Molecular Analysis of Canine Filaria and Its Wolbachia Endosymbionts in Domestic Dogs Collected from Two Animal University Hospitals in Bangkok Metropolitan Region, Thailand
Abstract
1. Introduction
2. Results
2.1. Morphological Characteristics of Filarial Worm
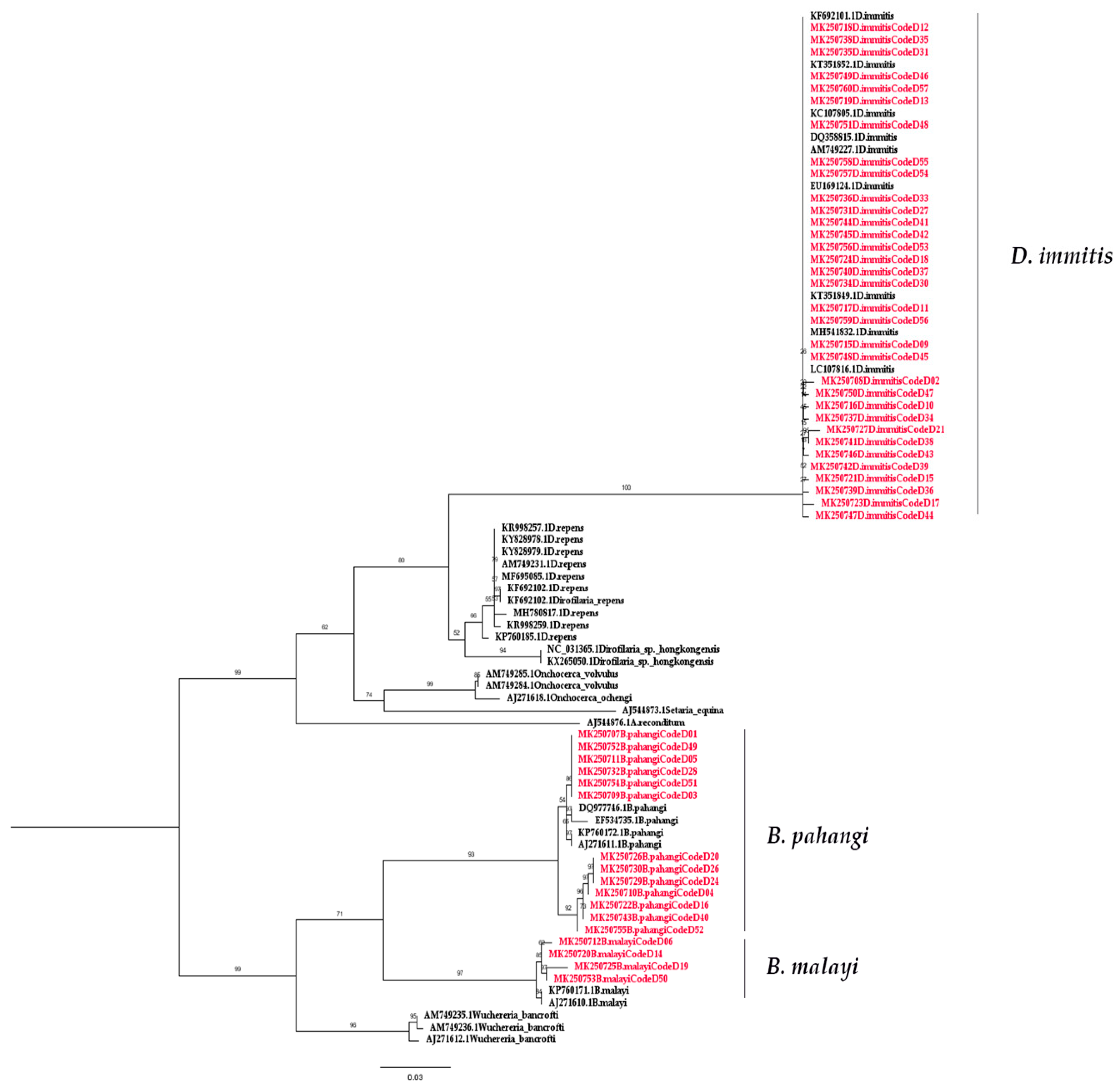
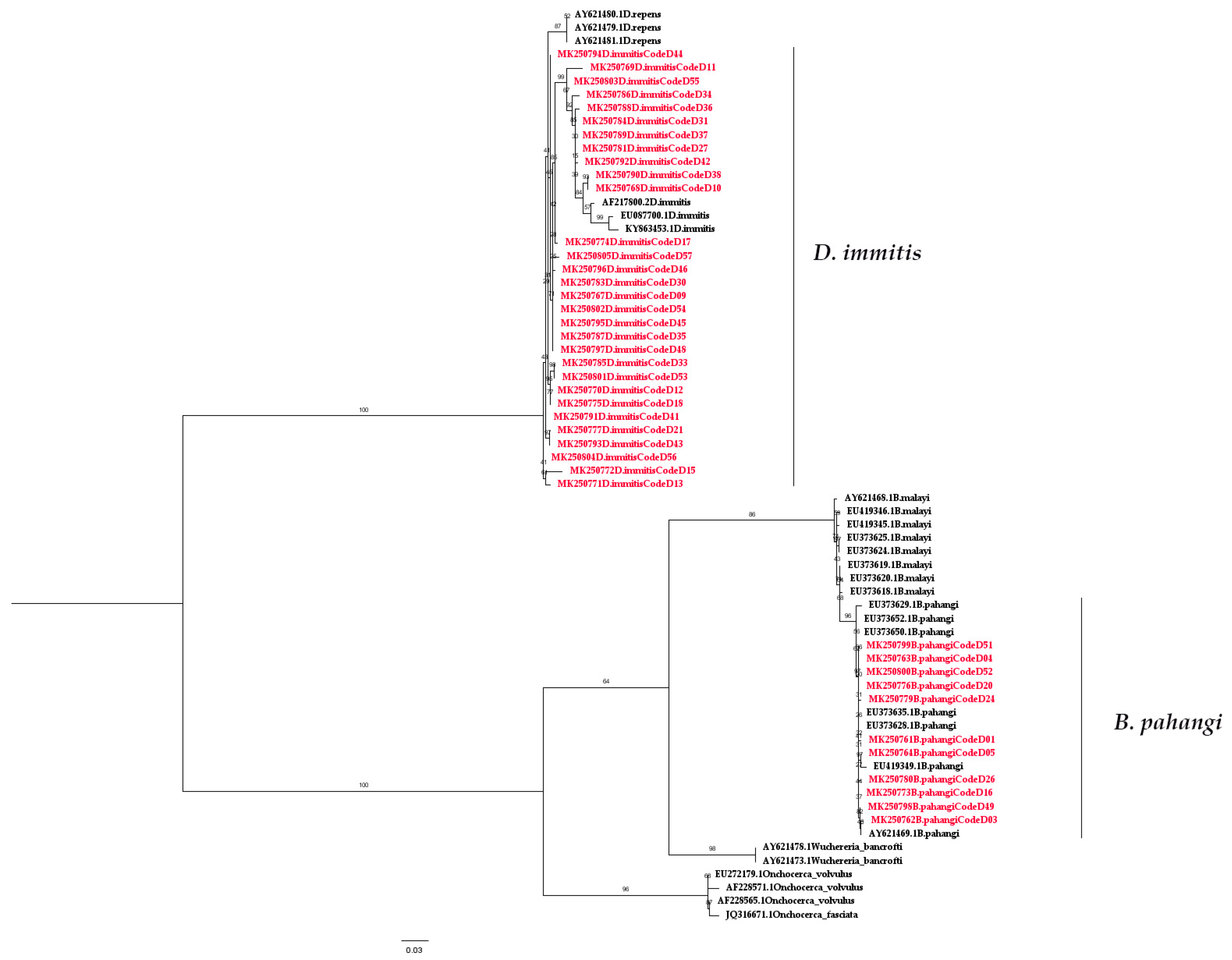
2.2. Molecular Detection of Filarial Nematode
2.3. Wolbachia Detection Using Nested PCR
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Sample Collection
4.3. Microscopic Examination
4.4. DNA Extraction
4.5. Molecular Identification of Canine Filarial
4.6. Wolbachia Bacterial Detection
4.7. Cloning and Sequencing
4.8. Sequence Analysis and Phylogenetic Tree Construction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Lymphatic Filariasis. Available online: https://www.who.int/news-room/fact-sheets/detail/lymphatic-filariasis (accessed on 28 May 2019).
