Transcriptomic Analysis of the Early Strobilar Development of Echinococcus granulosus
Abstract
1. Introduction
2. Results
2.1. Summary of the RNA Sequencing Data
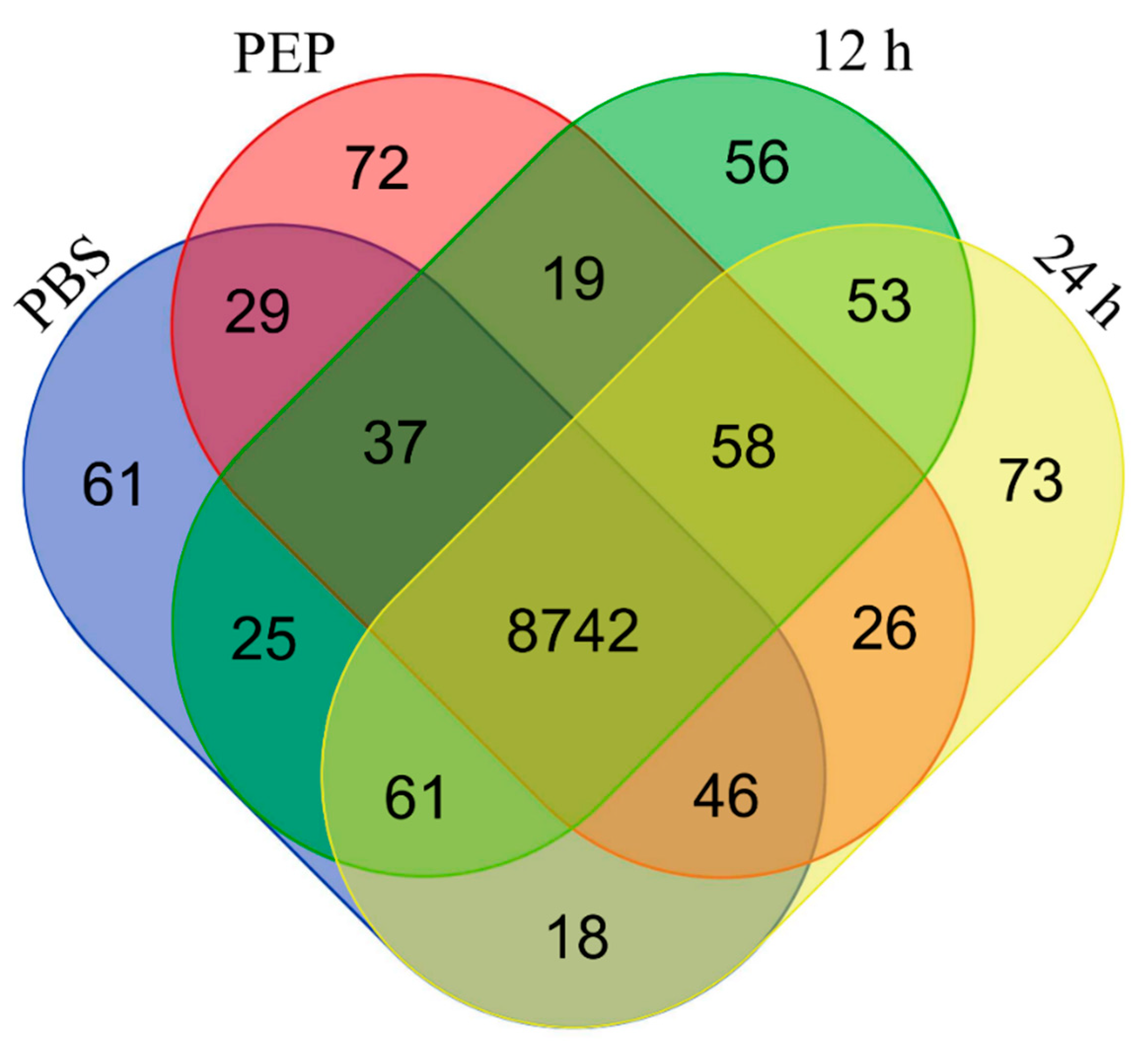
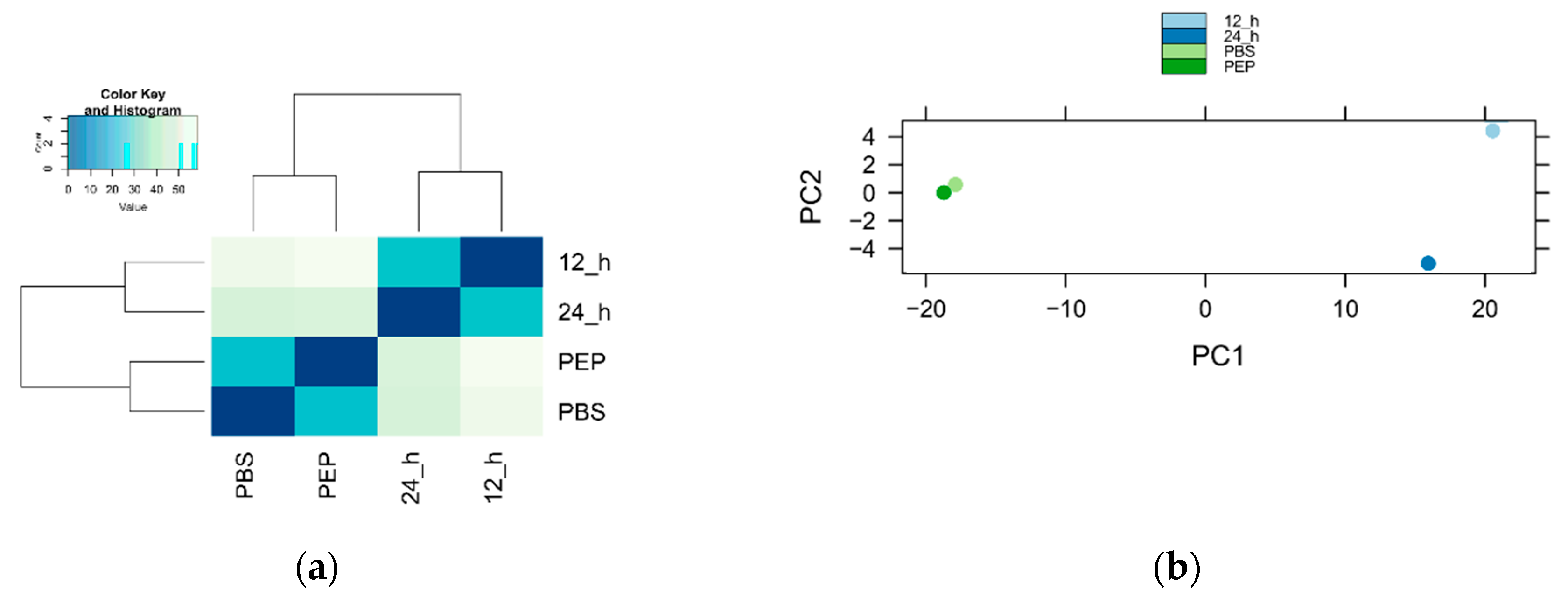
2.2. Identification and Analyses of Transcripts Detected in the Transcriptome
2.2.1. Number of Identified Genes
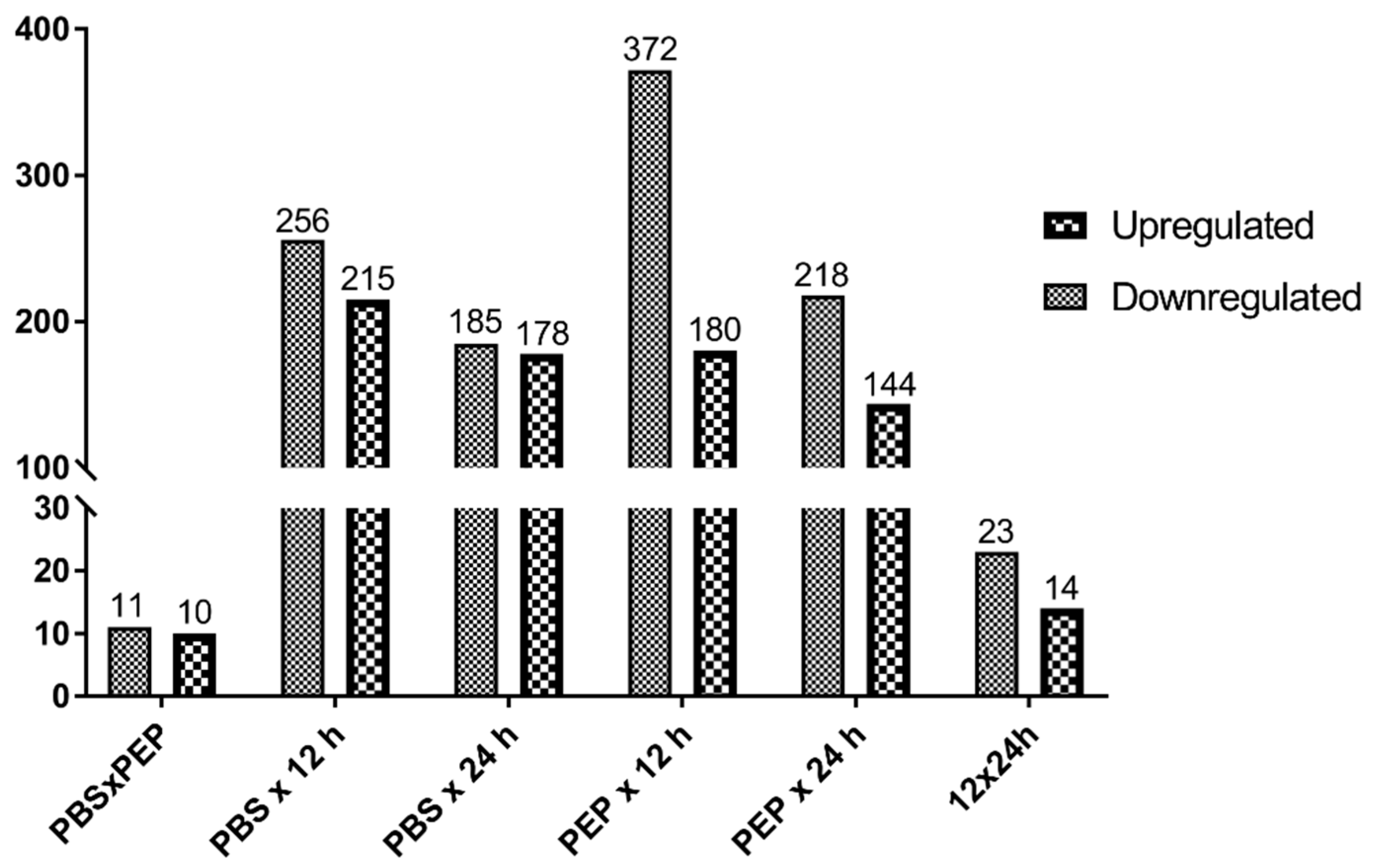
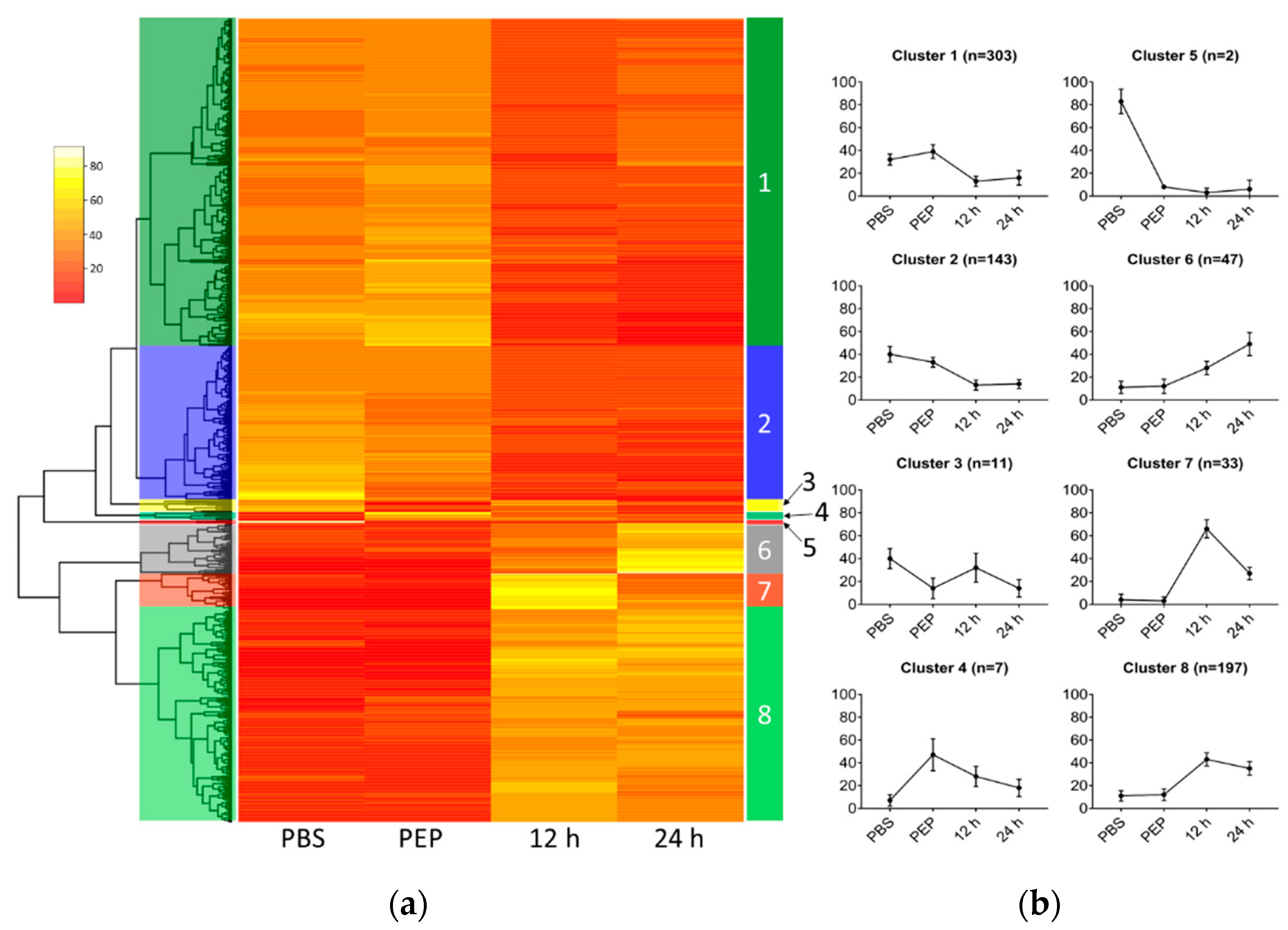
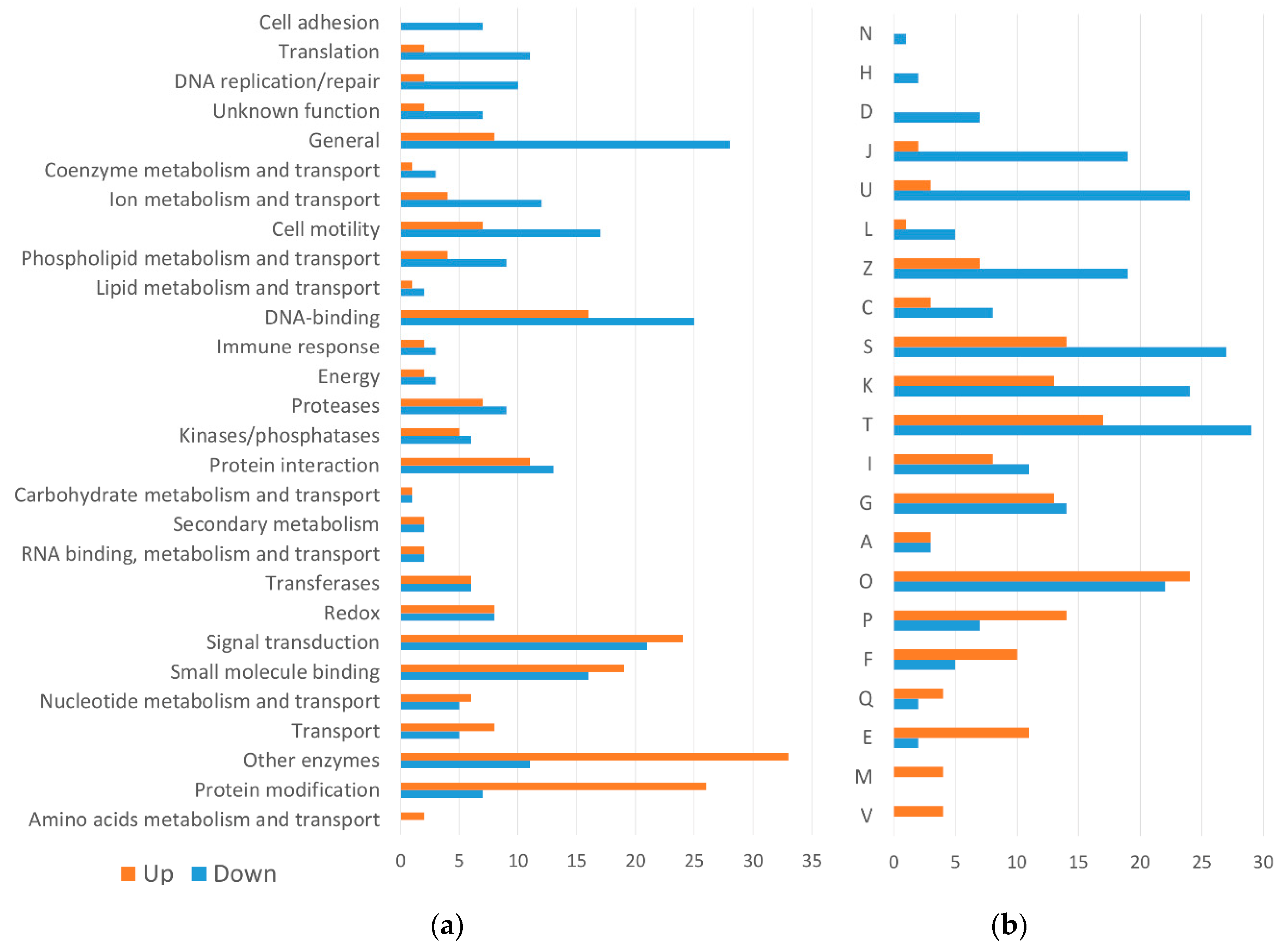
2.2.2. Differentially Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Parasite Material and In Vitro Cultivation
4.2. RNA Extraction
4.3. cDNA Library Construction and Sequencing
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- da Silva, A.M. Human echinococcosis: A neglected disease. Gastroenterol. Res. Pract. 2010, 2010, 583297. [Google Scholar] [CrossRef] [PubMed]
- WHO. Investing to Overcome the Global Impact of Neglected Tropical Diseases; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Eckert, J.; Deplazes, P. Biological, Epidemiological, and Clinical Aspects of Echinococcosis, a Zoonosis of Increasing Concern. Clin. Microbiol. Rev. 2004, 17, 107–135. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.C.A.; Jenkins, D.J. Echinococcus as a model system: Biology and epidemiology. Int. J. Parasitol. 2014, 44, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Casulli, A.; Siles-Lucas, M.; Tamarozzi, F. Echinococcus granulosus sensu lato. Trends Parasitol. 2019, 35, 663–664. [Google Scholar] [CrossRef] [PubMed]
