Evaluation of Nucleoside Analogs as Antimicrobials Targeting Unique Enzymes in Borrelia burgdorferi
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. B. burgdorferi Culture
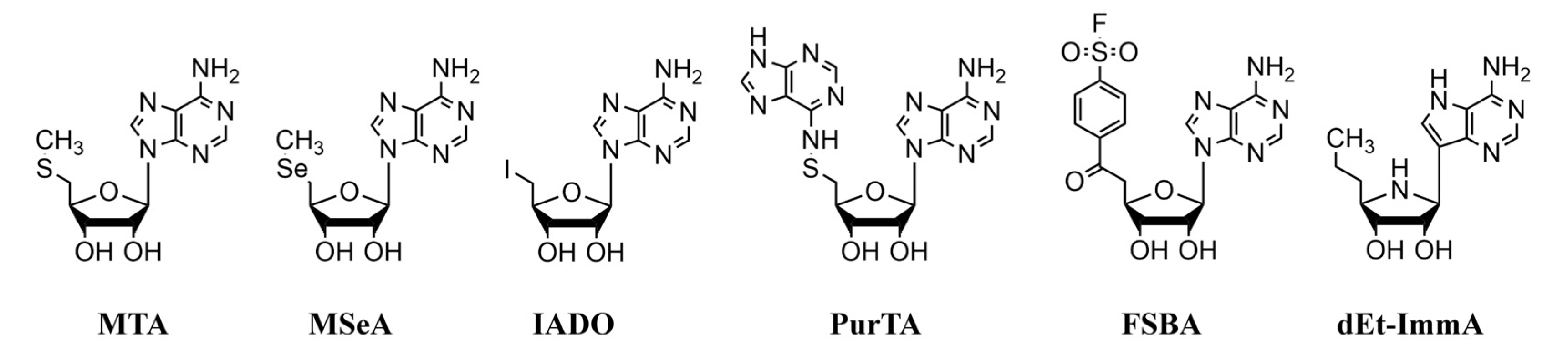
2.3. Substrates and Inhibitors
2.4. Expression and Purification of Recombinant MTANs
2.5. Enzyme Activity and Kinetics
2.6. Luciferase Assay for B. burgdorferi Survival
2.7. Assessment of Nucleosides/Nucleoside Analogs as Antimicrobials
2.8. Assessment of Bacteriostatic Versus Bactericidal Activities of Nucleosides and Nucleoside Analogs
3. Results
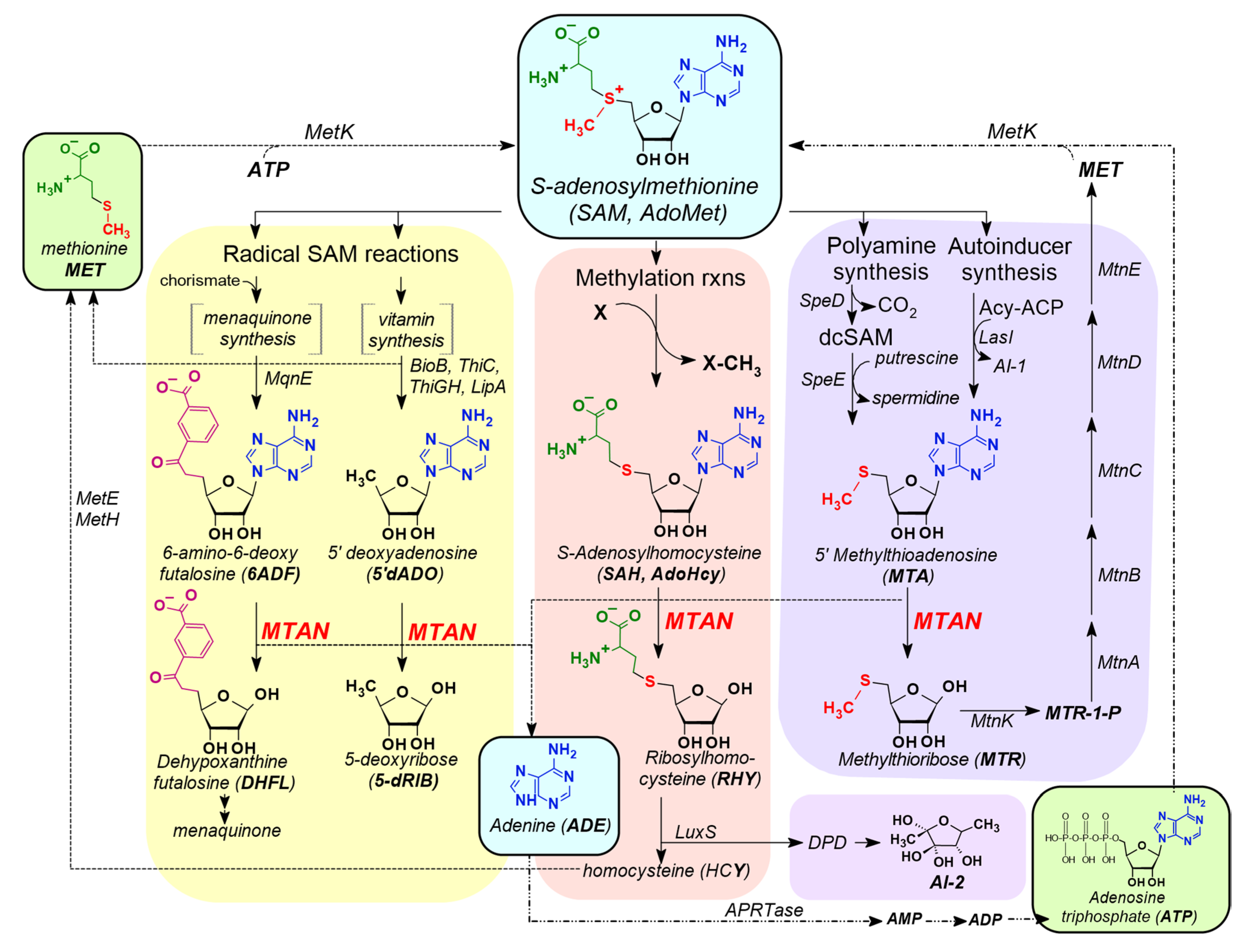
3.1. MTAN is a Critical Enzyme in the Salvage Pathways of Auxotrophic Bacteria
3.2. Kinetic Analysis of Purified Recombinant MTANs with Nucleosides and Nucleoside Analogs
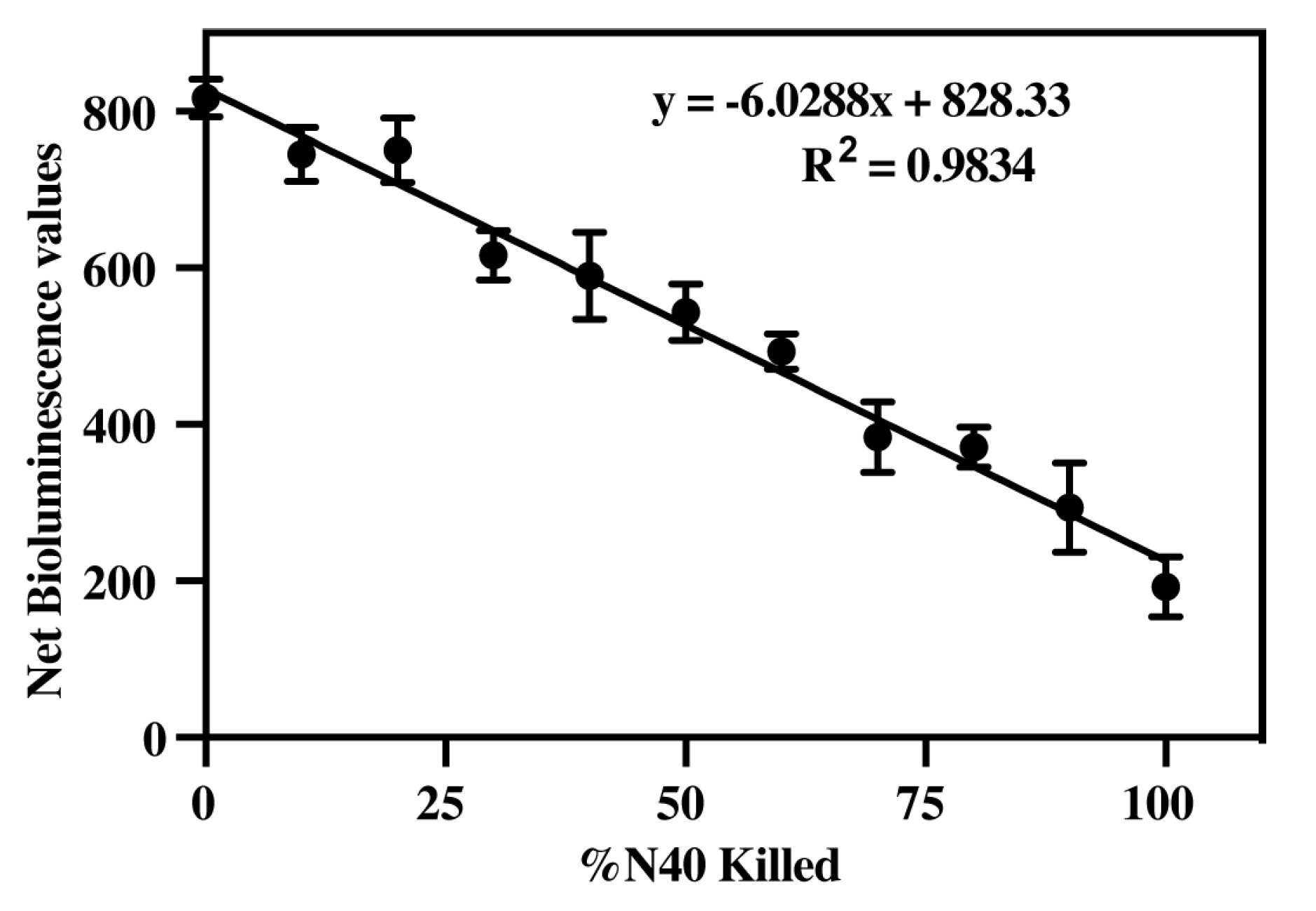