- Kaikuntod, M.; Thongkorn, K.; Tiwananthagorn, S.; Boonyapakorn, C. Filarial worms in dogs in Southeast Asia. Vet. Integr. Sci. 2018, 16, 1–17. [Google Scholar]
- Mak, J.W.; Yen, P.K.; Lim, K.C.; Ramiah, N. Zoonotic implications of cats and dogs in filarial transmission in Peninsular Malaysia. Trop. Geogr. Med. 1980, 32, 259–264. [Google Scholar]
- Irwin, P.J. Companion animal parasitology: A clinical perspective. Int. J. Parasitol. Res. 2002, 32, 581–593. [Google Scholar] [CrossRef]
- Kelly, J.D. Canine heart worm disease. In Current Veterinary Therapy VII; W. B. Saunders: Philadelphia, PA, USA, 1979; pp. 326–335. [Google Scholar]
- Rishniw, M.; Barr, S.C.; Simpson, K.W.; Frongillo, M.F.; Franz, M.; Dominguez Alpizar, J.L. Discrimination between six species of canine microfilariae by a single polymerase chain reaction. Vet. Parasitol. 2006, 135, 303–314. [Google Scholar] [CrossRef]
- Kanjanopas, K.; Choochote, W.; Jitpakdi, A.; Suvannadabba, S.; Loymak, S.; Chungpivat, S.; Nithiuthai, S. Brugia malayi in a naturally infected cat from Narathiwat province, southern Thailand. Southeast. Asian J. Trop. Med. Public Health 2001, 32, 585–587. [Google Scholar]
- Kamyingkird, K.; Junsiri, W.; Chimnoi, W.; Kengradomkij, C.; Saengow, S.; Sangchuto, K.; Ka jeerum, W.; Pangjai, D.; Nimsuphan, B.; Inpankeaw, T.; et al. Prevalence and risk factors associated with Dirofilaria immitis infection in dogs and cats in Songkhla and Satun provinces, Thailand. Agric. Nat. Resour. 2017, 51, 299–302. [Google Scholar] [CrossRef]
- Wongkamchai, S.; Nochotea, H.; Foongladda, S.; Dekumyoy, P.; Thammapalo, S.; Boitanoa, J.J.; Choochotee, W. A high resolution melting real time PCR for mapping of filaria infection in domestic cats living in brugian filariosis-endemic areas. Vet. Parasitol. 2014, 201, 120–127. [Google Scholar] [CrossRef]
- Simón, F.; Morchón, R.; González-Miguel, J.; Marcos-Atxutegi, C.; Siles-Lucas, M. What is new about animal and human dirofilariosis? Trends. Parasitol. 2009, 25, 404–409. [Google Scholar] [CrossRef]
- Ichimori, K.; King, J.D.; Engels, D.; Yajima, A.; Mikhailov, A.; Lammie, P.; Ottesen, E.A. Global programme to eliminate lymphatic filariasis: The processes underlying programme success. PLoS. Negl. Trop. Dis. 2014, 8, e3328. [Google Scholar] [CrossRef]
- World Health Organization. Essential Medicines Donated to Control, Eliminate and Eradicate Neglected Tropical Diseases; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- World Health Organization. Lymphatic Filariasis: A Handbook of Practical Entomology for National Lymphatic Filariasis Elimination Programmes; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- Famakinde, D.O. Mosquitoes and the Lymphatic Filarial Parasites: Research Trends and Budding Roadmaps to Future Disease Eradication. Trop. Med. Infect. Dis. 2018, 43, 4. [Google Scholar] [CrossRef]
- Pradatsundarasar, A. Dirofilaria infection in man: Report of a case. J. Med. Assoc. Thailand 1955, 38, 378–379. [Google Scholar]
