An Acute Stress Model in New Zealand White Rabbits Exhibits Altered Immune Response to Infection with West Nile Virus
Abstract
1. Introduction
2. Results
2.1. Characterization of the Acute Stress Model Using Bolus Administration of Dexamethasone
2.2. Use of Acute Stress Model for WNV Infection
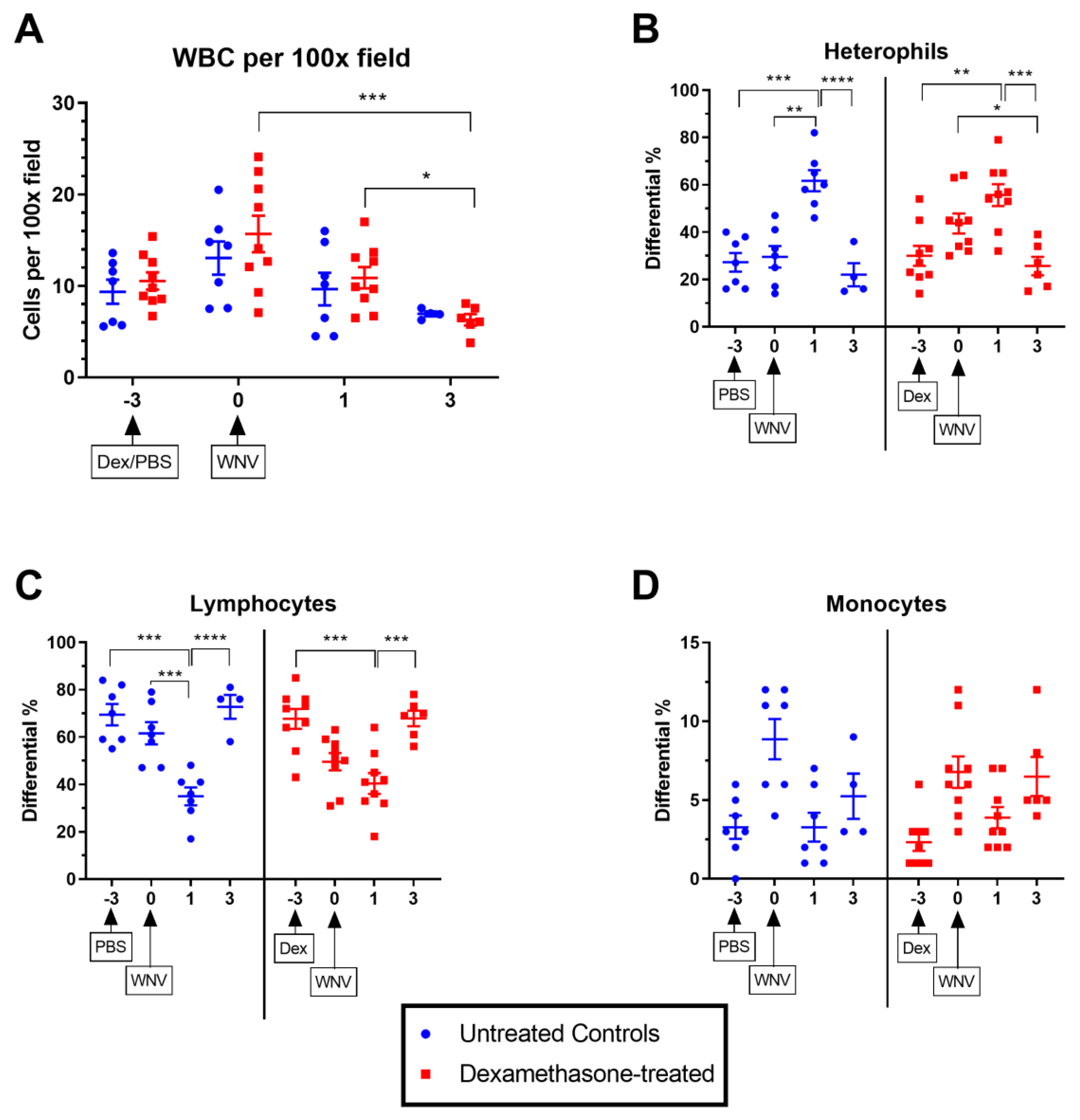
2.2.1. Hematological Parameters in Untreated and Dexamethasone-Treated WNV-Infected Rabbits.
2.2.2. Virus Kinetics in Untreated & Dexamethasone-Treated WNV-Infected Rabbits