3.3. Determine Efficacy of Luciferase Activity to Measure B. burgdorferi Viability
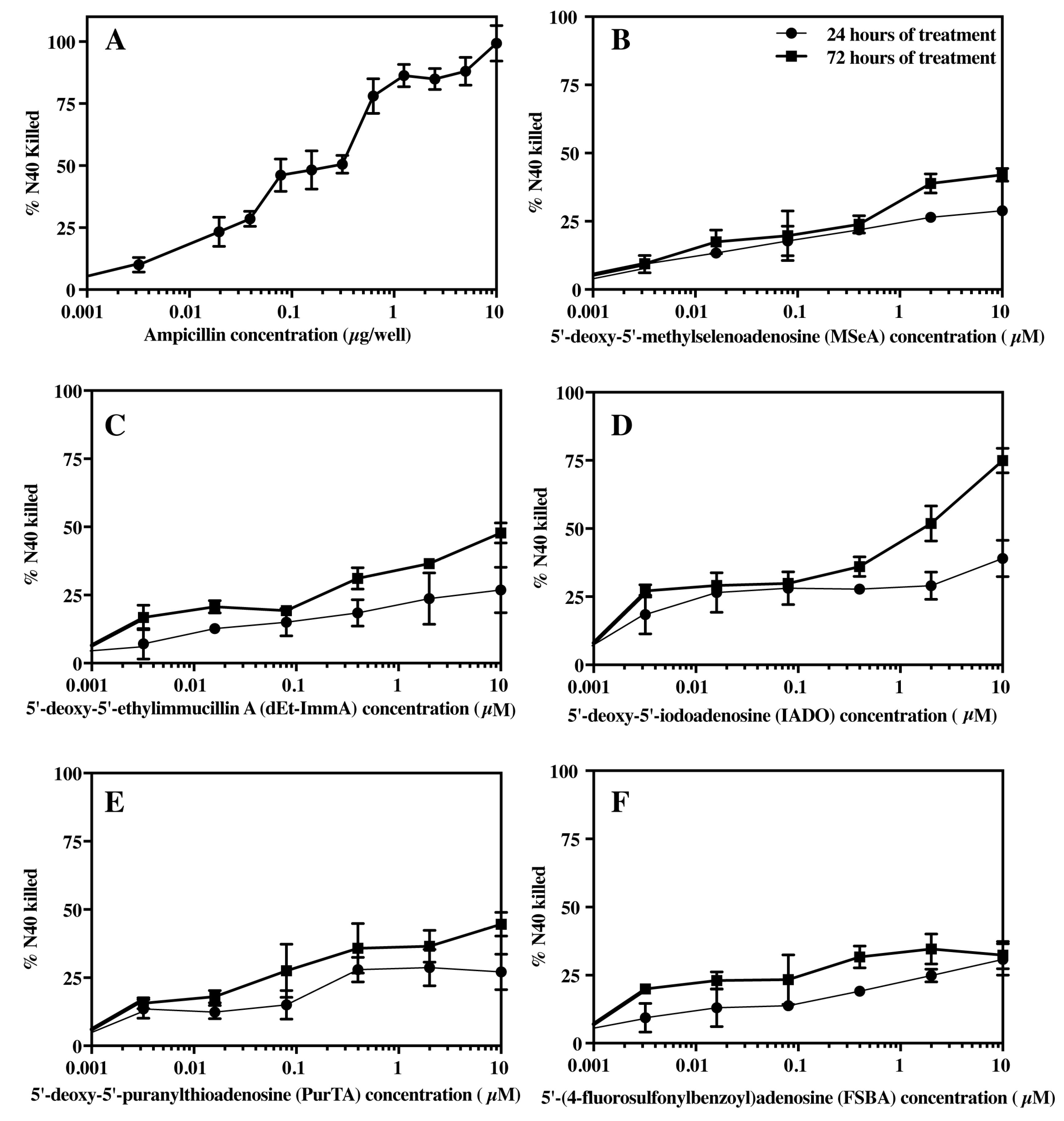
3.4. Effect of Ampicillin and Selected Inhibitors of MTANs on the Growth of B. burgdorferi
3.5. Bacteriostatic or Bactericidal Activities of MTAN Inhibitors Against B. burgdorferi N40
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moore, A.; Nelson, C.; Molins, C.; Mead, P.; Schriefer, M. Current Guidelines, Common Clinical Pitfalls, and Future Directions for Laboratory Diagnosis of Lyme Disease, United States. Emerg. Infect. Dis. 2016, 22, 1169–1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strle, F.; Ruzic-Sabljic, E.; Cimperman, J.; Lotric-Furlan, S.; Maraspin, V. Comparison of findings for patients with Borrelia garinii and Borrelia afzelii isolated from cerebrospinal fluid. Clin. Infect. Dis. 2006, 43, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Steere, A.C. Lyme disease. N. Engl. J. Med. 2001, 345, 115–125. [Google Scholar] [CrossRef]
- Wormser, G.P.; Dattwyler, R.J.; Shapiro, E.D.; Halperin, J.J.; Steere, A.C.; Klempner, M.S.; Krause, P.J.; Bakken, J.S.; Strle, F.; Stanek, G.; et al. The clinical assessment, treatment, and prevention of Lyme disease, human granulocytic anaplasmosis, and babesiosis: Clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 2006, 43, 1089–1134. [Google Scholar] [CrossRef]
- Hu, L.T. Patient education: Lyme disease symptoms and diagnosis (Beyond the Basics). 2020. UpTodate. Available online: https://www.uptodate.com/contents/lyme-disease-symptoms-and-diagnosis-beyond-the-basics (accessed on 19 August 2020).
- Centers for Disease Control and Prevention. Lyme disease treatment. Available online: https://www.cdc.gov/lyme/treatment/index.html (accessed on 17 December 2019).
- Dattwyler, R.J.; Luft, B.J.; Kunkel, M.J.; Finkel, M.F.; Wormser, G.P.; Rush, T.J.; Grunwaldt, E.; Agger, W.A.; Franklin, M.; Oswald, D.; et al. Ceftriaxone compared with doxycycline for the treatment of acute disseminated Lyme disease. N. Engl. J. Med. 1997, 337, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Dattwyler, R.J.; Wormser, G.P.; Rush, T.J.; Finkel, M.F.; Schoen, R.T.; Grunwaldt, E.; Franklin, M.; Hilton, E.; Bryant, G.L.; Agger, W.A.; et al. A comparison of two treatment regimens of ceftriaxone in late Lyme disease. Wien. Klin. Wochenschr. 2005, 117, 393–397. [Google Scholar] [CrossRef]
- Hu, L.T. Treatment of Lyme disease. 2020. UpTodate. Available online: https://www.uptodate.com/contents/treatment-of-lyme-disease (accessed on 15 July 2020).
