
4.6.3. Data Acquisition
4.7. Data Analysis
4.7.1. RT-qPCR Analysis
4.7.2. Metabolomic Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mancini, M.V.; Damiani, C.; Accoti, A.; Tallarita, M.; Nunzi, E.; Cappelli, A.; Bozic, J.; Catanzani, R.; Rossi, P.; Valzano, M.; et al. Estimating bacteria diversity in different organs of nine species of mosquito by next generation sequencing. BMC Microbiol. 2018, 18, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osei-Poku, J.; Mbogo, C.M.; Palmer, W.J.; Jiggins, F.M. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol. Ecol. 2012, 21, 5138–5150. [Google Scholar] [CrossRef] [PubMed]
- Straif, S.C.; Mbogo, C.N.M.; Toure, A.M.; Walker, E.D.; Kaufman, M.; Toure, Y.T.; Beier, J.C. Midgut Bacteria in Anopheles gambiae and An. funestus (Diptera: Culicidae) from Kenya and Mali. J. Med. Entomol. 1998, 35, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Boissière, A.; Tchioffo, M.T.; Bachar, D.; Abate, L.; Marie, A.; Nsango, S.E.; Shahbazkia, H.R.; Awono-Ambene, P.H.; Levashina, E.A.; Christen, R.; et al. Midgut Microbiota of the Malaria Mosquito Vector Anopheles gambiae and Interactions with Plasmodium falciparum Infection. PLoS Pathog. 2012, 8, e1002742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Manfredini, F.; Dimopoulos, G. Implication of the Mosquito Midgut Microbiota in the Defense against Malaria Parasites. PLoS Pathog. 2009, 5, e1000423. [Google Scholar] [CrossRef] [Green Version]
- Akorli, J.; Gendrin, M.; Pels, N.A.P.; Yeboah-Manu, D.; Christophides, G.K.; Wilson, M.D. Seasonality and Locality Affect the Diversity of Anopheles gambiae and Anopheles coluzzii Midgut Microbiota from Ghana. PLoS ONE 2016, 11, e0157529. [Google Scholar] [CrossRef] [Green Version]
- Bascuñán, P.; Niño-Garcia, J.P.; Galeano-Castañeda, Y.; Serre, D.; Correa, M.M. Factors shaping the gut bacterial community assembly in two main Colombian malaria vectors. Microbiome 2018, 6, 148. [Google Scholar] [CrossRef]
- Krajacich, B.J.; Huestis, D.L.; Dao, A.; Yaro, A.S.; Diallo, M.; Krishna, A.; Xu, J.; Lehmann, T. Investigation of the seasonal microbiome of Anopheles coluzzii mosquitoes in Mali. PLoS ONE 2018, 13, e0194899. [Google Scholar] [CrossRef] [Green Version]
- Gimonneau, G.; Tchioffo, M.T.; Abate, L.; Boissière, A.; Awono-Ambéné, P.H.; Nsango, S.E.; Christen, R.; Morlais, I. Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect. Genet. Evol. 2014, 28, 715–724. [Google Scholar] [CrossRef]
- Pumpuni, C.B.; Demaio, J.; Kent, M.; Davis, J.R.; Beier, J.C. Bacterial Population Dynamics in Three Anopheline Species: The Impact on Plasmodium Sporogonic Development. Am. J. Trop. Med. Hyg. 1996, 54, 214–218. [Google Scholar] [CrossRef]
- Wang, Y.; Gilbreath, T.M., III; Kukutla, P.; Yan, G.; Xu, J. Dynamic Gut Microbiome across Life History of the Malaria Mosquito Anopheles gambiae in Kenya. PLoS ONE 2011, 6, e24767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coon, K.L.; Vogel, K.J.; Brown, M.R.; Strand, M.R. Mosquitoes rely on their gut microbiota for development. Mol. Ecol. 2014, 23, 2727–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Dhayal, D.; Singh, O.P.; Adak, T.; Bhatnagar, R.K. Gut microbes influence fitness and malaria transmission potential of Asian malaria vector Anopheles stephensi. Acta Trop. 2013, 128, 41–47. [Google Scholar] [CrossRef]
- Gendrin, M.; Rodgers, F.H.; Yerbanga, R.S.; Ouédraogo, J.B.; Basáñez, M.-G.; Cohuet, A.; Christophides, G.K. Antibiotics in ingested human blood affect the mosquito microbiota and capacity to transmit malaria. Nat. Commun. 2015, 6, 5921. [Google Scholar] [CrossRef] [Green Version]