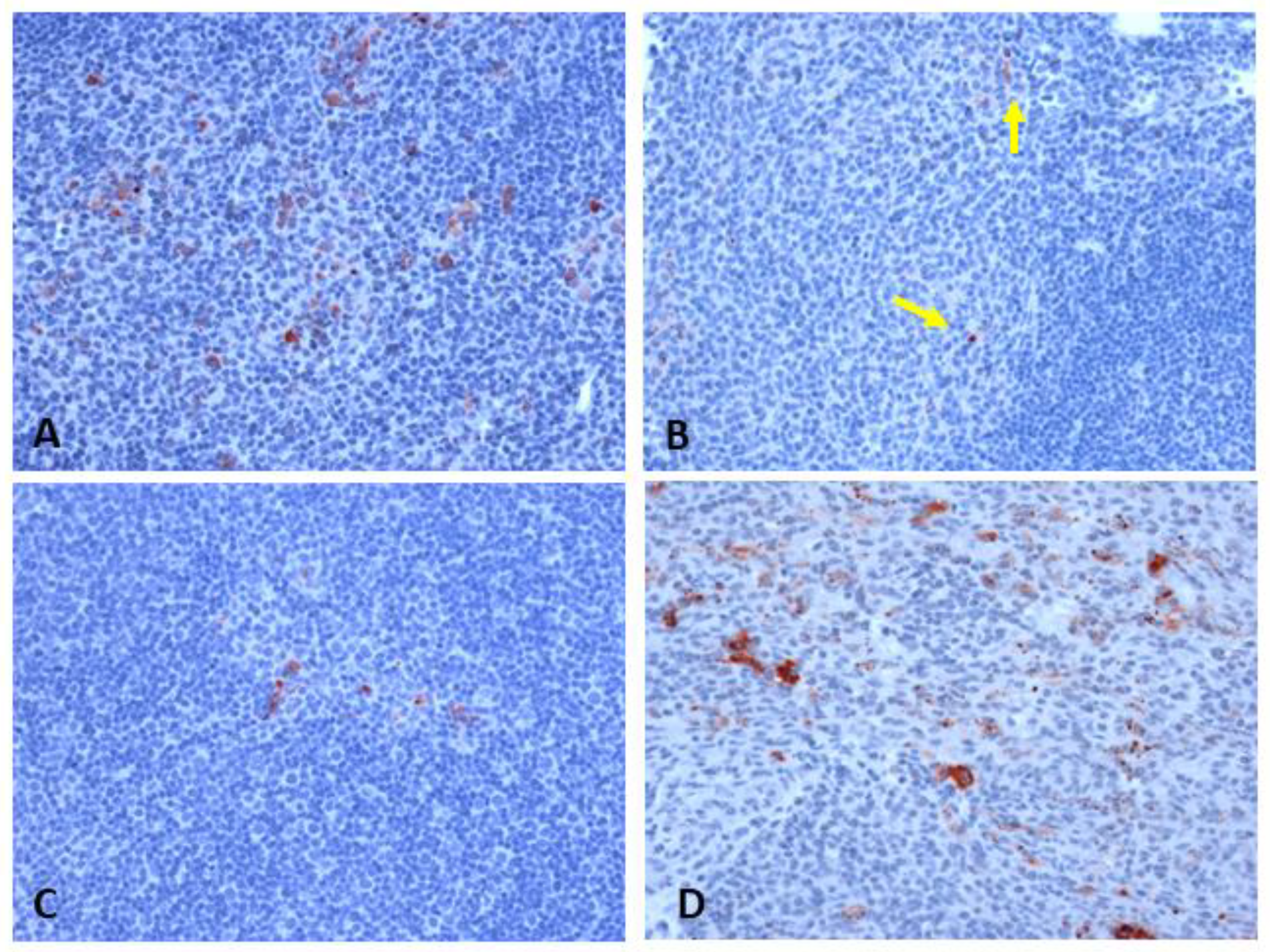
2.2.3. Histopathology and Immunophenotyping of Lymphoid Organs
2.2.4. Antibody Responses
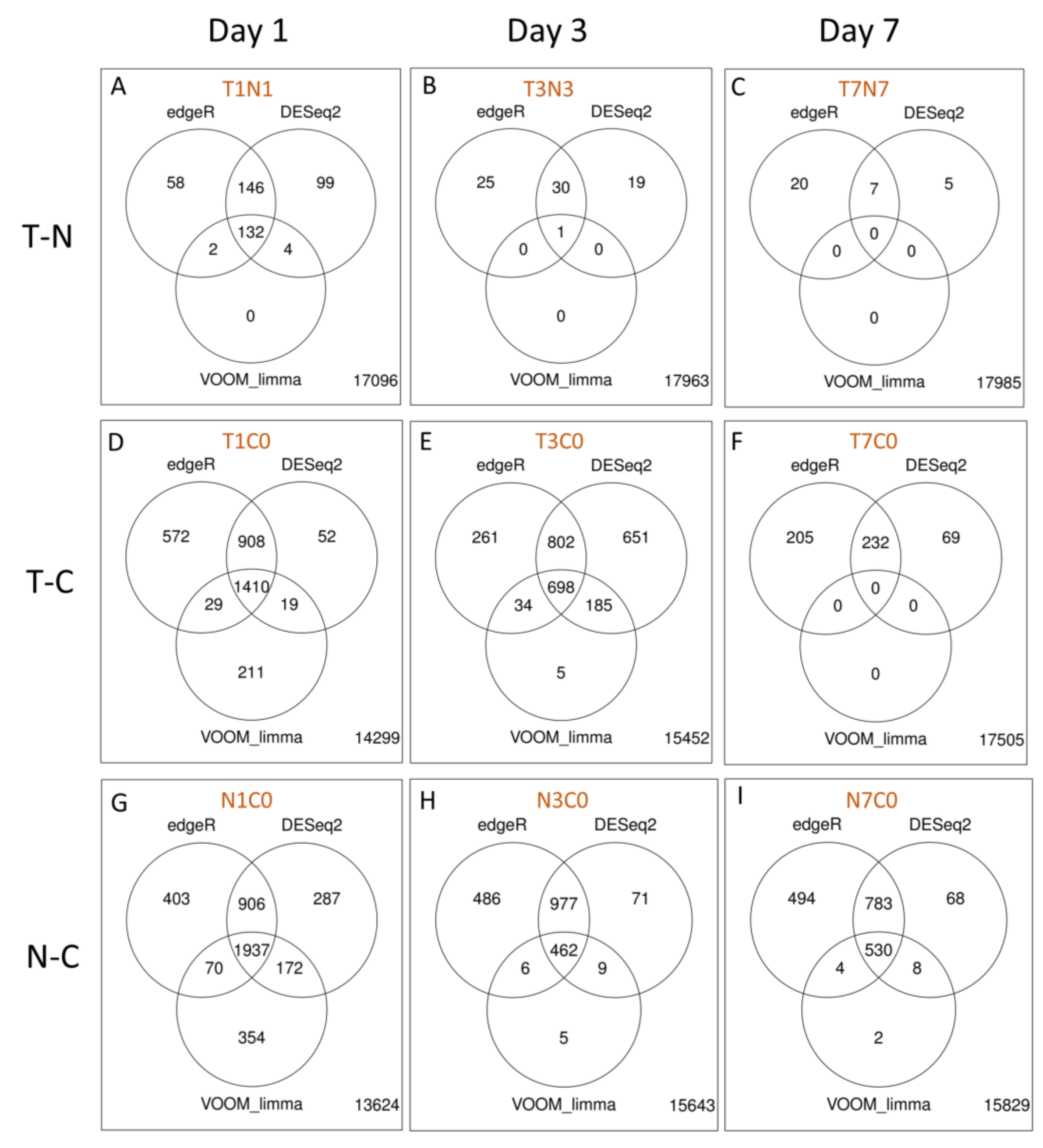
2.2.5. Differential Gene Expression Analyses of RNA Extracted from Fraining Lymph Nodes
2.2.6. Gene Ontology Analyses
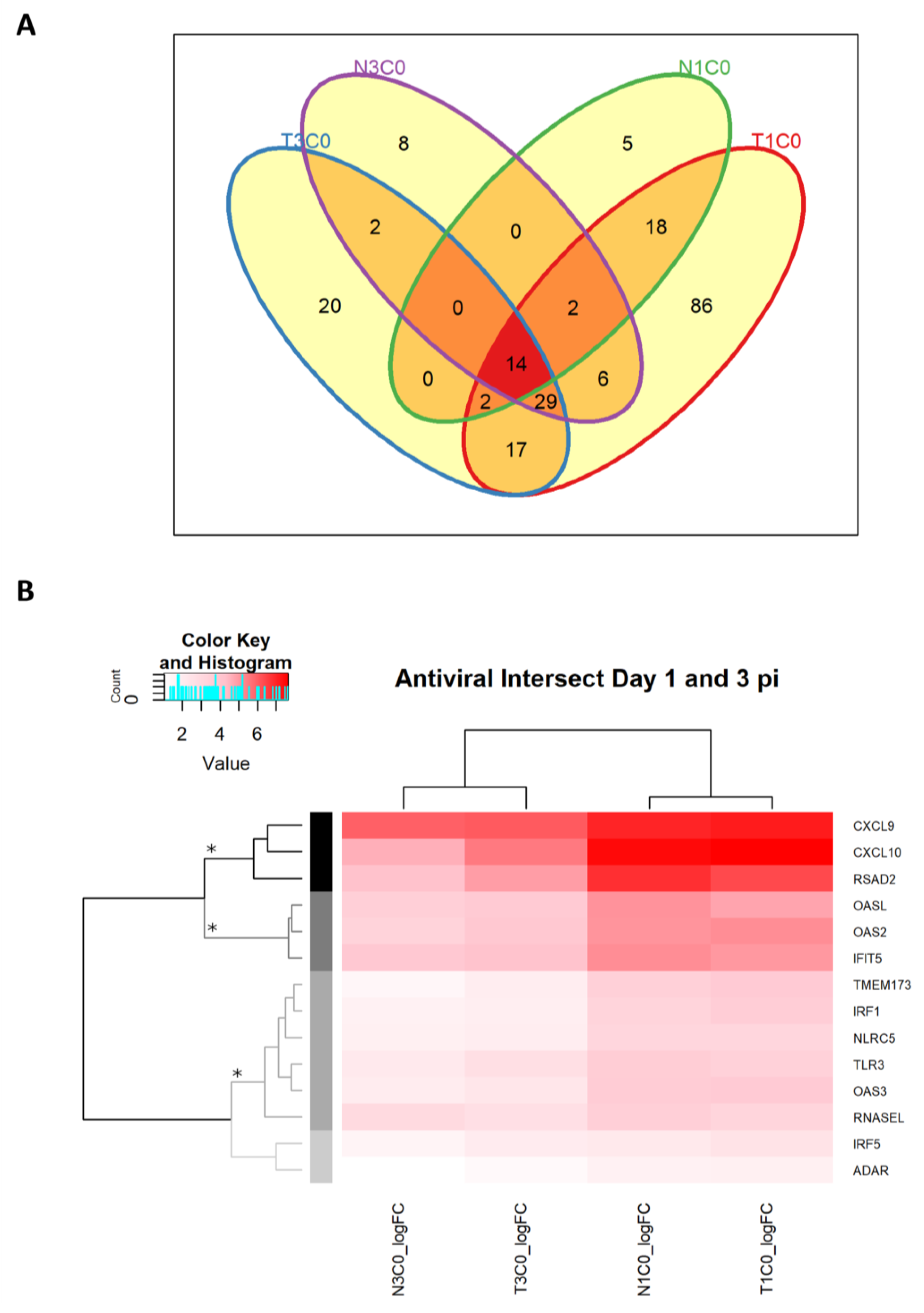
2.2.7. Antiviral DEGs in Rabbits Infected with WNVNSW2011
2.2.8. Antiviral Genes Across Day 1 and 3 p.i. Unaffected by Dexamethasone Treatment