- Cucher, M.; Prada, L.; Mourglia-Ettlin, G.; Dematteis, S.; Camicia, F.; Asurmendi, S.; Rosenzvit, M. Identification of Echinococcus granulosus microRNAs and their expression in different life cycle stages and parasite genotypes. Int. J. Parasitol. 2011, 41, 439–448. [Google Scholar] [CrossRef]
- Thompson, R.C.A. Biology and systematics of echinococcus. In Echinococcus and Hydatid Disease; CAB International: Wallingford, UK, 1995; pp. 1–50. [Google Scholar]
- Smyth, J.D. Parasites as biological models. Parasitology 1969, 59, 73. [Google Scholar] [CrossRef]
- Smyth, J.D.; Miller, H.J.; Howkins, A.B. Further analysis of the factors controlling strobilization, differentiation, and maturation of Echinococcus granulosus in vitro. Exp. Parasitol. 1967, 21, 31–41. [Google Scholar] [CrossRef]
- Smyth, J.; Howkins, A.; Barton, M. Factors controlling the differentiation of the hydatid organism, Echinococcus granulosus, into cystic or strobilar stages in vitro. Nature 1966, 211, 1374–1377. [Google Scholar] [CrossRef]
- Morseth, D.J. Fine structure of the hydatid cyst and protoscolex of Echinococcus granulosus. J. Parasitol. 1967, 53, 312–325. [Google Scholar] [CrossRef]