- Dada, N.; Sheth, M.; Liebman, K.; Pinto, J.; Lenhart, A. Whole metagenome sequencing reveals links between mosquito microbiota and insecticide resistance in malaria vectors. Sci. Rep. 2018, 8, 2084. [Google Scholar] [CrossRef] [PubMed]
- Graumans, W.; Jacobs, E.; Bousema, T.; Sinnis, P. When Is a Plasmodium-Infected Mosquito an Infectious Mosquito? Trends Parasitol. 2020, 36, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Cirimotich, C.M.; Dong, Y.; Clayton, A.M.; Sandiford, S.L.; Souza-Neto, J.A.; Mulenga, M.; Dimopoulos, G. Natural Microbe-Mediated Refractoriness to Plasmodium Infection in Anopheles gambiae. Science 2011, 332, 855. [Google Scholar] [CrossRef] [Green Version]
- Gendrin, M.; Turlure, F.; Rodgers, F.H.; Cohuet, A.; Morlais, I.; Christophides, G.K. The Peptidoglycan Recognition Proteins PGRPLA and PGRPLB Regulate Anopheles Immunity to Bacteria and Affect Infection by Plasmodium. J. Innate Immun. 2017, 9, 333–342. [Google Scholar] [CrossRef]
- Meister, S.; Agianian, B.; Turlure, F.; Relógio, A.; Morlais, I.; Kafatos, F.C.; Christophides, G.K. Anopheles gambiae PGRPLC-Mediated Defense against Bacteria Modulates Infections with Malaria Parasites. PLoS Pathog. 2009, 5, e1000542. [Google Scholar] [CrossRef] [Green Version]
- Stecher, B.; Hardt, W.-D. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 2011, 14, 82–91. [Google Scholar] [CrossRef]
- Rodgers, F.H.; Gendrin, M.; Wyer, C.A.S.; Christophides, G.K. Microbiota-induced peritrophic matrix regulates midgut homeostasis and prevents systemic infection of malaria vector mosquitoes. PLoS Pathog. 2017, 13, e1006391. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Behrends, V.; Bundy, J.G.; Crisanti, A.; Nolan, T. Phenylalanine Metabolism Regulates Reproduction and Parasite Melanization in the Malaria Mosquito. PLoS ONE 2014, 9, e84865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Champion, C.J.; Kukutla, P.; Glennon, E.K.K.; Wang, B.; Luckhart, S.; Xu, J. Anopheles gambiae: Metabolomic Profiles in Sugar-Fed, Blood-Fed, and Plasmodium falciparum-Infected Midgut. Dataset Pap. Sci. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Benzylpenicillin (DrugBank). Available online: https://www.drugbank.ca/drugs/DB01053 (accessed on 14 July 2020).
- Streptomycin (DrugBank). Available online: https://www.drugbank.ca/drugs/DB01082 (accessed on 14 July 2020).
- Gentamicin (DrugBank). Available online: https://www.drugbank.ca/drugs/DB00798 (accessed on 14 July 2020).
- Amphotericin B (DrugBank). Available online: https://www.drugbank.ca/drugs/DB00681 (accessed on 14 July 2020).
- Nephrotoxic agents (DrugBank). Available online: https://www.drugbank.ca/categories/DBCAT003959 (accessed on 14 July 2020).
- Montali, R.J.; Bush, M.; Smeller, J.M. The Pathology of Nephrotoxicity of Gentamicin in Snakes. Vet. Pathol 1979, 16, 108–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakumar, P.; WitnessKoe, W.E.; Gan, Y.S.; JemayPuah, S.M.; Kuganesswari, S.; Prajapati, S.K.; Varatharajan, R.; Jayachristy, S.A.; Sundram, K.; Bahari, M.B. Effects of pre and post-treatments with dipyridamole in gentamicin-induced acute nephrotoxicity in the rat. Regul. Toxicol. Pharmacol. 2017, 84, 35–44. [Google Scholar] [CrossRef]
- Liberopoulos, E.N.; Kei, A.A.; Elisaf, M.S. Lysis syndrome during therapy of visceral leishmaniasis. Infection 2012, 40, 121–123. [Google Scholar] [CrossRef]
- Rodgers, F.H.; Gendrin, M.; Wyer, C.A.S.; Christophides, G.K. RNA-Seq Experiment Antibiotic Treatment of Midgut Microbiota during Blood Feeding (VectorBase). 2017. Available online: https://www.vectorbase.org/expression-browser/experiment/SRP106793 (accessed on 14 July 2020).