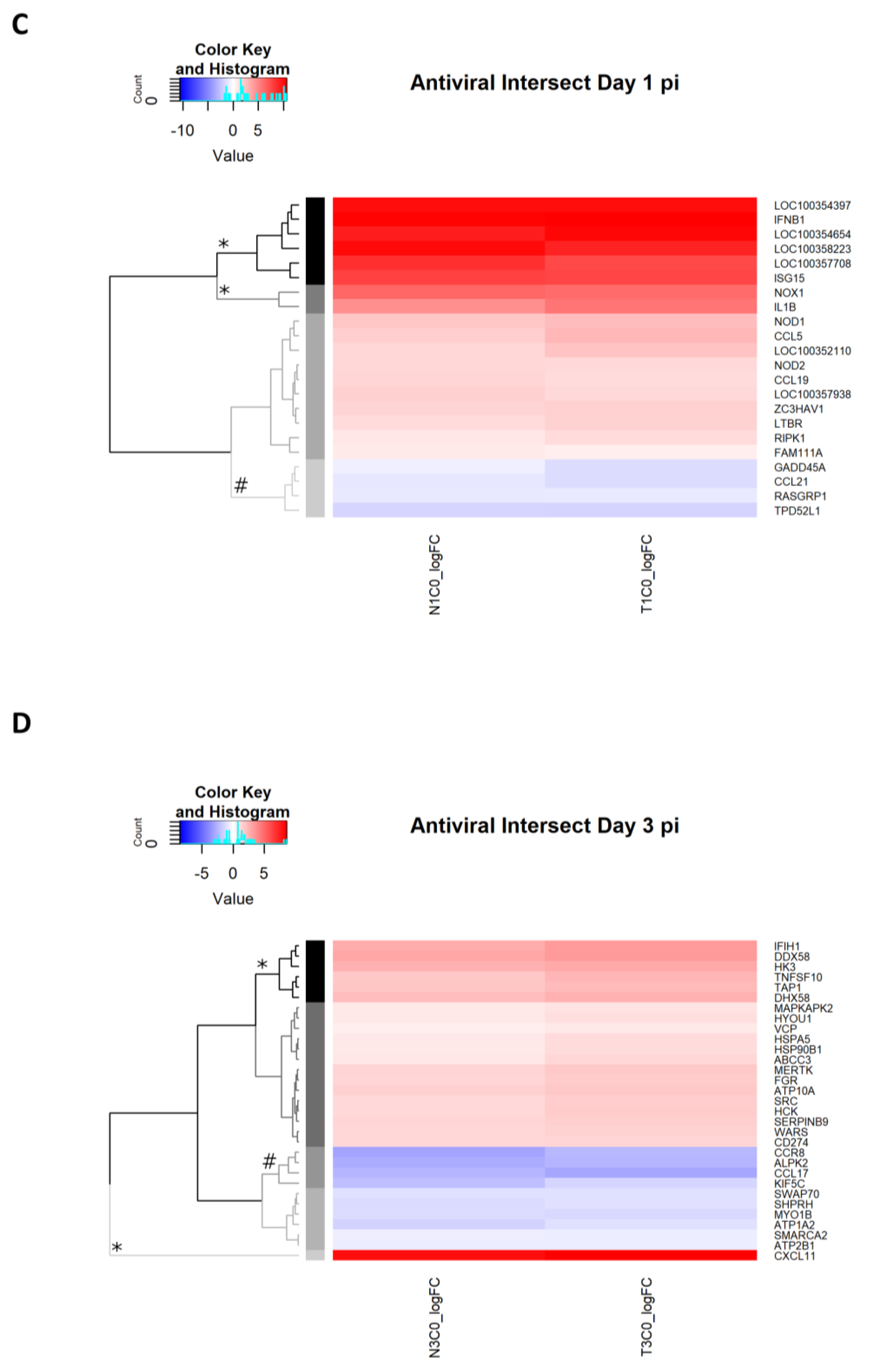
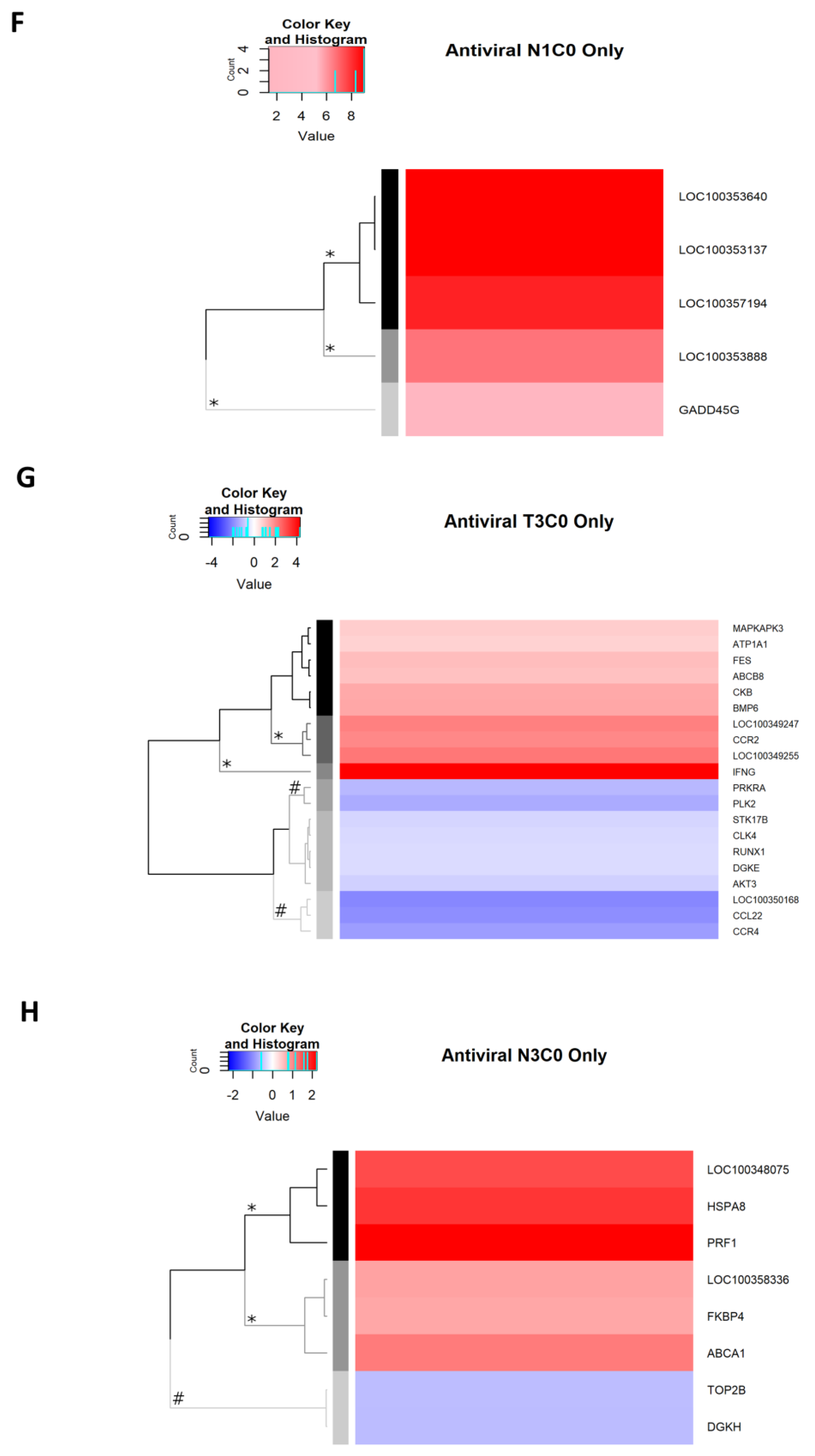
2.2.9. Antiviral Genes Uniquely Expressed on Day 1 p.i.
2.2.10. Antiviral Genes Uniquely Expressed on Day 3 p.i.
2.2.11. Antiviral Genes Expressed on Day 7 p.i.
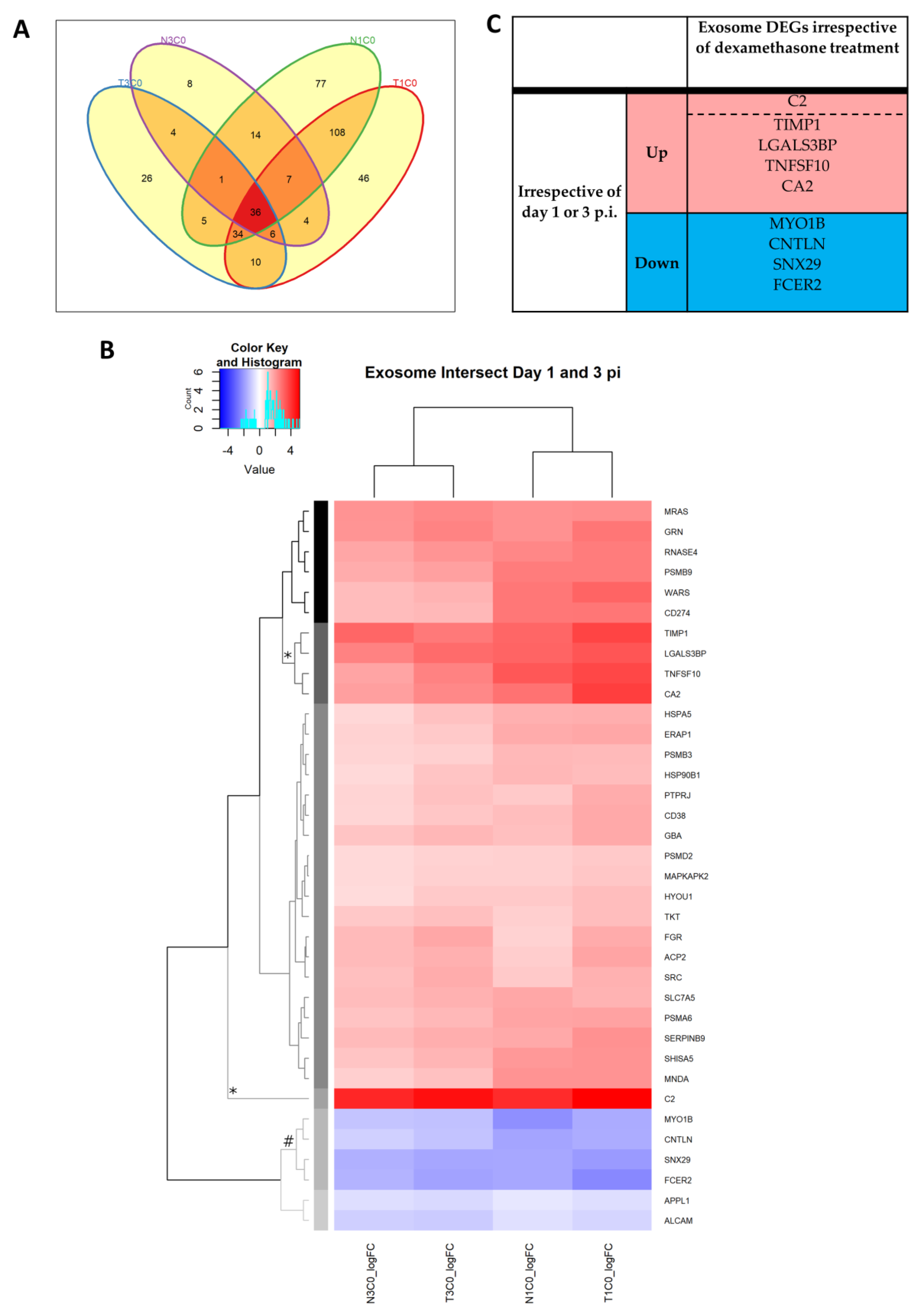
2.2.12. Extracellular Exosome
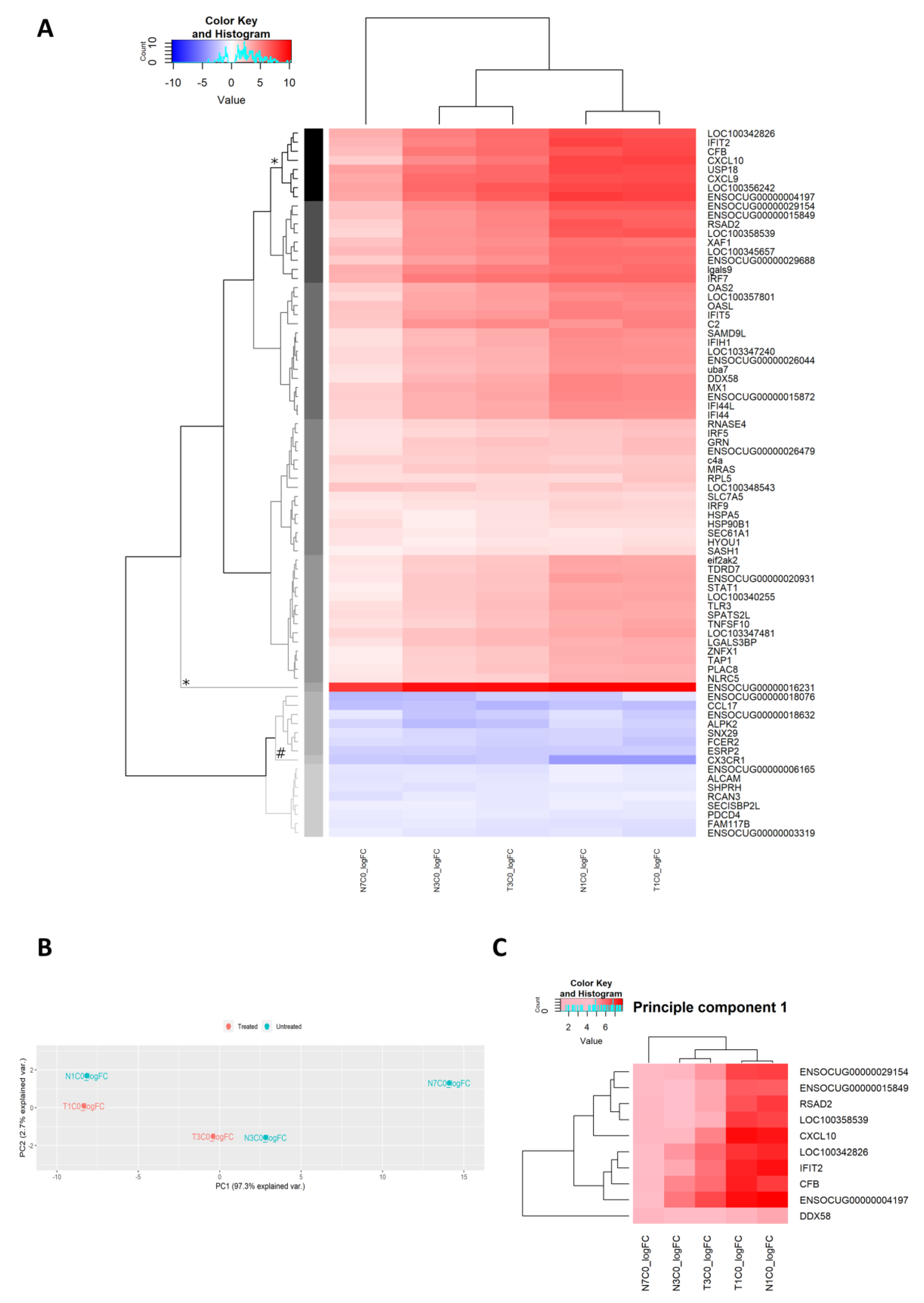
2.2.13. Unsupervised Analysis of Gene Expression
3. Discussion
4. Materials and Methods
4.1. Virus
4.2. Animals and Experimental Design
4.3. Hematology
4.4. Histopathology
4.5. Immunohistochemistry
4.6. Cell culture and Virus Isolation
4.7. RNA Extraction and Viral Transcript Quantitation
4.8. RNAseq
4.9. Differential Gene Expression Analysis
4.10. Serology
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control & Prevention (CDC). West Nile virus final cumulative maps & data for 1999–2018. Available online: https://www.cdc.gov/westnile/statsmaps/cumMapsData.html (accessed on 21 July 2019).