- Jariya, P.; Sucharit, S. Dirofilaria repens from the eyelid of a woman in Thailand. Am. J. Trop. Med. Hyg. 1983, 32, 1456–1457. [Google Scholar] [CrossRef]
- Sukudom, P.; Phumee, A.; Siriyasatien, P. First report on subconjunctival dirofilariasis in Thailand caused by a Dirofilaria sp. closely related to D. hongkongensis. Acad. J. Sci. Res. 2018, 6, 114–116. [Google Scholar] [CrossRef]
- Taylor, M.J.; Bandi, C.; Hoerauf, A. Wolbachia bacterial endosymbionts of filarial nematodes. Adv. Parasitol. 2005, 60, 245–284. [Google Scholar] [CrossRef]
- Taylor, M.J.; Voronin, D.; Johnston, K.L.; Ford, L. Wolbachia filarial interactions. Cell. Microbiol. 2013, 15, 520–526. [Google Scholar] [CrossRef]
- Bandi, C.; Trees, A.J.; Brattig, N.W. Wolbachia in filarial nematodes: Evolutionary aspects and implications for the pathogenesis and treatment of filarial diseases. Vet. Parasitol. 2001, 98, 215–238. [Google Scholar] [CrossRef]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Clark, E.L.; Karley, A.J.; Hubbard, S.F. Insect endosymbionts: Manipulators of insect herbivore trophic interactions? Protoplasma 2010, 244, 25–51. [Google Scholar] [CrossRef]
- Slatko, B.E.; Taylor, M.J.; Foster, J.M. The Wolbachia endosymbiont as an anti-filarial nematode target. Symbiosis 2010, 51, 55–65. [Google Scholar] [CrossRef]
- Pfarr, K.M.; Hoerauf, A.M. Antibiotics which target the Wolbachia endosymbionts of filarial parasites: A new strategy for control of filariasis and amelioration of pathology. Mini. Rev. Med. Chem. 2006, 6, 203–210. [Google Scholar] [CrossRef][Green Version]
- Ravindran, R.; Varghese, S.; Nair, S.N.; Balan, V.M.; Lakshmanan, B.; Ashruf, R.M.; Kumar, S.S.; Gopalan, A.K.; Nair, A.S.; Malayil, A.; et al. Canine filarial infections in a human Brugia malayi endemic area of India. Biomed. Res. Int. 2014, 2014, 630160. [Google Scholar] [CrossRef]
- Megat Abd Rani, P.A.; Irwin, P.J.; Gatne, M.; Coleman, G.T.; McInnes, L.M.; Traub, R.J. A survey of canine filarial diseases of veterinary and public health significance in India. Parasit. Vectors 2010, 3, 30. [Google Scholar] [CrossRef]
- Saseendranath, M.R.; Varghese, C.G.; Jayakumar, K.M. Incidence of canine dirofilariosis in Trichur, Kerala. Indian J. Vet. Med. 1986, 6, 139. [Google Scholar]
- Otranto, D.; Dantas-Torres, F.; Breitschwerdt, E.B. Managing canine vector-borne diseases of zoonotic concern: Part one. Trends Parasitol. 2009, 25, 157–163. [Google Scholar] [CrossRef]
- Rosenblatt, J.E. Laboratory diagnosis of infections due to blood and tissue parasites. Clin. Infect. Dis. 2009, 49, 1103–1108. [Google Scholar] [CrossRef]
- Ricciardi, A.; Ndao, M. Diagnosis of Parasitic Infections: What’s Going On? J. Biomol. Screen. 2015, 20, 6–21. [Google Scholar] [CrossRef]
- Nuchprayoon, S.; Junpee, A.; Poovorawan, Y.; Scott, A.L. Detection and differentiation of filarial parasites by universal primers and polymerase chain reaction-restriction fragment length polymorphism analysis. Am. J. Trop. Med. Hyg. 2005, 73, 895–900. [Google Scholar] [CrossRef]
