Dancing with the Dust Devil: Examining the Lung Mycobiome of Sonoran Desert Wild Mammals and the Effect of Coccidioides Presence
Abstract
1. Introduction
2. Methods
2.1. Samples
2.2. DNA Extraction
2.3. Molecular Detection of Coccidioides
2.4. Amplicon Sequencing
2.5. Microbiome Data Processing
2.6. Animal-Associated Fungal Genera
2.7. Statistical Analysis
3. Results
3.1. Coccidioides Detection
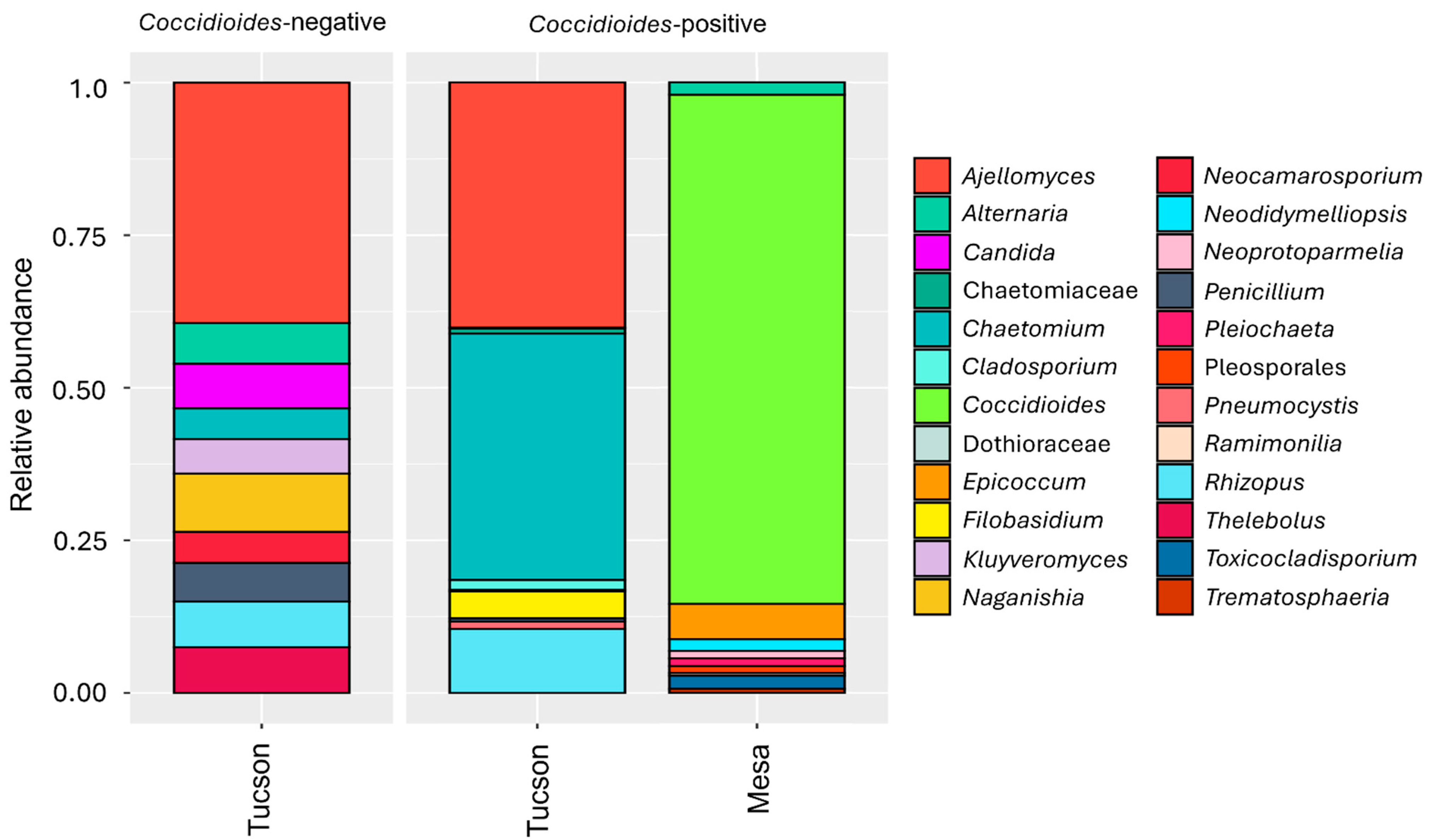
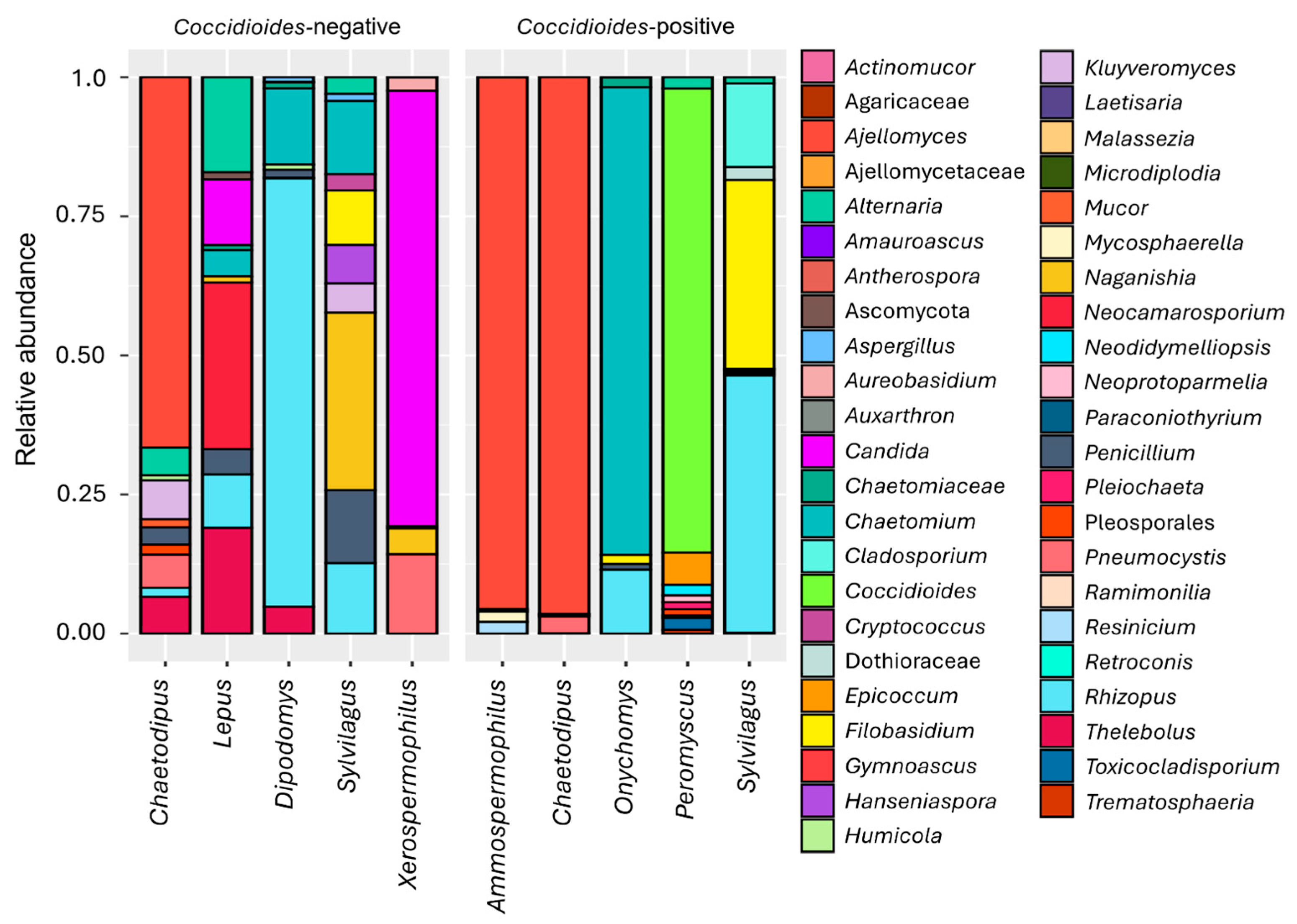
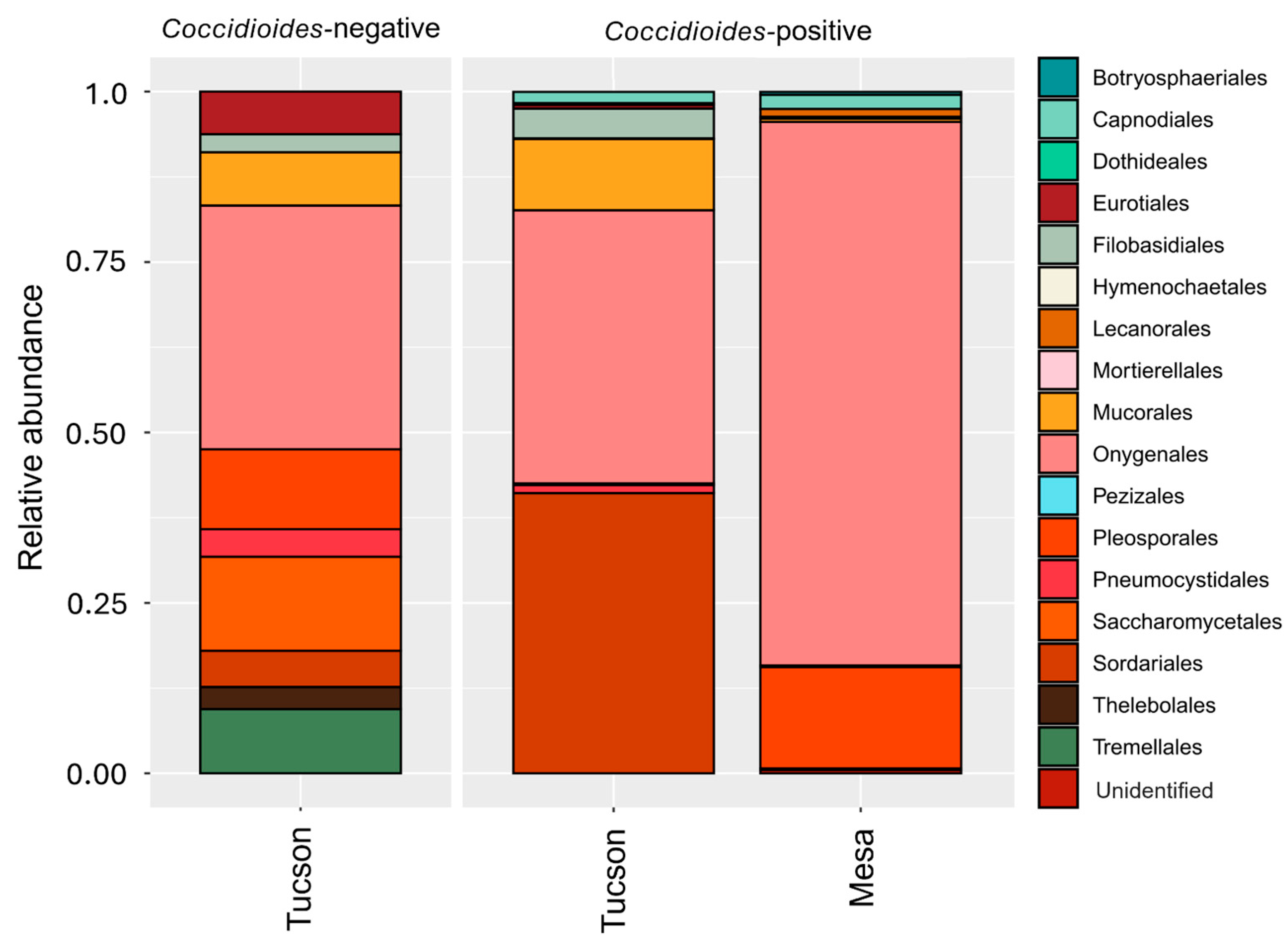
3.2. Fungal Microbiome
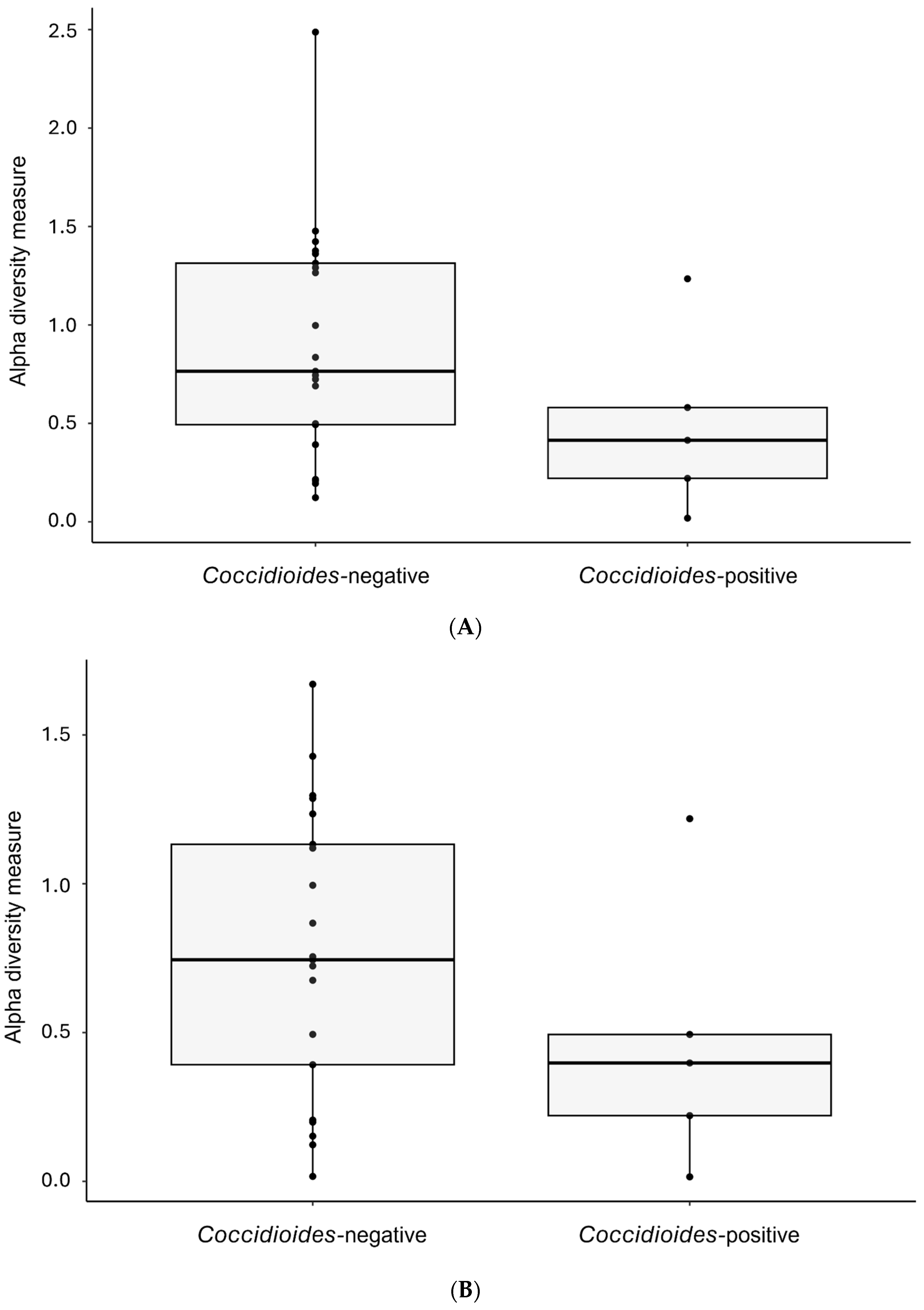
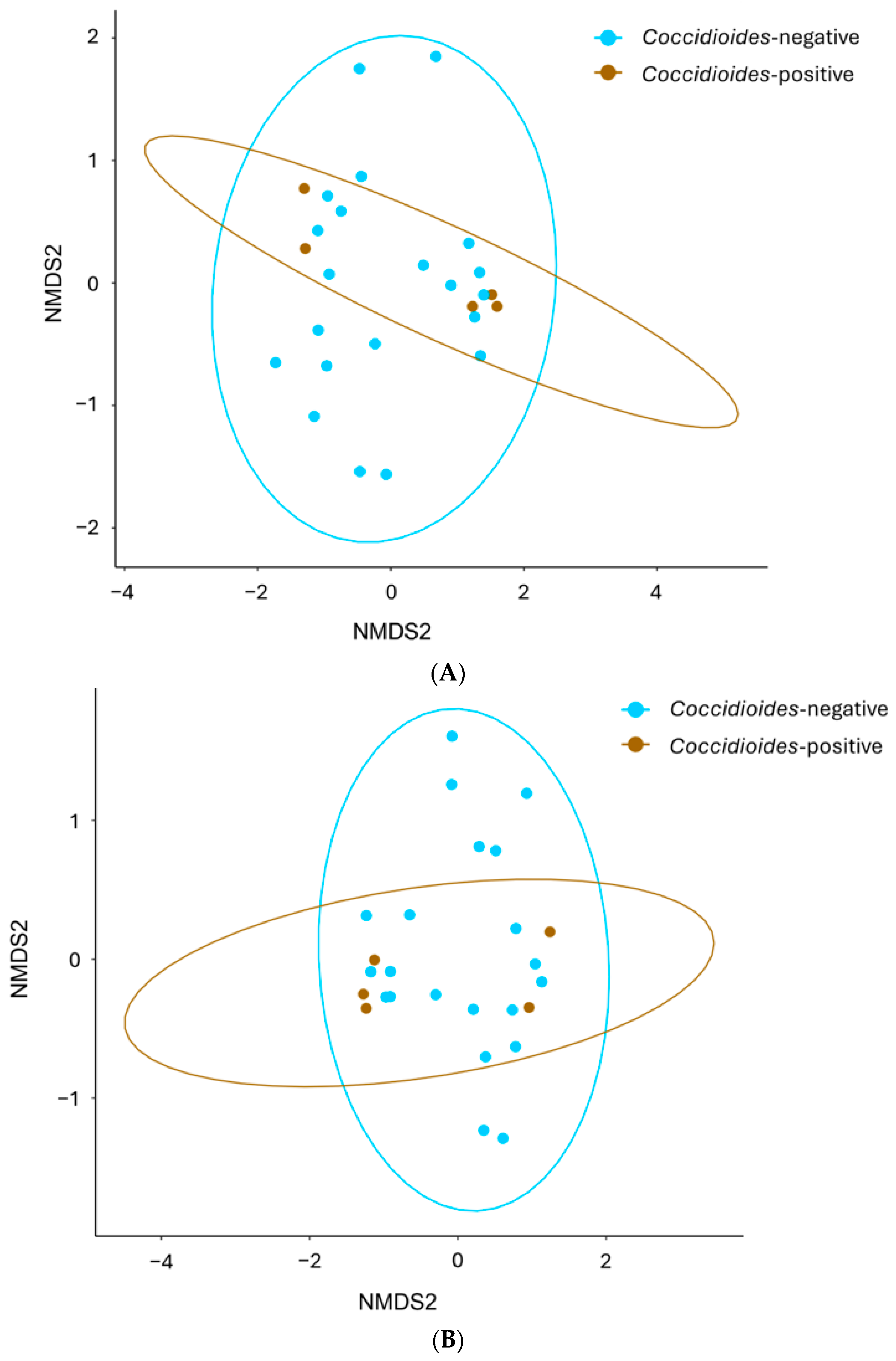
3.3. Comparisons of Alpha and Beta Diversity
3.4. Animal-Associated Fungal Community
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Melo-Neto, M.F.; Maranhão, F.C.A.; Guimarães, P.S.; Silva, D.M.W. Mycobiota recovered from the trachea and lungs of pigeons (Columba livia) captured in a grain mill. Res. Soc. Dev. 2022, 11, e57211326802. [Google Scholar] [CrossRef]
- Salazar-Hamm, P.S.; Gadek, C.R.; Mann, M.A.; Steinberg, M.; Montoya, K.N.; Behnia, M.; Gyllenhaal, E.F.; Brady, S.S.; Takano, O.M.; Williamson, J.L.; et al. Phylogenetic and ecological drivers of the avian lung mycobiome and its potentially pathogenic component. Commun. Biol. 2025, 8, 634. [Google Scholar] [CrossRef]
- Salazar-Hamm, P.S.; Montoya, K.N.; Montoya, L.; Cook, K.; Liphardt, S.; Taylor, J.W.; Cook, J.A.; Natvig, D.O. Breathing can be dangerous: Opportunistic fungal pathogens and the diverse community of the small mammal lung mycobiome. Front. Fungal Biol. 2022, 3, 996574. [Google Scholar] [CrossRef] [PubMed]