- Smyth, J.D.; Gemmell, M.; Smyth, M.M. Establishment of Echinococcus Granuiosus in the Intestine of Normal and Vaccinated Dogs; Indian Veterinary Research Institute: Bareilly, India, 1970. [Google Scholar]
- Smyth, J.D. Cestoda. In In Vitro Cultivation of Parasitic Helminthes; Smyth, J.D., Ed.; CRC Press: London, UK, 1990; pp. 123–137. [Google Scholar]
- Parkinson, J.; Wasmuth, J.D.; Salinas, G.; Bizarro, C.V.; Sanford, C.; Berriman, M.; Ferreira, H.B.; Zaha, A.; Blaxter, M.L.; Maizels, R.M.; et al. A Transcriptomic Analysis of Echinococcus granulosus Larval Stages: Implications for Parasite Biology and Host Adaptation. PLoS Negl. Trop. Dis. 2012, 6, e1897. [Google Scholar] [CrossRef]
- Tsai, I.J.; Zarowiecki, M.; Holroyd, N.; Garciarrubio, A.; Sanchez-Flores, A.; Brooks, K.L.; Tracey, A.; Bobes, R.J.; Fragoso, G.; Sciutto, E.; et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 2013, 496, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, W.; Zhang, L.; Zhang, Z.; Li, J.; Lu, G.; Zhu, Y.; Wang, Y.; Huang, Y.; Liu, J.; et al. The genome of the hydatid tapeworm Echinococcus granulosus. Nat. Genet. 2013, 45, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Dang, Z.; Suzuki, Y.; Horiuchi, T.; Yagi, K.; Kouguchi, H.; Irie, T.; Kim, K.; Oku, Y. Analysis on Gene Expression Profile in Oncospheres and Early Stage Metacestodes from Echinococcus multilocularis. PLoS Negl. Trop. Dis. 2016, 10, e0004634. [Google Scholar] [CrossRef]