- Kondrashov, F.A.; Koonin, E.V.; Morgunov, I.G.; Finogenova, T.V.; Kondrashova, M.N. Evolution of glyoxylate cycle enzymes in Metazoa: Evidence of multiple horizontal transfer events and pseudogene formation. Biol. Direct 2006, 1, 31. [Google Scholar] [CrossRef] [Green Version]
- Lavazec, C.; Boudin, C.; Lacroix, R.; Bonnet, S.; Diop, A.; Thiberge, S.; Boisson, B.; Tahar, R.; Bourgouin, C. Carboxypeptidases B of Anopheles gambiae as Targets for a Plasmodium falciparum Transmission-Blocking Vaccine. Infect. Immun. 2007, 75, 1635. [Google Scholar] [CrossRef] [Green Version]
- de O Gaio, A.; Gusmão, D.S.; Santos, A.V.; Berbert-Molina, M.A.; Pimenta, P.F.; Lemos, F.J. Contribution of midgut bacteria to blood digestion and egg production in Aedes aegypti (diptera: Culicidae) (L.). Parasites Vectors 2011, 4, 105. [Google Scholar] [CrossRef] [Green Version]
- Hughes, G.L.; Dodson, B.L.; Johnson, R.M.; Murdock, C.C.; Tsujimoto, H.; Suzuki, Y.; Patt, A.A.; Cui, L.; Nossa, C.W.; Barry, R.M.; et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. USA 2014, 111, 12498. [Google Scholar] [CrossRef] [Green Version]
- Consuegra, J.; Grenier, T.; Baa-Puyoulet, P.; Rahioui, I.; Akherraz, H.; Gervais, H.; Parisot, N.; da Silva, P.; Charles, H.; Calevro, F.; et al. Drosophila-associated bacteria differentially shape the nutritional requirements of their host during juvenile growth. PLoS Biol. 2020, 18, e3000681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirimotich, C.M.; Ramirez, J.L.; Dimopoulos, G. Native Microbiota Shape Insect Vector Competence for Human Pathogens. Cell Host Microbe 2011, 10, 307–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beier, M.S.; Pumpuni, C.B.; Beier, J.C.; Davis, J.R. Effects of Para-Aminobenzoic Acid, Insulin, and Gentamicin on Plasmodium falciparum Development in Anopheline Mosquitoes (Diptera: Culicidae). J. Med. Entomol. 1994, 31, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Mancio-Silva, L.; Slavic, K.; Grilo Ruivo, M.T.; Grosso, A.R.; Modrzynska, K.K.; Vera, I.M.; Sales-Dias, J.; Gomes, A.R.; MacPherson, C.R.; Crozet, P.; et al. Nutrient sensing modulates malaria parasite virulence. Nature 2017, 547, 213–216. [Google Scholar] [CrossRef]
- Wang, M.; An, Y.; Dong, S.; Feng, Y.; Gao, L.; Wang, P.; Dimopoulus, G.; Tang, H.; Wang, J. Glucose-mediated expansion of a gut commensal bacterium promotes Plasmodium infection through alkalizing mosquito midgut. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Hayward, R.E. Plasmodium falciparum phosphoenolpyruvate carboxykinase is developmentally regulated in gametocytes. Mol. Biochem. Parasitol. 2000, 107, 227–240. [Google Scholar] [CrossRef]
- Downie, M.J.; Kirk, K.; Mamoun, C.B. Purine Salvage Pathways in the Intraerythrocytic Malaria Parasite Plasmodium falciparum. Eukaryot Cell 2008, 7, 1231. [Google Scholar] [CrossRef] [Green Version]
- El Bissati, K.; Zufferey, R.; Witola, W.H.; Carter, N.S.; Ullman, B.; Ben Mamoun, C. The plasma membrane permease PfNT1 is essential for purine salvage in the human malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 2006, 103, 9286. [Google Scholar] [CrossRef] [Green Version]
- Sinden, R.; Talman, A.; Marques, S.; Wass, M.; Sternberg, M. The flagellum in malarial parasites. Curr. Opin. Microbiol. 2010, 13, 491–500. [Google Scholar] [CrossRef]
- Lee, K.-A.; Kim, S.-H.; Kim, E.-K.; Ha, E.-M.; You, H.; Kim, B.; Kim, M.-J.; Kwon, Y.; Ryu, J.-H.; Lee, W.-J. Bacterial-Derived Uracil as a Modulator of Mucosal Immunity and Gut-Microbe Homeostasis in Drosophila. Cell 2013, 153, 797–811. [Google Scholar] [CrossRef] [Green Version]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass Spectral and Retention Index Libraries for Metabolomics Based on Quadrupole and Time-of-Flight Gas Chromatography/Mass Spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behrends, V.; Tredwell, G.D.; Bundy, J.G. A software complement to AMDIS for processing GC-MS metabolomic data. Anal. Biochem. 2011, 415, 206–208. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chabanol, E.; Behrends, V.; Prévot, G.; Christophides, G.K.; Gendrin, M. Antibiotic Treatment in Anopheles coluzzii Affects Carbon and Nitrogen Metabolism. Pathogens 2020, 9, 679. https://doi.org/10.3390/pathogens9090679
Chabanol E, Behrends V, Prévot G, Christophides GK, Gendrin M. Antibiotic Treatment in Anopheles coluzzii Affects Carbon and Nitrogen Metabolism. Pathogens. 2020; 9(9):679. https://doi.org/10.3390/pathogens9090679
Chicago/Turabian StyleChabanol, Estelle, Volker Behrends, Ghislaine Prévot, George K. Christophides, and Mathilde Gendrin. 2020. "Antibiotic Treatment in Anopheles coluzzii Affects Carbon and Nitrogen Metabolism" Pathogens 9, no. 9: 679. https://doi.org/10.3390/pathogens9090679
APA StyleChabanol, E., Behrends, V., Prévot, G., Christophides, G. K., & Gendrin, M. (2020). Antibiotic Treatment in Anopheles coluzzii Affects Carbon and Nitrogen Metabolism. Pathogens, 9(9), 679. https://doi.org/10.3390/pathogens9090679