- Babb-Biernacki, S.J.; Esselstyn, J.A.; Doyle, V.P. Predicting Species Boundaries and Assessing Undescribed Diversity in Pneumocystis, an Obligate Lung Symbiont. J. Fungi 2022, 8, 799. [Google Scholar] [CrossRef] [PubMed]
- Allender, M.C.; Baker, S.; Britton, M.; Kent, A.D. Snake fungal disease alters skin bacterial and fungal diversity in an endangered rattlesnake. Sci. Rep. 2018, 8, 12147. [Google Scholar] [CrossRef]
- Jani, A.J.; Briggs, C.J. The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc. Natl. Acad. Sci. USA 2014, 111, E5049–E5058. [Google Scholar] [CrossRef]
- Lloyd, M.M.; Pespeni, M.H. Microbiome shifts with onset and progression of Sea Star Wasting Disease revealed through time course sampling. Sci. Rep. 2018, 8, 16476. [Google Scholar] [CrossRef]
- Richardson, H.; Dicker, A.J.; Barclay, H.; Chalmers, J.D. The microbiome in bronchiectasis. Eur. Respir. Rev. 2019, 28, 190048. [Google Scholar] [CrossRef]
- Richardson, M.; Bowyer, P.; Sabino, R. The human lung and Aspergillus: You are what you breathe in? Med. Mycol. 2019, 57, S145–S154. [Google Scholar] [CrossRef]
- Li, R.; Li, J.; Zhou, X. Lung microbiome: New insights into the pathogenesis of respiratory diseases. Signal Transduct. Target. Ther. 2024, 9, 19. [Google Scholar] [CrossRef]
- Natalini, J.G.; Singh, S.; Segal, L.N. The dynamic lung microbiome in health and disease. Nat. Rev. Microbiol. 2023, 21, 222–235. [Google Scholar] [CrossRef]