- Logigian, E.L.; Kaplan, R.F.; Steere, A.C. Successful treatment of Lyme encephalopathy with intravenous ceftriaxone. J. Infect. Dis. 1999, 180, 377–383. [Google Scholar] [CrossRef] [Green Version]
- Parveen, N.; Cornell, K.A. Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol. Microbiol. 2011, 79, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Cornell, K.A.; Swarts, W.E.; Barry, R.D.; Riscoe, M.K. Characterization of recombinant Escherichia coli 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase: Analysis of enzymatic activity and substrate specificity. Biochem. Biophys. Res. Commun. 1996, 228, 724–732. [Google Scholar] [CrossRef]
- Choi-Rhee, E.; Cronan, J.E. A nucleosidase required for in vivo function of the S-adenosyl-L-methionine radical enzyme, biotin synthase. Chem. Biol. 2005, 12, 589–593. [Google Scholar] [CrossRef] [Green Version]
- Challand, M.R.; Martins, F.T.; Roach, P.L. Catalytic activity of the anaerobic tyrosine lyase required for thiamine biosynthesis in Escherichia coli. J. Biol. Chem. 2010, 285, 5240–5248. [Google Scholar] [CrossRef] [Green Version]
- Challand, M.R.; Ziegert, T.; Douglas, P.; Wood, R.J.; Kriek, M.; Shaw, N.M.; Roach, P.L. Product inhibition in the radical S-adenosylmethionine family. FEBS Lett. 2009, 583, 1358–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Apel, D.; Gaynor, E.C.; Tanner, M.E. 5’-methylthioadenosine nucleosidase is implicated in playing a key role in a modified futalosine pathway for menaquinone biosynthesis in Campylobacter jejuni. J. Biol. Chem. 2011, 286, 19392–19398. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, Y.; Ogasawara, Y.; Matsumoto, A.; Dairi, T. Aplasmomycin and boromycin are specific inhibitors of the futalosine pathway. J. Antibiot. (Tokyo) 2018, 71, 968–970. [Google Scholar] [CrossRef] [PubMed]
- Cornell, K.A.; Knippel, R.J.; Cortright, G.R.; Fonken, M.; Guerrero, C.; Hall, A.R.; Mitchell, K.A.; Thurston, J.H.; Erstad, P.; Tao, A.; et al. Characterization of 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidases from Borrelia burgdorferi: Antibiotic targets for Lyme disease. Biochim. Biophys. Acta. Gen. Subj. 2020, 1864, 129455. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, J.A.; Crowder, T.; Rinaldo-Matthis, A.; Ho, M.C.; Almo, S.C.; Schramm, V.L. Transition state analogs of 5’-methylthioadenosine nucleosidase disrupt quorum sensing. Nat. Chem. Biol. 2009, 5, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Parveen, N.; Leong, J.M. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000, 35, 1220–1234. [Google Scholar] [CrossRef]
- Cornell, K.A.; Primus, S.; Martinez, J.A.; Parveen, N. Assessment of methylthioadenosine/S-adenosylhomocysteine nucleosidases of Borrelia burgdorferi as targets for novel antimicrobials using a novel high-throughput method. J. Antimicrob. Chemother. 2009, 63, 1163–1172. [Google Scholar] [CrossRef] [Green Version]
- Rogers, E.A.; Terekhova, D.; Zhang, H.M.; Hovis, K.M.; Schwartz, I.; Marconi, R.T. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol. Microbiol. 2009, 71, 1551–1573. [Google Scholar] [CrossRef] [Green Version]
- Mongodin, E.F.; Casjens, S.R.; Bruno, J.F.; Xu, Y.; Drabek, E.F.; Riley, D.R.; Cantarel, B.L.; Pagan, P.E.; Hernandez, Y.A.; Vargas, L.C.; et al. Inter- and intra-specific pan-genomes of Borrelia burgdorferi sensu lato: Genome stability and adaptive radiation. BMC Genom. 2013, 14, 693. [Google Scholar] [CrossRef] [Green Version]
- Parveen, N.; Leong, J.M. Genetic Analysis of Attachment of Borrelia Burgdorferi to Host Cells and Extracellular Matrix; NATO Science Series; IOS Press: Prague, Czech Republic, 2006; Volume 373, p. 400. [Google Scholar]