- Camargo de Lima, J.; Monteiro, K.M.; Basika Cabrera, T.N.; Paludo, G.P.; Moura, H.; Barr, J.R.; Zaha, A.; Ferreira, H.B. Comparative proteomics of the larval and adult stages of the model cestode parasite Mesocestoides corti. J. Proteom. 2018, 175, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Basika, T.; Paludo, G.P.; Araujo, F.M.; Salim, A.C.; Pais, F.; Maldonado, L.; Macchiaroli, N.; Camargo de Lima, J.; Rosenzvit, M.; Oliveira, G.C.; et al. Transcriptomic profile of two developmental stages of the cestode parasite Mesocestoides corti. Mol. Biochem. Parasitol. 2019, 229, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Basika, T.; Macchiaroli, N.; Cucher, M.; Espínola, S.; Kamenetzky, L.; Zaha, A.; Rosenzvit, M.; Ferreira, H.B. Identification and profiling of microRNAs in two developmental stages of the model cestode parasite Mesocestoides corti. Mol. Biochem. Parasitol. 2016, 210, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Debarba, J.A.; Monteiro, K.M.; Moura, H.; Barr, J.R.; Ferreira, H.B.; Zaha, A. Identification of Newly Synthesized Proteins by Echinococcus granulosus Protoscoleces upon Induction of Strobilation. PLoS Negl. Trop. Dis. 2015, 9, e0004085. [Google Scholar] [CrossRef]