- Magnis, J.; Lorentz, S.; Guardone, L.; Grimm, F.; Magi, M.; Naucke, T.J.; Deplazes, P. Morphometric analyses of canine blood microfilariae isolated by the Knott’s test enables Dirofilaria immitis and D. repens species-specific and Acanthocheilonema (syn. Dipetalonema) genus-specific diagnosis. Parasit. Vectors 2013, 6, 48. [Google Scholar] [CrossRef]
- Hoch, H.; Strickland, K. Canine and feline dirofilariasis: Life cycle, pathophysiology, and diagnosis. Compend. Contin. Educ. Vet. 2008, 30, 133–140. [Google Scholar]
- Casiraghi, M.; Bazzocchi, C.; Mortarino, M.; Ottina, E.; Genchi, C. A simple molecular method for discriminating common filarial nematodes of dogs (Canis familiaris). Vet. Parasitol. 2006, 141, 368–372. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, N.; Zhang, Z.; Wang, D.; Yao, Z.; Zhang, H.; Ma, J.; Zheng, B.; Ren, H.; Liu, S. Prevalence of Dirofilaria immitis infection in dogs in Henan province, central China. Parasite 2016, 23, 43. [Google Scholar] [CrossRef]
- Byeon, K.H.; Kim, B.J.; Kim, S.M.; Yu, H.S.; Jeong, H.J.; Ock, M.S. A serological survey of Dirofilaria immitis infection in pet dogs of Busan, Korea, and effects of chemoprophylaxis. Korean J. Parasitol. 2007, 45, 27–32. [Google Scholar] [CrossRef]
- Khedri, J.; Radfar, M.H.; Borji, H.; Azizzadeh, M.; Akhtardanesh, B. Canine Heartworm in Southeastern of Iran with Review of disease distribution. Iran J. Parasitol. 2014, 9, 560–567. [Google Scholar]
- Alho, A.M.; Landum, M.; Ferreira, C.; Meireles, J.; Goncalves, L.; de Carvalho, L.M.; Belo, S. Prevalence and seasonal variations of canine dirofilariosis in Portugal. Vet. Parasitol. 2014, 206, 99–105. [Google Scholar] [CrossRef]
- Bacsadi, A.; Papp, A.; Szeredi, L.; Toth, G.; Nemes, C.; Imre, V.; Tolnai, Z.; Szell, Z.; Sreter, T. Retrospective study on the distribution of Dirofilaria immitis in dogs in Hungary. Vet. Parasitol. 2016, 220, 83–86. [Google Scholar] [CrossRef]
- Tiawsirisup, S.; Thanapaisarnkit, T.; Varatorn, E.; Apichonpongsa, T.; Bumpenkiattikun, N.; Rattanapuchpong, S.; Chungpiwat, S.; Sanprasert, V.; Nuchprayoon, S. Canine Heartworm (Dirofilaria immitis) Infection and Immunoglobulin G Antibodies Against Wolbachia (Rickettsiales: Rickettsiaceae) in Stray Dogs in Bangkok, Thailand. Thai. J. Vet. Med. 2015, 40, 165–170. [Google Scholar]
- Ambily, V.R.; Pillai, U.N.; Arun, R.; Pramod, S.; Jayakumar, K.M. Detection of human filarial parasite Brugia malayi in dogs by histochemical staining and molecular techniques. Vet. Parasitol. 2011, 181, 210–214. [Google Scholar] [CrossRef]
- Chungpivat, S.; Taweethavonsawat, P. The differentiation of microfilariae in dogs and cats using Giemsa’s staining and the detection of acid phosphatase activity. J. Thai Vet. Pract. 2008, 20, 47–55. [Google Scholar]