- Hamm, P.S.; Taylor, J.W.; Cook, J.A.; Natvig, D.O. Decades-old studies of fungi associated with mammalian lungs and modern DNA sequencing approaches help define the nature of the lung mycobiome. PLoS Pathog. 2020, 16, e1008684. [Google Scholar] [CrossRef]
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; van den Brandt, P.A.; Stobberingh, E.E. Factors Influencing the Composition of the Intestinal Microbiota in Early Infancy. Pediatrics 2006, 118, 511–521. [Google Scholar] [CrossRef]
- Mitchell, C.M.; Mazzoni, C.; Hogstrom, L.; Bryant, A.; Bergerat, A.; Cher, A.; Pochan, S.; Herman, P.; Carrigan, M.; Sharp, K.; et al. Delivery Mode Affects Stability of Early Infant Gut Microbiota. Cell Rep. Med. 2020, 1, 100156. [Google Scholar] [CrossRef]
- Reyman, M.; van Houten, M.A.; van Baarle, D.; Bosch, A.A.T.M.; Man, W.H.; Chu, M.L.J.N.; Arp, K.; Watson, R.L.; Sanders, E.A.M.; Fuentes, S.; et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019, 10, 4997. [Google Scholar] [CrossRef]
- Badal, V.D.; Vaccariello, E.D.; Murray, E.R.; Yu, K.E.; Knight, R.; Jeste, D.V.; Nguyen, T.T. The Gut Microbiome, Aging, and Longevity: A Systematic Review. Nutrients 2020, 12, 3759. [Google Scholar] [CrossRef]
- Bartley, J.M.; Zhou, X.; Kuchel, G.A.; Weinstock, G.M.; Haynes, L. Impact of Age, Caloric Restriction, and Influenza Infection on Mouse Gut Microbiome: An Exploratory Study of the Role of Age-Related Microbiome Changes on Influenza Responses. Front. Immunol. 2017, 8, 1164. [Google Scholar] [CrossRef]