- Kramer, S. Developmental regulation of gene expression in the absence of transcriptional control: The case of kinetoplastids. Mol. Biochem. Parasitol. 2012, 181, 61–72. [Google Scholar] [CrossRef]
- Thorson, R.E. Environmental Stimuli and the Responses of Parasitic Helminths. Bioscience 1969, 19, 126–130. [Google Scholar] [CrossRef]
- Haile, S.; Papadopoulou, B. Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr. Opin. Microbiol. 2007, 10, 569–577. [Google Scholar] [CrossRef]
- Thompson, R.C.A.; Lymbery, A.J. Let’s not forget the thinkers. Trends Parasitol. 2013, 29, 581–584. [Google Scholar] [CrossRef] [PubMed]
- Olson, P.D.; Littlewood, D.T.J.; Bray, R.A.; Mariaux, J.; Olson, P.D.; Bray, R.A.; Mariaux, J. Interrelationships and evolution of the tapeworms (Platyhelminthes: Cestoda). Mol. Phylogenet. Evol. 2001, 19, 443–467. [Google Scholar] [CrossRef] [PubMed]
- Obal, G.; Ramos, A.L.; Silva, V.; Lima, A.; Batthyany, C.; Bessio, M.I.; Ferreira, F.; Salinas, G.; Ferreira, A.M. Characterisation of the Native Lipid Moiety of Echinococcus granulosus Antigen B. PLoS Negl. Trop. Dis. 2012, 6, e1642. [Google Scholar] [CrossRef] [PubMed]
- Pórfido, J.L.; Herz, M.; Kiss, F.; Kamenetzky, L.; Brehm, K.; Rosenzvit, M.C.; Córsico, B.; Franchini, G.R. Fatty acid-binding proteins in Echinococcus spp.: The family has grown. Parasitol. Res. 2020, 119, 1401–1408. [Google Scholar] [CrossRef] [PubMed]