- Schlachter, S.; Seshu, J.; Lin, T.; Norris, S.; Parveen, N. The Borrelia burgdorferi Glycosaminoglycan Binding Protein Bgp in the B31 Strain Is Not Essential for Infectivity despite Facilitating Adherence and Tissue Colonization. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, K.B.; von Meyenburg, K. Charges of nicotinamide adenine nucleotides and adenylate energy charge as regulatory parameters of the metabolism in Escherichia coli. J. Biol. Chem. 1977, 252, 4151–4156. [Google Scholar] [PubMed]
- Chapman, A.G.; Atkinson, D.E. Adenine nucleotide concentrations and turnover rates. Their correlation with biological activity in bacteria and yeast. Adv. Microb. Physiol. 1977, 15, 253–306. [Google Scholar] [CrossRef] [PubMed]
- Cekic, C.; Linden, J. Purinergic regulation of the immune system. Nat. Rev. Immunol. 2016, 16, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Collins, J.J. Microbial environments confound antibiotic efficacy. Nat. Chem. Biol. 2011, 8, 6–9. [Google Scholar] [CrossRef]
- Cantoni, G.L. The Centrality of S-Adenosylhomocysteinase in the Regulation of the Biological Utilization of S-Adenosylmethionine.; Humana Press: Clifton, NJ, USA, 1986; pp. 227–238. [Google Scholar]
- Beeston, A.L.; Surette, M.G. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2002, 184, 3450–3456. [Google Scholar] [CrossRef] [Green Version]
- Halliday, N.M.; Hardie, K.R.; Williams, P.; Winzer, K.; Barrett, D.A. Quantitative liquid chromatography-tandem mass spectrometry profiling of activated methyl cycle metabolites involved in LuxS-dependent quorum sensing in Escherichia coli. Anal. Biochem. 2010, 403, 20–29. [Google Scholar] [CrossRef]
- Apsel, B.; Blair, J.A.; Gonzalez, B.; Nazif, T.M.; Feldman, M.E.; Aizenstein, B.; Hoffman, R.; Williams, R.L.; Shokat, K.M.; Knight, Z.A. Targeted polypharmacology: Discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat. Chem. Biol. 2008, 4, 691–699. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; Li, Y.; Jiang, Q.; Zhao, L.; Xue, T.; Hu, B.; Sun, B. Methylthioadenosine/S-adenosylhomocysteine nucleosidase (Pfs) of Staphylococcus aureus is essential for the virulence independent of LuxS/AI-2 system. Int. J. Med. Microbiol. 2013, 303, 190–200. [Google Scholar] [CrossRef]
- Fan, F.; Wood, K.V. Bioluminescent assays for high-throughput screening. Assay Drug Dev. Technol. 2007, 5, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Barman, T.K.; Rao, M.; Bhati, A.; Kishore, K.; Shukla, G.; Kumar, M.; Mathur, T.; Pandya, M.; Upadhyay, D.J. Non invasive real-time monitoring of bacterial infection & therapeutic effect of anti-microbials in five mouse models. Indian J. Med. Res. 2011, 134, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Bishai, W.R.; Grosset, J.H.; Nuermberger, E.L. Rapid assessment of antibacterial activity against Mycobacterium ulcerans by using recombinant luminescent strains. Antimicrob. Agents. Chemother. 2010, 54, 2806–2813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuts, F.; Carow, B.; Wigzell, H.; Rottenberg, M.E. Use of non-invasive bioluminescent imaging to assess mycobacterial dissemination in mice, treatment with bactericidal drugs and protective immunity. Microbes Infect. 2009, 11, 1114–1121. [Google Scholar] [CrossRef] [PubMed]
- Manning, A.J.; Ovechkina, Y.; McGillivray, A.; Flint, L.; Roberts, D.M.; Parish, T. A high content microscopy assay to determine drug activity against intracellular Mycobacterium tuberculosis. Methods 2017, 127, 3–11. [Google Scholar] [CrossRef]
- Siciliano, G.; Santha Kumar, T.R.; Bona, R.; Camarda, G.; Calabretta, M.M.; Cevenini, L.; Davioud-Charvet, E.; Becker, K.; Cara, A.; Fidock, D.A.; et al. A high susceptibility to redox imbalance of the transmissible stages of Plasmodium falciparum revealed with a luciferase-based mature gametocyte assay. Mol. Microbiol. 2017, 104, 306–318. [Google Scholar] [CrossRef] [Green Version]