- Prow, N.A.; Hewlett, E.K.; Faddy, H.M.; Coiacetto, F.; Wang, W.; Cox, T.; Hall, R.A.; Bielefeldt-Ohmann, H. The Australian public is still vulnerable to emerging virulent strains of WNV. Front. Public Health. 2014, 2, 146. [Google Scholar] [CrossRef]
- Prow, N.A.; Edmonds, J.H.; William, D.T.; Setoh, Y.X.; Bielefeldt-Ohmann, H.; Suen, W.W.; Hobson-Peters, J.; Van den Hurk, A.F.; Pyke, A.F.; Hall-Mendelin, S.; et al. Virulence and evolution of West Nile virus, Australia, 1960–2012. Emerg. Infect. Dis. 2016, 22, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Frost, M.J.; Zhang, J.; Edmonds, J.H.; Prow, N.A.; Gu, X.; Davis, R.; Hornitzky, C.; Arzey, K.E.; Finlaison, D.; Hick, P.; et al. Characterization of virulent WNV Kunjin strain, Australia, 2011. Emerg. Infect. Dis. 2012, 18, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, N.P.; Staples, J.E.; Lehman, J.A.; Fischer, M. Surveillance for human WNV disease—United States, 1999–2008. MMWR Surveill. Summ. 2010, 59, 1–17. [Google Scholar] [PubMed]
- Sejvar, J.J.; Lindsay, N.P.; Campbell, G.L. Primary causes of death in reported cases of fatal West Nile fever, United States, 2002–2006. Vector Borne Zoonotic Dis. 2011, 11, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Jean, C.M.; Honarmand, S.; Louie, J.K.; Glaser, C.A. Risk factors for West Nile virus neuroinvasive disease, California, 2005. Emerg. Infect. Dis. 2007, 13, 1918–1920. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, N.P.; Staples, J.E.; Lehman, J.A.; Fischer, M. Medical risk factors for severe West Nile virus disease, United States, 2008–2010. Am. J. Trop. Med. Hyg. 2012, 87, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Grubaugh, N.D.; Massey, A.; Shives, K.D.; Stenglein, M.D.; Ebel, G.D.; Beckham, J.D. West Nile Virus Population structure, injury, and interferon-stimulated gene expression in the brain from a fatal case of encephalitis. Open Forum Infect. Dis. 2015, 3, ofv182. [Google Scholar] [CrossRef]
- Suen, W.W.; Prow, N.A.; Hall, R.A.; Bielefeldt-Ohmann, H. Mechanism of West Nile virus neuroinvasion: A critical appraisal. Viruses 2014, 6, 2796–2825. [Google Scholar] [CrossRef]
- Gause, K.T.; Wheatley, A.K.; Cui, J.; Yan, Y.; Kent, S.J.; Caruso, F. Immunological principles guiding the rational design of particles for vaccine delivery. ACS Nano 2017, 11, 54–68. [Google Scholar] [CrossRef]
- McClain, M.T.; Henao, R.; Williams, J.; Nicholson, B.; Veldman, T.; Hudson, L.; Tsalik, E.L.; Lambkin-Williams, R.; Gilbert, A.; Mann, A.; et al. Differential evolution of peripheral cytokine levels in symptomatic and asymptomatic responses to experimental influenza virus challenge. Clin. Exp. Immunol. 2016, 183, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Lopez, C.B. The innate immune response to RSV: Advances in our understanding of critical viral & host factors. Vaccine 2017, 35, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Werner, J.M.; Heller, T.; Gordon, A.M.; Sheets, A.; Sherker, A.H.; Kessler, E.; Bean, K.S.; Stevens, M.; Schmitt, J.; Rehermann, B. Innate immune responses in hepatitis C virus-exposed healthcare workers who do not develop acute infection. Hepatology 2013, 58, 1621–1631. [Google Scholar] [CrossRef] [PubMed]
- Prow, N.A.; Setoh, Y.X.; Biron, R.M.; Sester, D.P.; Kim, K.S.; Hobson-Peters, J.; Hall, R.A.; Bielefeldt-Ohmann, H. The West Nile virus-like flavivirus Koutango is highly virulent in mice due to delayed viral clearance and the induction of a poor neutralizing antibody response. J. Virol. 2014, 88, 9947–9962. [Google Scholar] [CrossRef]
- Suen, W.W.; Prow, N.A.; Setoh, Y.X.; Hall, R.A.; Bielefeldt-Ohmann, H. End-point disease investigation for virus strains of intermediate virulence as illustrated by flavivirus infections. J. Gen. Virol. 2016, 97, 366–377. [Google Scholar] [CrossRef]
- Bielefeldt-Ohmann, H.; Bosco-Lauth, A.; Hartwig, A.E.; Uddin, M.J.; Barcelon, J.; Suen, W.W.; Wang, W.; Hall, R.A.; Bowen, R.A. Characterization of non-lethal West Nile Virus (WNV) infection in horses: Subclinical pathology and innate immune response. Microb. Pathog. 2017, 103, 71–79. [Google Scholar] [CrossRef]
- Graham, J.B.; Thomas, S.; Swarts, J.; McMillan, A.A.; Ferris, M.T.; Suthar, M.S.; Treuting, P.M.; Ireton, R.; Gale, M., Jr.; Lund, J.M. Genetic diversity in the Collaborative Cross model recapitulates human WNV disease outcomes. mBio 2015, 6, e00493–e00515. [Google Scholar] [CrossRef]
- Seok, J.; Warren, H.S.; Cuenca, A.G.; Mindrinos, M.N.; Baker, H.V.; Xu, W.; Richards, D.R.; McDonald-Smith, G.P.; Gao, H.; Hennessy, L.; et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2013, 110, 3507–3512. [Google Scholar] [CrossRef]
- Suen, W.W.; Uddin, M.J.; Wang, W.; Brown, V.; Adney, D.R.; Broad, N.; Prow, N.A.; Bowen, R.A.; Hall, R.A.; Bielefeldt-Ohmann, H. Experimental West Nile virus infection in rabbits: an alternative model for studying induction of disease and virus control. Pathogens 2015, 4, 529–558. [Google Scholar] [CrossRef]
- Suen, W.W.; Uddin, M.J.; Prow, N.A.; Bowen, R.A.; Hall, R.A.; Bielefeldt-Ohmann, H. Tissue-specific transcription profile of cytokine and chemokine genes associated with flavivirus control and non-lethal neuropathogenesis in rabbits. Virology 2016, 494, 1–14. [Google Scholar] [CrossRef]
- Kirschman, L.J.; Crespi, E.J.; Warne, R.W. Critical disease windows shaped by stress exposure alter allocation trade-offs between development and immunity. J. Anim. Ecol. 2018, 87, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Aljebab, F.; Choonara, I.; Conroy, S. Systematic review of the toxicity of short-course oral corticosteroids in children. Arch. Dis. Child. 2016, 101, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Singanayagam, A.; Glanville, N.; Girkin, J.L.; Ching, Y.M.; Marcellini, A.; Porter, J.D.; Toussaint, M.; Walton, R.P.; Finney, L.J.; Aniscenko, J.; et al. Corticosteroid suppression of antiviral immunity increases bacterial loads and mucus production in COPD exacerbations. Nat. Commun. 2018, 9, 2229. [Google Scholar] [CrossRef] [PubMed]