- Emmons, C.W.; Ashburn, L.L. The Isolation of Haplosporangium parvum n. sp. and Coccidioides immitis from Wild Rodents. Their Relationship to Coccidioidomycosis. Public Health Rep. (1896–1970) 1942, 57, 1715–1727. [Google Scholar] [CrossRef]
- Emmons, C.W. Coccidioidomycosis in Wild Rodents. A Method of Determining the Extent of Endemic Areas. Public Health Rep. (1896–1970) 1943, 58, 1–5. [Google Scholar] [CrossRef]
- Bakerspigel, A. Haplosporangium in Saskatchewan Rodents. Mycologia 1956, 48, 568–572. [Google Scholar] [CrossRef]
- Jellison, W.L. Haplomycosis in Montana Rabbits, Rodents, and Carnivores. Public Health Rep. (1896–1970) 1950, 65, 1057–1063. [Google Scholar] [CrossRef]
- Delhaes, L.; Monchy, S.; Fréalle, E.; Hubans, C.; Salleron, J.; Leroy, S.; Prevotat, A.; Wallet, F.; Wallaert, B.; Dei-Cas, E.; et al. The Airway Microbiota in Cystic Fibrosis: A Complex Fungal and Bacterial Community—Implications for Therapeutic Management. PLoS ONE 2012, 7, e36313. [Google Scholar] [CrossRef]
- Kollath, D.R.; Dolby, G.A.; Webster, T.H.; Barker, B.M. Characterizing fungal communities on Mojave desert tortoises (Gopherus agassizii) in Arizona to uncover potential pathogens. J. Arid Environ. 2023, 219, 105089. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, K.; Ma, H.; Deng, J.; Fang, C.; Zhao, H.; An, X.; Zhang, J.; Wang, Q.; Jiang, W.; et al. Seasonality Has Greater Influence on Amphibian Cutaneous Mycobiome than Host Species. J. Fungi 2025, 11, 473. [Google Scholar] [CrossRef]