- Espínola, S.M.; Ferreira, H.B.; Zaha, A. Validation of suitable reference genes for expression normalization in Echinococcus spp. larval stages. PLoS ONE 2014, 9, e102228. [Google Scholar] [CrossRef] [PubMed]
- Mamuti, W.; Sako, Y.; Xiao, N.; Nakaya, K.; Nakao, M.; Yamasaki, H.; Lightowlers, M.W.; Craig, P.S.; Ito, A. Echinococcus multilocularis: Developmental stage-specific expression of Antigen B 8-kDa-subunits. Exp. Parasitol. 2006, 113, 75–82. [Google Scholar] [CrossRef][Green Version]
- Roberts, A.J.; Kon, T.; Knight, P.J.; Sutoh, K.; Burgess, S.A. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 713–726. [Google Scholar] [CrossRef]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar] [CrossRef]
- Cantacessi, C.; Seddon, J.M.; Miller, T.L.; Leow, C.Y.; Thomas, L.; Mason, L.; Willis, C.; Walker, G.; Loukas, A.; Gasser, R.B.; et al. A genome-wide analysis of annexins from parasitic organisms and their vectors. Sci. Rep. 2013, 3, 2893. [Google Scholar] [CrossRef]
- Siozios, S.; Ioannidis, P.; Klasson, L.; Andersson, S.G.E.; Braig, H.R.; Bourtzis, K. The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS ONE 2013, 8, e55390. [Google Scholar] [CrossRef]
- Stankewich, M.C.; Moeckel, G.W.; Ji, L.; Ardito, T.; Morrow, J.S. Isoforms of Spectrin and Ankyrin Reflect the Functional Topography of the Mouse Kidney. PLoS ONE 2016, 11, e0142687. [Google Scholar] [CrossRef][Green Version]