- Alfakeekh, K.; Azar, M.; Sowailmi, B.A.; Alsulaiman, S.; Makdob, S.A.; Omair, A.; Albanyan, E.; Bawazeer, M.S. Immunosuppressive burden and risk factors of infection in primary childhood nephrotic syndrome. J. Infect. Public Health 2019, 12, 90–94. [Google Scholar] [CrossRef]
- Jeklova, E.; Leva, L.; Jaglic, Z.; Faldyna, M. Dexamethasone-induced immunosuppression: A rabbit model. Vet. Immunol. Immunopathol. 2008, 122, 231–240. [Google Scholar] [CrossRef]
- Nakagawa, M.; Terashima, T.; D’yachkova, Y.; Bondy, G.P.; Hogg, J.C.; Van Eeden, S.F. Glucocorticoid-induced granulocytosis: Contribution of marrow release and demargination of intravascular granulocytes. Circulation 1998, 98, 2307–2313. [Google Scholar] [CrossRef]
- Lazear, H.M.; Diamond, M.S. New insights into innate immune restriction of West Nile virus infection. Curr. Opin. Virol. 2015, 11, 1–6. [Google Scholar] [CrossRef]
- Buckwold, V.E.; Wei, J.; Huang, Z.; Huang, C.; Nalca, A.; Wells, J.; Russell, J.; Collins, B.; Ptak, R.; Lang, W.; et al. Antiviral activity of CHO-SS cell-derived human omega interferon and other human interferons against HCV RNA replicons and related viruses. Antivir. Res. 2007, 73, 118–125. [Google Scholar] [CrossRef]
- Garcia-Sastre, A. Ten strategies of interferon evasion by viruses. Cell Host Microbe 2017, 22, 176–184. [Google Scholar] [CrossRef]
- Schoggins, J.W.; MacDuff, D.A.; Imanaka, N.; Gainey, M.D.; Shrestha, B.; Eitson, J.L.; Mar, K.B.; Richardson, R.B.; Ratushny, A.V.; Litvak, V.; et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014, 505, 691–695. [Google Scholar] [CrossRef]
- Zhu, C.; Xiao, F.; Hong, J.; Wang, K.; Liu, X.; Cai, D.; Fusco, D.N.; Zhao, L.; Jeong, S.W.; Brisac, C.; et al. EFTUD2 is a novel innate immune regulator restricting hepatitis C virus infection through the RIG-I/MDA5 Pathway. J. Virol. 2015, 89, 6608–6618. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Michal, J.J.; Zhang, L.; Ding, B.; Lunney, J.K.; Liu, B.; Jiang, Z. Interferon induced IFIT family genes in host anitiviral defense. Int. J. Biol. Sci. 2013, 9, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.Y.; Yaneva, R.; Cresswell, P. Viperin: A multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 2011, 10, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Gizzi, A.S.; Grove, T.L.; Arnold, J.J.; Jose, J.; Jangra, R.K.; Garforth, S.J.; Du, Q.; Cahill, S.M.; Dulyaninova, N.G.; Love, J.D.; et al. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 2018, 558, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Dumbrepatil, A.B.; Ghosh, S.; Zegalia, K.A.; Malec, P.A.; Hoff, J.D.; Kennedy, R.T.; Marsh, E.N.G. Viperin interacts with the kinase IRAK1 and the E3 ubiquitin ligase TRAF6, coupling innate immune signaling to antiviral ribonucleotide synthesis. J. Biol. Chem. 2019, 294, 6888–6898. [Google Scholar] [CrossRef]
- Panayiotou, C.; Lindqvist, R.; Kurhade, C.; Vonderstein, K.; Pasto, J.; Edlund, K.; Upadhyay, A.S.; Overby, A.K. Viperin restricts Zika virus and tick-borne encephalitis virus replication by targeting NS3 for proteasomal degradation. J. Virol. 2018, 92, e02054–e02117. [Google Scholar] [CrossRef]
- Vonderstein, K.; Nilsson, E.; Hubel, P.; Nygard Skalman, L.; Upadhyay, A.; Pasto, J.; Pichlmair, A.; Lundmark, R.; Overby, A.K. Viperin targets flavivirus virulence by inducing assembly of noninfectious capsid particles. J. Virol. 2017, 92, e01751–e01817. [Google Scholar] [CrossRef]
- Szretter, K.J.; Brien, J.D.; Thackray, L.B.; Virgin, H.W.; Cresswell, P.; Diamond, M.S. The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J. Virol. 2011, 85, 11557–11566. [Google Scholar] [CrossRef]
- Morales, D.J.; Lenschow, D.J. The antiviral activities of ISG15. J. Mol. Biol. 2013, 425, 4995–5008. [Google Scholar] [CrossRef]
- Bielefeldt-Ohmann, H.; Smirnova, N.P.; Tolnay, A.E.; Webb, B.T.; Antoniazzi, A.Q.; Van Campen, H.; Hansen, T.R. Neuro-invasion by a ‘Trojan Horse’ strategy and vasculopathy during intrauterine flavivirus infection. Int. J. Exp. Pathol. 2012, 93, 24–33. [Google Scholar] [CrossRef]
- Dai, J.; Pan, W.; Wang, P. ISG15 facilitates cellular antiviral response to dengue and West Nile virus infection in vitro. Virol. J. 2011, 8, 468. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, E.W.; Lehner, P.J. The ABC-transporter signature motif is required for peptide translocation but not peptide binding by TAP. Eur. J. Immunol. 2003, 33, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Sanchez, D.J.; Aliyari, R.; Lu, S.; Cheng, G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. USA 2012, 109, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Quicke, K.M.; Suthar, M.S. The innate immune playbook for restricting West Nile virus infection. Viruses 2013, 5, 2643–2658. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.J.; Suen, W.W.; Prow, N.A.; Hall, R.A.; Bielefeldt-Ohmann, H. West Nile virus challenge alters the transcription profiles of innate immune genes in rabbit peripheral blood mononuclear cells. Front. Vet. Sci. 2015, 2, 76. [Google Scholar] [CrossRef]
- Uddin, M.J.; Suen, W.W.; Bosco-Lauth, A.; Hartwig, A.E.; Hall, R.A.; Bowen, R.A.; Bielefeldt-Ohmann, H. Kinetics of the West Nile virus induced transcripts of selected cytokines and Toll-like receptors in equine peripheral blood mononuclear cells. Vet. Res. 2016, 47, 61. [Google Scholar] [CrossRef]
- McGuckin Wuertz, K.; Treuting, P.M.; Hemann, E.A.; Esser-Nobis, K.; Snyder, A.G.; Graham, J.B.; Daniels, B.P.; Wilkins, C.; Snyder, J.M.; Voss, K.M.; et al. STING is required for host defense against neuropathological West Nile virus infection. PLoS Pathog. 2019, 15, e1007899. [Google Scholar] [CrossRef]