- Thanchomnang, T.; Intapan, P.M.; Chungpivat, S.; Lulitanond, V.; Maleewong, W. Differential detection of Brugia malayi and Brugia pahangi by real-time fluorescence resonance energy transfer PCR and its evaluation for diagnosis of B. pahangi-infected dogs. Parasitol. Res. 2010, 106, 621–625. [Google Scholar] [CrossRef]
- Chansiri, K.; Tejangkura, T.; Kwaosak, P.; Sarataphan, N.; Phantana, S.; Sukhumsirichart, W. PCR based method for identification of zoonostic Brugia malayi microfilariae in domestic cats. Mol. Cell. Probes 2002, 16, 129–135. [Google Scholar] [CrossRef]
- Oh, I.Y.; Kim, K.T.; Sung, H.J. Molecular Detection of Dirofilaria immitis Specific Gene from Infected Dog Blood Sample Using Polymerase Chain Reaction. Iran. J. Parasitol. 2017, 12, 433–440. [Google Scholar]
- Lo, N.; Casiraghi, M.; Salati, E.; Bazzocchi, C.; Bandi, C. How many Wolbachia supergroups exist? Mol. Biol Evol. 2002, 19, 341–346. [Google Scholar] [CrossRef]
- Casiraghi, M.; Bordenstein, S.R.; Baldo, L.; Lo, N.; Beninati, T.; Wernegreen, J.J.; Werren, J.H.; Bandi, C. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: Clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology 2005, 151, 4015–4022. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Paraskevopoulos, C.; Hotopp, J.C.; Sapountzis, P.; Lo, N.; Bandi, C.; Tettelin, H.; Werren, J.H.; Bourtzis, K. Parasitism and mutualism in Wolbachia: What the phylogenomic trees can and cannot say. Mol. Biol. Evol. 2009, 26, 231–241. [Google Scholar] [CrossRef]
- Baldo, L.; Werren, J.H. Revisiting Wolbachia supergroup typing based on WSP: Spurious lineages and discordance with MLST. Curr. Microbiol. 2007, 55, 81–87. [Google Scholar] [CrossRef]
- Bandi, C.; Anderson, T.J.; Genchi, C.; Blaxter, M.L. Phylogeny of Wolbachia in filarial nematodes. Proc. Biol. Sci. 1998, 265, 2407–2413. [Google Scholar] [CrossRef]
- Darby, A.C.; Armstrong, S.D.; Bah, G.S.; Kaur, G.; Hughes, M.A.; Kay, S.M.; Koldkjær, P.; Rainbow, L.; Radford, A.D.; Blaxter, M.L.; et al. Analysis of gene expression from the Wolbachia genome of a filarial nematode supports both metabolic and defensive roles within the symbiosis. Genome Res. 2012, 22, 2467–2477. [Google Scholar] [CrossRef]
- Lefoulon, E.; Bain, O.; Makepeace, B.L.; d’Haese, C.; Uni, S.; Martin, C.; Gavotte, L. Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ 2016, 4, e1840. [Google Scholar] [CrossRef]
- Fenn, K.; Blaxter, M. Quantification of Wolbachia bacteria in Brugia malayi through the nematode lifecycle. Mol. Biochem. Parasitol. 2004, 137, 361–364. [Google Scholar] [CrossRef]
- McGarry, H.F.; Egerton, G.L.; Taylor, M.J. Population dynamics of Wolbachia bacterial endosymbionts in Brugia malayi. Mol. Biochem. Parasitol. 2004, 135, 57–67. [Google Scholar] [CrossRef]
- Nelson, G.S. The identification of infective filarial larvae in mosquitoes: With a note on the species found in “wild” mosquitoes on the Kenya coast. J. Helmintho. 1959, 33, 233–256. [Google Scholar] [CrossRef]