- Silva, L.L.; Marcet-Houben, M.; Nahum, L.A.; Zerlotini, A.; Gabaldón, T.; Oliveira, G. The Schistosoma mansoni phylome: Using evolutionary genomics to gain insight into a parasite’s biology. BMC Genom. 2012, 13, 617. [Google Scholar] [CrossRef]
- Hu, D.; Song, X.; Xie, Y.; Zhong, X.; Wang, N.; Zheng, Y.; Gu, X.; Wang, T.; Peng, X.; Yang, G. Molecular insights into a tetraspanin in the hydatid tapeworm Echinococcus granulosus. Parasit. Vectors 2015, 8, 311. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.M.; Afgar, A.; Mohammadi, M.A.; Mortezaei, S.; Faridi, A.; Sadeghi, B.; Fasihi Harandi, M. Biological and morphological consequences of dsRNA-induced suppression of tetraspanin mRNA in developmental stages of Echinococcus granulosus. Parasit. Vectors 2020, 13, 190. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.J.; Xu, L.L.; Zhang, T.; Xu, M.; Yao, J.; Fang, C.Y.; Feng, Z.; Yang, P.Y.; Hu, W.; Liu, F. Proteomic characterization of larval and adult developmental stages in Echinococcus granulosus reveals novel insight into host-parasite interactions. J. Proteom. 2013, 84, 158–175. [Google Scholar] [CrossRef]
- Paludo, G.P.; Thompson, C.E.; Miyamoto, K.N.; Guedes, R.L.M.; Zaha, A.; Vasconcelos, A.T.; Sehabiague, M.C.; Ferreira, H.B. Cestode strobilation: Prediction of developmental genes and pathways. BMC Genom. (under review).