- Ding, Q.; Gaska, J.M.; Douam, F.; Wei, L.; Kim, D.; Balev, M.; Heller, B.; Ploss, A. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc. Natl. Acad. Sci. USA 2018, 115, E6310–E6318. [Google Scholar] [CrossRef]
- Robbins, G.R.; Truax, A.D.; Davis, B.K.; Zhang, L.; Brickey, W.J.; Ting, J.P. Regulation of class I major histocompatibility complex (MHC) by nucleotide-binding domain, leucine-rich repeat-containing (NLR) proteins. J. Biol. Chem. 2012, 287, 24294–24303. [Google Scholar] [CrossRef]
- Guo, X.; Liu, T.; Shi, H.; Wang, J.; Ji, P.; Wang, H.; Hou, Y.; Tan, R.X.; Li, E. Respiratory syncytial virus infection upregulates NLRC5 and Major Histocompatibility Complex Class I expression through RIG-I induction in airway epithelial cells. J. Virol. 2015, 89, 7636–7645. [Google Scholar] [CrossRef]
- Ranjan, P.; Singh, N.; Kumar, A.; Neerincx, A.; Kremmer, E.; Cao, W.; Davis, W.G.; Katz, J.M.; Gangappa, S.; Lin, R.; et al. NLRC5 interacts with RIG-I to induce a robust antiviral response against influenza virus infection. Eur. J. Immunol. 2015, 45, 758–772. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.; Xu, R.H.; Rubio, D.; Lev, A.; Stotesbury, C.; Fang, M.; Sigal, L.J. Migratory dendritic cells, group 1 innate lymphoid cells, and inflammatory monocytes collaborate to recruit NK cells to the virus-infected lymph node. Cell Rep. 2018, 24, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; Carter, S.; Hofer, M.J.; Campbell, I.L. Review: The chemokine receptor CXCR3 and its ligands CXCL9, CXCL10 and CXCL11 in neuroimmunity—A tale of conflict and conundrum. Neuropathol. Appl. Neurobiol. 2010, 36, 368–387. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Khan, A.A.; Chilukuri, S.; Syed, S.A.; Tran, T.T.; Furness, J.; Bahraoui, E.; BenMohamed, L. CXCL10/CXCR3-dependent mobilization of herpes simplex virus-specific CD8+ TEM and CD8+ TRM cells within infected tissues allows efficient protection against recurrent herpesvirus infection and disease. J. Virol. 2017, 91, e00278–e00317. [Google Scholar] [CrossRef]
- Fujino, M.; Sato, H.; Okamura, T.; Uda, A.; Takeda, S.; Ahmed, N.; Shichino, S.; Shiino, T.; Saito, Y.; Watanabe, S.; et al. Simian immunodeficiency virus targeting of CXCR3+ CD4+ T Cells in secondary lymphoid organs is associated with robust CXCL10 expression in monocyte/macrophage subsets. J. Virol. 2017, 91, e00439–e00517. [Google Scholar] [CrossRef]
- Lu, Y.; Lin, L.Y.; Tan, J.G.; Deng, H.P.; Li, X.H.; Zhang, Z.; Li, Y.; Zhou, Z.; Xu, X.; Xie, X.; et al. A correlation study between gene polymorphism of Th cell expressed chemokine receptor CXCR3 and its ligand levels with HCV infection prognosis. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1290–1295. [Google Scholar]
- Oelmann, E.; Herbst, H.; Zuhlsdorf, M.; Albrecht, O.; Nolte, A.; Schmitmann, C.; Manzke, O.; Diehl, V.; Stein, H.; Berdel, W.E. Tissue inhibitor of metalloproteinases 1 is an autocrine and paracrine survival factor, with additional immune-regulatory functions, expressed by Hodgkin/Reed-Sternberg cells. Blood 2002, 99, 258–267. [Google Scholar] [CrossRef]
- Zanotti, L.; Angioni, R.; Cali, B.; Soldani, C.; Ploia, C.; Moalli, F.; Gargesha, M.; D’Amico, G.; Elliman, S.; Tedeschi, G.; et al. Mouse mesenchymal stem cells inhibit high endothelial cell activation and lymphocyte homing to lymph nodes by releasing TIMP-1. Leukemia 2016, 30, 1143–1154. [Google Scholar] [CrossRef]
- Jones, R.S.; Tu, C.; Zhang, M.; Qu, J.; Morris, M.E. Characterization and proteomic-transcriptomic investigation of monocarboxylate transporter 6 knockout mice: evidence of a potential role in glucose and lipid metabolism. Mol. Pharmacol. 2019, 96, 364–376. [Google Scholar] [CrossRef]
- Oudshoorn, D.; Van Boheemen, S.; Sanchez-Aparicio, M.T.; Rajsbaum, R.; Garcia-Sastre, A.; Versteeg, G.A. HERC6 is the main E3 ligase for global ISG15 conjugation in mouse cells. PLoS ONE 2012, 7, e29870. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; Van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.A.; Broom, A.K.; Hartnett, A.C.; Howard, M.J.; Mackenzie, J.S. Immunodominant epitopes on the NS1 protein of MVE and KUN viruses serve as targets for a blocking ELISA to detect virus-specific antibodies in sentinel animal serum. J. Virol. Methods 1995, 51, 201–210. [Google Scholar] [CrossRef]
- Prow, N.A.; Tan, C.S.; Wang, W.; Hobson-Peters, J.; Kidd, L.; Barton, A.; Wright, J.; Hall, R.A.; Bielefeldt-Ohmann, H. Natural exposure of horses to mosquito-borne flaviviruses in south-east Queensland, Australia. Int. J. Environ. Res. Public Health 2013, 10, 4432–4443. [Google Scholar] [CrossRef] [PubMed]
| Pre-Treat | Day p.i. | Rabbit ID | L. Ln Popl | R. Ln Popl | Spleen | Thymus | Ln. Mes | Left Footpad | Right Footpad | Brain | Liver |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dex | 1 | 2101 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | 0 |
| 2102 | + | 0 | 0 | 0 | 0 | 0 | 0 | ND | 0 | ||
| 2103 | + | 0 | 0 | 0 | 0 | 0 | 0 | ND | 0 | ||
| 3 | 2104 | + | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND | |
| 2105 | ++ | +++ | + | 0 | ++ | 0 | 0 | ND | ND | ||
| 2106 | + | 0 | + | 0 | 0 | 0 | 0 | ND | ND | ||
| 7 | 2107 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | |
| 2108 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND | ||
| 2109 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND | ||
| PBS | 1 | 2110 | +++ | 0 | 0 | 0 | 0 | 0 | 0 | ND | 0 |
| 2111 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | ||
| 2112 | ++ | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND | ||
| 3 | 2115 | + | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND | |
| 2116 | + | 0 | 0 | 0 | 0 | 0 | 0 | ND | 0 | ||
| 7 | 2113 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND | |
| 2114 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | 0 |