- Kearns, P.J.; Fischer, S.; Fernández-Beaskoetxea, S.; Gabor, C.R.; Bosch, J.; Bowen, J.L.; Tlusty, M.F.; Woodhams, D.C. Fight Fungi with Fungi: Antifungal Properties of the Amphibian Mycobiome. Front. Microbiol. 2017, 8, 2494. [Google Scholar] [CrossRef] [PubMed]
- Kearns, P.J.; Winter, A.S.; Woodhams, D.C.; Northup, D.E. The Mycobiome of Bats in the American Southwest Is Structured by Geography, Bat Species, and Behavior. Microb. Ecol. 2023, 86, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, S.B.; Stephens, W.Z.; Greenhalgh, R.; Round, J.L.; Dearing, M.D. Wild herbivorous mammals (genus Neotoma) host a diverse but transient assemblage of fungi. Symbiosis 2022, 87, 45–58. [Google Scholar] [CrossRef]
- Vargas-Gastélum, L.; Romer Alexander, S.; Ghotbi, M.; Dallas Jason, W.; Alexander, N.R.; Moe Kylie, C.; McPhail Kerry, L.; Neuhaus George, F.; Shadmani, L.; Spatafora Joseph, W.; et al. Herptile gut microbiomes: A natural system to study multi-kingdom interactions between filamentous fungi and bacteria. mSphere 2024, 9, e00475-23. [Google Scholar] [CrossRef]
- Hathaway, J.J.M.; Salazar-Hamm, P.S.; Caimi, N.A.; Natvig, D.O.; Buecher, D.C.; Northup, D.E. Comparison of Fungal and Bacterial Microbiomes of Bats and Their Cave Roosting Environments at El Malpais National Monument, New Mexico, USA. Geomicrobiol. J. 2024, 41, 82–97. [Google Scholar] [CrossRef]