- Brissette, C.A.; Cooley, A.E.; Burns, L.H.; Riley, S.P.; Verma, A.; Woodman, M.E.; Bykowski, T.; Stevenson, B. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. Int. J. Med. Microbiol. 2008, 298, 257–267. [Google Scholar] [CrossRef] [Green Version]
- Deheyn, D.D.; Latz, M.I. Bioluminescence characteristics of a tropical terrestrial fungus (Basidiomycetes). Luminescence 2007, 22, 462–467. [Google Scholar] [CrossRef]
- Chan, K.; Alter, L.; Barthold, S.W.; Parveen, N. Disruption of bbe02 by insertion of a luciferase gene increases transformation efficiency of Borrelia burgdorferi and allows live imaging in Lyme disease susceptible C3H mice. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.; Awan, M.; Barthold, S.W.; Parveen, N. Comparative molecular analyses of Borrelia burgdorferi sensu stricto strains B31 and N40D10/E9 and determination of their pathogenicity. BMC Microbiol. 2012, 12, 157. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.; Casjens, S.; Parveen, N. Detection of established virulence genes and plasmids to differentiate Borrelia burgdorferi strains. Infect. immun. 2012, 80, 1519–1529. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.; Lee, J.E.; Nunez, S.; Howell, P.L.; Schramm, V.L. Transition state structure of 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli and its similarity to transition state analogues. Biochemistry 2005, 44, 11647–11659. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Wang, S.; Meredith, H.R.; Zhuang, B.; Dai, Z.; You, L. Robust, linear correlations between growth rates and beta-lactam-mediated lysis rates. Proc. Natl. Acad. Sci. USA 2018, 115, 4069–4074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.E.; Cornell, K.A.; Riscoe, M.K.; Howell, P.L. Structure of E. coli 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases. Structure 2001, 9, 941–953. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.E.; Cornell, K.A.; Riscoe, M.K.; Howell, P.L. Expression, purification, crystallization and preliminary X-ray analysis of Escherichia coli 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase. Acta. Crystallogr. D. Biol. Crystallogr. 2001, 57, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Settembre, E.C.; Cornell, K.A.; Riscoe, M.K.; Sufrin, J.R.; Ealick, S.E.; Howell, P.L. Structural comparison of MTA phosphorylase and MTA/AdoHcy nucleosidase explains substrate preferences and identifies regions exploitable for inhibitor design. Biochemistry 2004, 43, 5159–5169. [Google Scholar] [CrossRef]
- Cacciapuoti, G.; Gorassini, S.; Mazzeo, M.F.; Siciliano, R.A.; Carbone, V.; Zappia, V.; Porcelli, M. Biochemical and structural characterization of mammalian-like purine nucleoside phosphorylase from the Archaeon Pyrococcus furiosus. FEBS J. 2007, 274, 2482–2495. [Google Scholar] [CrossRef]
- Cacciapuoti, G.; Fusco, S.; Caiazzo, N.; Zappia, V.; Porcelli, M. Heterologous expression of 5’-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus: Characterization of the recombinant protein and involvement of disulfide bonds in thermophilicity and thermostability. Protein Expr. Purif. 1999, 16, 125–135. [Google Scholar] [CrossRef]
- Cacciapuoti, G.; Bertoldo, C.; Brio, A.; Zappia, V.; Porcelli, M. Purification and characterization of 5’-methylthioadenosine phosphorylase from the hyperthermophilic archaeon Pyrococcus furiosus: Substrate specificity and primary structure analysis. Extremophiles 2003, 7, 159–168. [Google Scholar] [CrossRef]