- Yen, P.K.F.; Zaman, V.; Mak, J.W. Identification of some common infective filarial larvae in Malaysia. J. Helminthol. 1982, 56, 69–80. [Google Scholar] [CrossRef]
- Orihel, T.C.; Ash, L.R.; Ramachandran, C.P.; Ottesen, E.A. Bench Aids for the Diagnosis of Filarial Infections; World Health Organization: Geneva, Switzerland, 1997. [Google Scholar]
- Casiraghi, M.; Anderson, T.J.; Bandi, C.; Bazzocchi, C.; Genchi, C. A phylogenetic analysis of filarial nematodes: Comparison with the phylogeny of Wolbachia endosymbionts. Parasitology 2001, 122, 93–103. [Google Scholar] [CrossRef]
- Turba, M.E.; Zambon, E.; Zannoni, A.; Russo, S.; Gentilini, F. Detection of Wolbachia DNA in blood for diagnosing filaria-associated syndromes in cats. J. Clin. Microbiol. 2012, 50, 2624–2630. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic. Acids. Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]


| Species | Microscopic Examination (%) | Molecular Detection (%) | |
|---|---|---|---|
| COI | ITS1 | ||
| Dirofilaria spp. | 11(19.30%) | ||
| Brugia spp. | 5 (8.77%) | ||
| D. immitis | 33 (57.89%) | 30 (52.63%) | |
| B. pahangi | 13 (22.81%) | 11 (19.30%) | |
| B. malayi | 4 (7.02%) | 0 | |
| Total | 16 (28.07%) | 50 (87.72%) | 41 (71.93%) |
| Species | Positive for Wolbachia (%) |
|---|---|
| D. immitis | 33 (57.89%) |
| B. pahangi | 11 (19.30%) |
| B. malayi | 3 (5.26%) |
| Total | 47 (82.45%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satjawongvanit, H.; Phumee, A.; Tiawsirisup, S.; Sungpradit, S.; Brownell, N.; Siriyasatien, P.; Preativatanyou, K. Molecular Analysis of Canine Filaria and Its Wolbachia Endosymbionts in Domestic Dogs Collected from Two Animal University Hospitals in Bangkok Metropolitan Region, Thailand. Pathogens 2019, 8, 114. https://doi.org/10.3390/pathogens8030114
Satjawongvanit H, Phumee A, Tiawsirisup S, Sungpradit S, Brownell N, Siriyasatien P, Preativatanyou K. Molecular Analysis of Canine Filaria and Its Wolbachia Endosymbionts in Domestic Dogs Collected from Two Animal University Hospitals in Bangkok Metropolitan Region, Thailand. Pathogens. 2019; 8(3):114. https://doi.org/10.3390/pathogens8030114
Chicago/Turabian StyleSatjawongvanit, Hathaithip, Atchara Phumee, Sonthaya Tiawsirisup, Sivapong Sungpradit, Narisa Brownell, Padet Siriyasatien, and Kanok Preativatanyou. 2019. "Molecular Analysis of Canine Filaria and Its Wolbachia Endosymbionts in Domestic Dogs Collected from Two Animal University Hospitals in Bangkok Metropolitan Region, Thailand" Pathogens 8, no. 3: 114. https://doi.org/10.3390/pathogens8030114
APA StyleSatjawongvanit, H., Phumee, A., Tiawsirisup, S., Sungpradit, S., Brownell, N., Siriyasatien, P., & Preativatanyou, K. (2019). Molecular Analysis of Canine Filaria and Its Wolbachia Endosymbionts in Domestic Dogs Collected from Two Animal University Hospitals in Bangkok Metropolitan Region, Thailand. Pathogens, 8(3), 114. https://doi.org/10.3390/pathogens8030114