- Zapanta, K.; Kavanagh, M.; Keller, K.; Nguyen, L.; Rosenkrantz, W.; Krumbeck, J.A. The cutaneous microbiota and Nannizziomycosis in bearded dragons (Pogona vitticeps): Associations between infectious Nannizziopsis species and common bacterial pathogens. Vet. Dermatol. 2025, 36, 506–515. [Google Scholar] [CrossRef]
- Insuk, C.; Cheeptham, N.; Lausen, C.; Xu, J. DNA metabarcoding analyses reveal fine-scale microbiome structures on Western Canadian bat wings. Microbiol. Spectr. 2024, 12, e00376-24. [Google Scholar] [CrossRef]
- Fisher, M.C.; Koenig, G.L.; White, T.J.; Taylor, J.W. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 2002, 94, 73–84. [Google Scholar] [CrossRef]
- de Perio, M.; Niemeier, R.T.; Burr, G. Coccidioides Exposure and Coccidioidomycosis among Prison Employees, California, United States. Emerg. Infect. Dis. J. 2015, 21, 1031. [Google Scholar] [CrossRef] [PubMed]
- Wack, E.E.; Ampel, N.M.; Sunenshine, R.H.; Galgiani, J.N. The Return of Delayed-Type Hypersensitivity Skin Testing for Coccidioidomycosis. Clin. Infect. Dis. 2015, 61, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Drips, W., Jr.; Smith, C.E. Epidemiology of Coccidioidomycosis: A Contemporary Military Experience. JAMA 1964, 190, 1010–1012. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.E.; Saito, M.T.; Simons, S.A. Pattern of 39,500 serologic tests in coccidioidomycosis. J. Am. Med. Assoc. 1956, 160, 546–552. [Google Scholar] [CrossRef]
- CDC, C.f.D.C.a.P. Valley Fever (Coccidioidomycosis). Available online: https://www.cdc.gov/valley-fever/php/statistics/index.html (accessed on 3 March 2025).