- Parveen, N.; Cornell, K.A.; Bono, J.L.; Chamberland, C.; Rosa, P.; Leong, J.M. Bgp, a secreted GAG-binding protein of B. burgdorferi strain N40, displays nucleosidase activity and is not essential for infection of immunodeficient mice. Infect. Immun. 2006, 74, 3016–3020. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.H.; Wright, S.N.; Hamblin, M.; McCloskey, D.; Alcantar, M.A.; Schrubbers, L.; Lopatkin, A.J.; Satish, S.; Nili, A.; Palsson, B.O.; et al. A White-Box Machine Learning Approach for Revealing Antibiotic Mechanisms of Action. Cell 2019, 177, 1649–1661.e9. [Google Scholar] [CrossRef]
- Manvar, D.; Singh, K.; Pandey, V.N. Affinity labeling of hepatitis C virus replicase with a nucleotide analogue: Identification of binding site. Biochemistry 2013, 52, 432–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savarese, T.M.; Chu, S.H.; Chu, M.Y.; Parks, R.E., Jr. 5’-Deoxy-5’-methylthioadenosine phosphorylase--III. Role of the enzyme in the metabolism and action of 5’-halogenated adenosine analogs. Biochem. Pharmacol. 1985, 34, 361–367. [Google Scholar] [CrossRef]
- Lee, J.E.; Cornell, K.A.; Riscoe, M.K.; Howell, P.L. Structure of Escherichia coli 5’-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis. J. Biol. Chem. 2003, 278, 8761–8770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Specific Activity (U/mg) * | % Max Specific Activity ** | |||||
|---|---|---|---|---|---|---|
| Enzyme | Enzyme | |||||
| Nucleoside | Bgp | MtnN | Pfs | Bgp | MtnN | Pfs |
| MTA | 1.00 | 1.00 | 1.70 | 100 | 100 | 100 |
| MSeA | 1.79 | 1.50 | 0.27 | 179 | 150 | 16 |
| IADO | 0.04 | 0.03 | 0.11 | 4 | 3 | 6 |
| PurTA | 0.14 | 0.04 | 0.04 | 14 | 4 | 2 |
| FSBA | 0.12 | 0.02 | 0.02 | 12 | 2 | 1 |
| Enzyme | |||
|---|---|---|---|
| Nucleoside | Bgp | MtnN | Pfs |
| MTA * | Km = 0.49 ± 0.10 µM | Km = 9.09 ± 1.62 µM | Km = 0.61 ± 0.16 µM |
| MSeA | Km = 1.62 ± 0.10 µM | Km = 1.50 ± 0.14 µM | Km = 2.12 ± 0.24 µM |
| IADO | Ki = 0.63 ± 0.12 µM | Ki = 2.47 ± 0.46 µM | Ki = 0.90 ± 0.07 µM |
| PurTA | Ki = 1.31 ± 0.18 µM | Ki = 2.05 ± 0.18 µM | Ki = 0.69 ± 0.09 µM |
| FSBA | Ki = 0.34 ± 0.03 µM | Ki = 2.04 ± 0.13 µM | Ki = 0.39 ± 0.07 µM |
| dEt-ImmA * | Ki = 2.48 ± 0.38 nM, Ki’ = 0.35 ± 0.02 nM | Ki = 1.48 ± 0.17 nM Ki’ = 0.58 ± 0.05 nM | Ki = 10.49 ± 2.01 nM Ki’ = 0.53 ± 0.15 nM |
| B. burgdorferi/mL | Equation | R2 |
|---|---|---|
| 1.0 × 108 | −0.5296x + 39.133 | 0.9188 |
| 1.5 × 108 | −0.7453x + 27.088 | 0.9237 |
| 2.0 × 108 | −0.7379x + 101.38 | 0.9834 |
| Nucleoside Inhibitor | Inhibitor Activity | Dose |
|---|---|---|
| MSeA | Bacteriostatic | All doses |
| IADO | Bactericidal | 2 µM, 10 µM |
| PurTA | Bacteriostatic | All doses |
| FSBA | Bacteriostatic | All doses |
| dEt-ImmA | Bactericidal | 2 µM, 10 µM |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborti, M.; Schlachter, S.; Primus, S.; Wagner, J.; Sweet, B.; Carr, Z.; Cornell, K.A.; Parveen, N. Evaluation of Nucleoside Analogs as Antimicrobials Targeting Unique Enzymes in Borrelia burgdorferi. Pathogens 2020, 9, 678. https://doi.org/10.3390/pathogens9090678
Chakraborti M, Schlachter S, Primus S, Wagner J, Sweet B, Carr Z, Cornell KA, Parveen N. Evaluation of Nucleoside Analogs as Antimicrobials Targeting Unique Enzymes in Borrelia burgdorferi. Pathogens. 2020; 9(9):678. https://doi.org/10.3390/pathogens9090678
Chicago/Turabian StyleChakraborti, Monideep, Samantha Schlachter, Shekerah Primus, Julie Wagner, Brandi Sweet, Zoey Carr, Kenneth A. Cornell, and Nikhat Parveen. 2020. "Evaluation of Nucleoside Analogs as Antimicrobials Targeting Unique Enzymes in Borrelia burgdorferi" Pathogens 9, no. 9: 678. https://doi.org/10.3390/pathogens9090678
APA StyleChakraborti, M., Schlachter, S., Primus, S., Wagner, J., Sweet, B., Carr, Z., Cornell, K. A., & Parveen, N. (2020). Evaluation of Nucleoside Analogs as Antimicrobials Targeting Unique Enzymes in Borrelia burgdorferi. Pathogens, 9(9), 678. https://doi.org/10.3390/pathogens9090678