| Group Abbreviation | Day Post-Infection | WNVNSW2011 Infected | Dexamethasone Treated | Rabbit IDs |
|---|---|---|---|---|
| C0 | N/A | N | N | 2001 and 2002 * |
| N1 | 1 | Y | N | 2110, 2111, and 2112 |
| N3 | 3 | Y | N | 2115 and 2116 |
| N7 | 7 | Y | N | 2113 and 2114 |
| T1 | 1 | Y | Y | 2101 and 2102 # |
| T3 | 3 | Y | Y | 2104, 2105, and 2106 |
| T7 | 7 | Y | Y | 2107, 2108, and 2109 |
| Selected Enriched GO Terms | Treated | Mock-Treated | |||
|---|---|---|---|---|---|
| Day 1 | Day 3 | Day 1 | Day 3 | Day 7 | |
| defense response to virus (GO:0051607, BP) | X | X | X | X | X |
| extracellular exosome (GO:0070062, CC) | X | X | X | X | X |
| ATP binding (GO:0005524, MF) | X | X | X | ||
| negative regulation of viral genome replication (GO:0045071, BP) | X | X | X | ||
| positive regulation of JNK cascade (GO:0046330, BP) | X | X | |||
| double-stranded RNA binding (GO:0003725, MF) | X | X | |||
| immune response (GO:0006955, BP) | X | X | X | ||
| Antiviral DEGs Irrespective of Dexamethasone Treatment |
|---|
| CXCL9 CXCL10 RSAD2 (viperin) |
| OASL OAS2 IFIT5 |
| TMEM173 (STING) IRF1 NLRC5 TLR3 OAS3 RNaseL |
| Day 1 p.i. | Antiviral Genes Affected by Dexamethasone Treatment | Antiviral DEGs Irrespective of Dexamethasone Treatment | |
|---|---|---|---|
| Expressed in Treated Group | Expressed in Mock-Treated Group | ||
| Up | LOC100355669 (IFN-α-21-like) | LOC100353640 (IFN-ω-1-like) | LOC100354397 (IFN-ω-1-like) |
| LOC100354910 (IFN-ω-1-like) | LOC100353137 (IFN-ω-1-like) | IFNβ1 | |
| LOC100355421 (IFN-ω-1-like) | LOC100357194 (IFN-α-21-like) | LOC100354654 (IFN-α-21-like) | |
| LOC100358223 (IFN-ω-1-like) | |||
| LOC100357708 (IFN-α-21-like) | |||
| ISG15 | |||
| LOC100346553 (multidrug resistance-associated protein 1) | LOC100353888 (IFN-ω-1-like) | NOX1 | |
| CCR1 | GADD45G | IL1β | |
| IL23A | |||
| CCL2 | |||
| Down | PDK4 | — | GADD45A |
| MYO6 | CCL21 | ||
| FGFR1 | RASGRP1 | ||
| TGFBR3 | TPD52L1 | ||
| KATNAL1 | |||
| CNNM2 | |||
| ABCA8 | |||
| PLCB1 | |||
| DYNC2H1 | |||
| ABCC5 | |||
| NPR1 | |||
| ACTA2 | |||
| EPHB3 | |||
| LOC100358984 (vascular endothelial growth factor receptor kdr-like) | |||
| Day 3 p.i. | Antiviral Genes Affected by Dexamethasone Treatment | Antiviral DEGs Irrespective of Dexamethasone Treatment | |
|---|---|---|---|
| Expressed in Treated Group | Expressed in Mock-Treated Group | ||
| Up | IFNγ | LOC100348075(tubulin alpha-1B chain-like) | CXCL11 |
| HSPA8 | |||
| PRF1 | |||
| CCR2 | LOC100358336 (actin-related protein 3-like) | IFIH1 (MDA5) | |
| LOC100349255 (lymphotactin) | FKBP4 | DDX58 (RIG-I) | |
| LOC100349247 (HLA class I histocompatibility antigen, B-7 alpha chain) | ABCA1 | DHX58 (LGP2) | |
| HK3 | |||
| TNFSF10 (TRAIL) | |||
| TAP1 | |||
| Down | CCL22 | TOP2B | CCR8 |
| CCR4 | DGKH | ALPK2 | |
| LOC100350168 (HLA class II histocompatibility antigen, DRB1-4 beta chain) | CCL17 | ||
| PRKRA | KIF5C | ||
| PLK2 | |||
| Gene ID | Weights | Gene Description |
|---|---|---|
| CXCL10 | −0.2492985 | C-X-C motif chemokine ligand 10 |
| LOC100358539 | −0.2233737 | guanylate-binding protein 1 |
| RSAD2 | −0.2079025 | radical S-adenosyl methionine domain containing 2 |
| IFIT2 | −0.1999001 | interferon induced protein with tetratricopeptide repeats 2 |
| ENSOCUG00000029154 | −0.191678 | interferon induced protein with tetratricopeptide repeats 3 |
| CFB | −0.1915019 | complement factor B |
| ENSOCUG00000004197 | −0.1883572 | novel gene (Human orthologue: IFIT1B [ENSG00000204010]) |
| LOC100342826 | −0.1697197 | interferon-induced protein with tetratricopeptide repeats 1-like |
| ENSOCUG00000015849 | −0.1678594 | novel gene (Human orthologue: HERC6 [ENSG00000138642]) |
| DDX58 | −0.1648038 | DExD/H-box helicase 58 |
| Day 1 p.i. | Day 3 p.i. | Day 7 p.i. | |
|---|---|---|---|
| Dexamethasone-treated | n = 3 (ID: 2101-3) | n = 3 (ID: 2104-6) | n = 3 (ID: 2107-9) |
| Mock-treated | n = 3 (ID: 2110-2) | n = 2 (ID: 2115-6) | n = 2 (ID: 2113-4) |
| Antibody | Source | Specificity | Antigen Retrieval | 1o Ab Incubation | 2o Ab (Envision Kit) |
|---|---|---|---|---|---|
| Anti-flavivirus NS1 (clone 4G4) | SCMB (UQ) | Flavivirus Non-Structural 1 (NS1) protein | EDTA, pH9 | 2 hours r.t. | Anti-Mouse Anti-rabbit |
| Anti-human myeloid/histiocyte antigen (clone Mac 387) | DAKO | Myeloid lineage cells | Proteinase K | ||
| CD 3 (clone F7.2.38) | DAKO | T lymphocytes | EDTA, pH9 | ||
| CD 79a (clone HM57) | DAKO | B lymphocytes | Citrate, pH6 | Overnight, 4 °C | |
| Ki67 | Abcam | Proliferation marker | EDTA, pH9 | 1–2 hours r.t. | |
| Activated caspase-3 | Abcam | Apoptosis marker | EDTA, pH9 | Overnight, 4 °C |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suen, W.W.; Imoda, M.; Thomas, A.W.; Nasir, N.N.B.M.; Tearnsing, N.; Wang, W.; Bielefeldt-Ohmann, H. An Acute Stress Model in New Zealand White Rabbits Exhibits Altered Immune Response to Infection with West Nile Virus. Pathogens 2019, 8, 195. https://doi.org/10.3390/pathogens8040195
Suen WW, Imoda M, Thomas AW, Nasir NNBM, Tearnsing N, Wang W, Bielefeldt-Ohmann H. An Acute Stress Model in New Zealand White Rabbits Exhibits Altered Immune Response to Infection with West Nile Virus. Pathogens. 2019; 8(4):195. https://doi.org/10.3390/pathogens8040195
Chicago/Turabian StyleSuen, Willy W., Mitchell Imoda, Albert W. Thomas, Nur N.B.M. Nasir, Nawaporn Tearnsing, Wenqi Wang, and Helle Bielefeldt-Ohmann. 2019. "An Acute Stress Model in New Zealand White Rabbits Exhibits Altered Immune Response to Infection with West Nile Virus" Pathogens 8, no. 4: 195. https://doi.org/10.3390/pathogens8040195
APA StyleSuen, W. W., Imoda, M., Thomas, A. W., Nasir, N. N. B. M., Tearnsing, N., Wang, W., & Bielefeldt-Ohmann, H. (2019). An Acute Stress Model in New Zealand White Rabbits Exhibits Altered Immune Response to Infection with West Nile Virus. Pathogens, 8(4), 195. https://doi.org/10.3390/pathogens8040195