- Grizzle, A.J.; Wilson, L.; Nix, D.E.; Galgiani, J.N. Clinical and Economic Burden of Valley Fever in Arizona: An Incidence-Based Cost-of-Illness Analysis. Open Forum Infect. Dis. 2021, 8, ofaa623. [Google Scholar] [CrossRef] [PubMed]
- Barker, B.M. Coccidioidomycosis in Animals. In Emerging and Epizootic Fungal Infections in Animals; Seyedmousavi, S., de Hoog, G.S., Guillot, J., Verweij, P.E., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 81–114. [Google Scholar] [CrossRef]
- Taylor, J.W.; Barker, B.M. The endozoan, small-mammal reservoir hypothesis and the life cycle of Coccidioides species. Med. Mycol. 2019, 57, S16–S20. [Google Scholar] [CrossRef]
- Andreote, F.D.; Gumiere, T.; Durrer, A. Exploring interactions of plant microbiomes. Sci. Agrícola 2014, 71, 528–539. [Google Scholar] [CrossRef]
- Elconin, A.F.; Egeberg, R.O.; Egeberg, M.C. Significance of soil salinity on the ecology of Coccidioides immitis. J. Bacteriol. 1964, 87, 500–503. [Google Scholar] [CrossRef]
- Lacy, G.H.; Swatek, F.E. Soil Ecology of Coccidioides immitis at Amerindian Middens in California. Appl. Microbiol. 1974, 27, 379–388. [Google Scholar] [CrossRef]
- Chow, N.A.; Kangiser, D.; Gade, L.; McCotter Orion, Z.; Hurst, S.; Salamone, A.; Wohrle, R.; Clifford, W.; Kim, S.; Salah, Z.; et al. Factors Influencing Distribution of Coccidioides immitis in Soil, Washington State, 2016. mSphere 2021, 6, e00598-21. [Google Scholar] [CrossRef]
- Kollath, D.R.; Teixeira, M.M.; Funke, A.; Miller, K.J.; Barker, B.M. Investigating the Role of Animal Burrows on the Ecology and Distribution of Coccidioides spp. in Arizona Soils. Mycopathologia 2020, 185, 145–159. [Google Scholar] [CrossRef]
- Ramsey, M.; Barker Bridget, M. Investigating Prevalence of Coccidioides Over 3 Years (2022–2025) at a Single Site in Mesa, Arizona, United States; Northern Arizona University: Flagstaff, AZ, USA, 2025; to be submitted. [Google Scholar]
- Lauer, A.; Baal, J.D.H.; Baal, J.C.H.; Verma, M.; Chen, J.M. Detection of Coccidioides immitis in Kern County, California, by multiplex PCR. Mycologia 2012, 104, 62–69. [Google Scholar] [CrossRef]
- Bowers, J.R.; Parise, K.L.; Kelley, E.J.; Lemmer, D.; Schupp, J.M.; Driebe, E.M.; Engelthaler, D.M.; Keim, P.; Barker, B.M. Direct detection of Coccidioides from Arizona soils using CocciENV, a highly sensitive and specific real-time PCR assay. Med. Mycol. 2019, 57, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Litvintseva, A.P.; Marsden-Haug, N.; Hurst, S.; Hill, H.; Gade, L.; Driebe, E.M.; Ralston, C.; Roe, C.; Barker, B.M.; Goldoft, M.; et al. Valley Fever: Finding New Places for an Old Disease: Coccidioides immitis Found in Washington State Soil Associated with Recent Human Infection. Clin. Infect. Dis. 2015, 60, e1–e3. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.L.; Walters William, A.; Lennon Niall, J.; Bochicchio, J.; Krohn, A.; Caporaso, J.G.; Pennanen, T. Accurate Estimation of Fungal Diversity and Abundance through Improved Lineage-Specific Primers Optimized for Illumina Amplicon Sequencing. Appl. Environ. Microbiol. 2016, 82, 7217–7226. [Google Scholar] [CrossRef] [PubMed]
- Colman, R.E.; Schupp, J.M.; Hicks, N.D.; Smith, D.E.; Buchhagen, J.L.; Valafar, F.; Crudu, V.; Romancenco, E.; Noroc, E.; Jackson, L.; et al. Detection of Low-Level Mixed-Population Drug Resistance in Mycobacterium tuberculosis Using High Fidelity Amplicon Sequencing. PLoS ONE 2015, 10, e0126626. [Google Scholar] [CrossRef]
- Martin, T.; Lu, S.-W.; van Tilbeurgh, H.; Ripoll, D.R.; Dixelius, C.; Turgeon, B.G.; Debuchy, R. Tracing the Origin of the Fungal α1 Domain Places Its Ancestor in the HMG-Box Superfamily: Implication for Fungal Mating-Type Evolution. PLoS ONE 2010, 5, e15199. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Van Nguyen, H.; Lavenier, D. PLAST: Parallel local alignment search tool for database comparison. BMC Bioinform. 2009, 10, 329. [Google Scholar] [CrossRef]
- Hibbett, D.; Abarenkov, K.; Kõljalg, U.; Öpik, M.; Chai, B.; Cole, J.; Wang, Q.; Crous, P.; Robert, V.; Helgason, T.; et al. Sequence-based classification and identification of Fungi. Mycologia 2016, 108, 1049–1068. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S. Microbiome R Package, Bioconductor. 2017. Available online: https://github.com/microbiome/microbiome (accessed on 6 August 2025).