- Zhang, R.; Aji, T.; Shao, Y.; Jiang, T.; Yang, L.; Lv, W.; Chen, Y.; Chen, X.; Wen, H. Nanosecond pulsed electric field (nsPEF) disrupts the structure and metabolism of human Echinococcus granulosus protoscolex in vitro with a dose effect. Parasitol. Res. 2017, 116, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Balbinotti, H.; Santos, G.B.; Badaraco, J.; Arend, A.C.; Graichen, D.Â.S.; Haag, K.L.; Zaha, A. Echinococcus ortleppi (G5) and Echinococcus granulosus sensu stricto (G1) loads in cattle from Southern Brazil. Vet. Parasitol. 2012, 188, 255–260. [Google Scholar] [CrossRef]
- FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 6 June 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Howe, K.L.; Bolt, B.J.; Shafie, M.; Kersey, P.; Berriman, M. WormBase ParaSite—A comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 2017, 215, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Meyer, C.A.; Wang, Q.; Liu, J.S.; Liu, X.S.; Zhang, Y. GFOLD: A generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 2012, 28, 2782–2788. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.; Pethica, R.; Zhou, Y.; Talbot, C.; Vogel, C.; Madera, M.; Chothia, C.; Gough, J. SUPERFAMILY--sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009, 37, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef]
| Samples | Q30 1 | Raw Reads | Filtered Reads | Mapped Reads |
|---|---|---|---|---|
| PBS | 0.984 | 6,994,764 | 6,965,344 (99.6%) | 4,909,354 (70.5%) |
| PEP | 0.983 | 7,348,062 | 7,309,726 (99.5%) | 5,090,978 (69.6%) |
| 12 h | 0.985 | 8,308,408 | 8,271,920 (99.6%) | 6,103,570 (73.8%) |
| 24 h | 0.982 | 8,315,180 | 8,274,926 (99.5%) | 6,085,915 (73.5%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Debarba, J.A.; Sehabiague, M.P.C.; Monteiro, K.M.; Gerber, A.L.; Vasconcelos, A.T.R.; Ferreira, H.B.; Zaha, A. Transcriptomic Analysis of the Early Strobilar Development of Echinococcus granulosus. Pathogens 2020, 9, 465. https://doi.org/10.3390/pathogens9060465
Debarba JA, Sehabiague MPC, Monteiro KM, Gerber AL, Vasconcelos ATR, Ferreira HB, Zaha A. Transcriptomic Analysis of the Early Strobilar Development of Echinococcus granulosus. Pathogens. 2020; 9(6):465. https://doi.org/10.3390/pathogens9060465
Chicago/Turabian StyleDebarba, João Antonio, Martín Pablo Cancela Sehabiague, Karina Mariante Monteiro, Alexandra Lehmkuhl Gerber, Ana Tereza Ribeiro Vasconcelos, Henrique Bunselmeyer Ferreira, and Arnaldo Zaha. 2020. "Transcriptomic Analysis of the Early Strobilar Development of Echinococcus granulosus" Pathogens 9, no. 6: 465. https://doi.org/10.3390/pathogens9060465
APA StyleDebarba, J. A., Sehabiague, M. P. C., Monteiro, K. M., Gerber, A. L., Vasconcelos, A. T. R., Ferreira, H. B., & Zaha, A. (2020). Transcriptomic Analysis of the Early Strobilar Development of Echinococcus granulosus. Pathogens, 9(6), 465. https://doi.org/10.3390/pathogens9060465