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; McGlinn, D.; Szoecs, E.; et al. Vegan: Community Ecology Package, 2.5-6. 2019. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 6 August 2025).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation. 2023. Available online: https://github.com/tidyverse/dplyr (accessed on 6 August 2025).
- Araujo, G.; Montoya, J.M.; Thomas, T.; Webster, N.S.; Lurgi, M. A mechanistic framework for complex microbe-host symbioses. Trends Microbiol. 2025, 33, 96–111. [Google Scholar] [CrossRef]
- Müller, L.; Gonçalves, G.L.; Cordeiro-Estrela, P.; Marinho, J.R.; Althoff, S.L.; Testoni, A.F.; González, E.M.; Freitas, T.R.O. DNA Barcoding of Sigmodontine Rodents: Identifying Wildlife Reservoirs of Zoonoses. PLoS ONE 2013, 8, e80282. [Google Scholar] [CrossRef]
- Madan, J.C.; Koestler, D.C.; Stanton, B.A.; Davidson, L.; Moulton, L.A.; Housman, M.L.; Moore, J.H.; Guill, M.F.; Morrison, H.G.; Sogin, M.L.; et al. Serial Analysis of the Gut and Respiratory Microbiome in Cystic Fibrosis in Infancy: Interaction between Intestinal and Respiratory Tracts and Impact of Nutritional Exposures. mBio 2012, 3, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Coburn, B.; Wang, P.W.; Diaz Caballero, J.; Clark, S.T.; Brahma, V.; Donaldson, S.; Zhang, Y.; Surendra, A.; Gong, Y.; Elizabeth Tullis, D.; et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci. Rep. 2015, 5, 10241. [Google Scholar] [CrossRef]
- Chen, R.; Wang, L.; Koch, T.; Curtis, V.; Yin-DeClue, H.; Handley, S.A.; Shan, L.; Holtzman, M.J.; Castro, M.; Wang, L. Sex effects in the association between airway microbiome and asthma. Ann. Allergy Asthma Immunol. 2020, 125, 652–657.e653. [Google Scholar] [CrossRef]
- Moitas, B.; Caldas, I.M.; Sampaio-Maia, B. Microbiology and postmortem interval: A systematic review. Forensic Sci. Med. Pathol. 2024, 20, 696–715. [Google Scholar] [CrossRef] [PubMed]
- Pechal, J.L.; Schmidt, C.J.; Jordan, H.R.; Benbow, M.E. Frozen: Thawing and Its Effect on the Postmortem Microbiome in Two Pediatric Cases. J. Forensic Sci. 2017, 62, 1399–1405. [Google Scholar] [CrossRef] [PubMed]


| Site | Host Species | ID | CT Value (CocciDx) | Amplicon Count 1 |
|---|---|---|---|---|
| Tucson | Chaetodipus | PM_4 | 31 | 56 (0.04%) |
| PM_5 | 32 | 111 (0.18%) | ||
| Sylvilagus | CT_6 | 38.7 | ||
| Onychomys | GH_M | 39.6 | ||
| Ammospermophilus | A_GS | 39.8 | ||
| Mesa | Peromyscus | 17L | 38.9 | |
| 24L | 34.5 | |||
| 29L | 38.5 | |||
| 31L | 28.6 | |||
| 33L | 36.3 | 9 (11.4%) | ||
| 34L | 31.5 | 431(60.4%) | ||
| 35L | 31.5 | 3606 (95.9%) | ||
| 36L | 33.7 | |||
| 37L | 34.1 | |||
| 38L | 36.4 | 193 (89.8%) | ||
| 39L | 31.7 | |||
| 40L | 34.1 | |||
| 41L | 33.3 | |||
| 43L | 33.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabio-Braga, A.; Salois, J.; Bryant, M.L.; Kollath, D.R.; Barker, B. Dancing with the Dust Devil: Examining the Lung Mycobiome of Sonoran Desert Wild Mammals and the Effect of Coccidioides Presence. Pathogens 2025, 14, 807. https://doi.org/10.3390/pathogens14080807
Fabio-Braga A, Salois J, Bryant ML, Kollath DR, Barker B. Dancing with the Dust Devil: Examining the Lung Mycobiome of Sonoran Desert Wild Mammals and the Effect of Coccidioides Presence. Pathogens. 2025; 14(8):807. https://doi.org/10.3390/pathogens14080807
Chicago/Turabian StyleFabio-Braga, Ana, Jaida Salois, Mitchell L. Bryant, Daniel R. Kollath, and Bridget Barker. 2025. "Dancing with the Dust Devil: Examining the Lung Mycobiome of Sonoran Desert Wild Mammals and the Effect of Coccidioides Presence" Pathogens 14, no. 8: 807. https://doi.org/10.3390/pathogens14080807
APA StyleFabio-Braga, A., Salois, J., Bryant, M. L., Kollath, D. R., & Barker, B. (2025). Dancing with the Dust Devil: Examining the Lung Mycobiome of Sonoran Desert Wild Mammals and the Effect of Coccidioides Presence. Pathogens, 14(8), 807. https://doi.org/10.3390/pathogens14080807








