Serotype Distribution of Aggregatibacter actinomycetemcomitans in Periodontitis Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Characterictics
2.2. PCR Detection of SEROTYPES
2.3. Statistical Analysis
2.4. Ethical Considerations
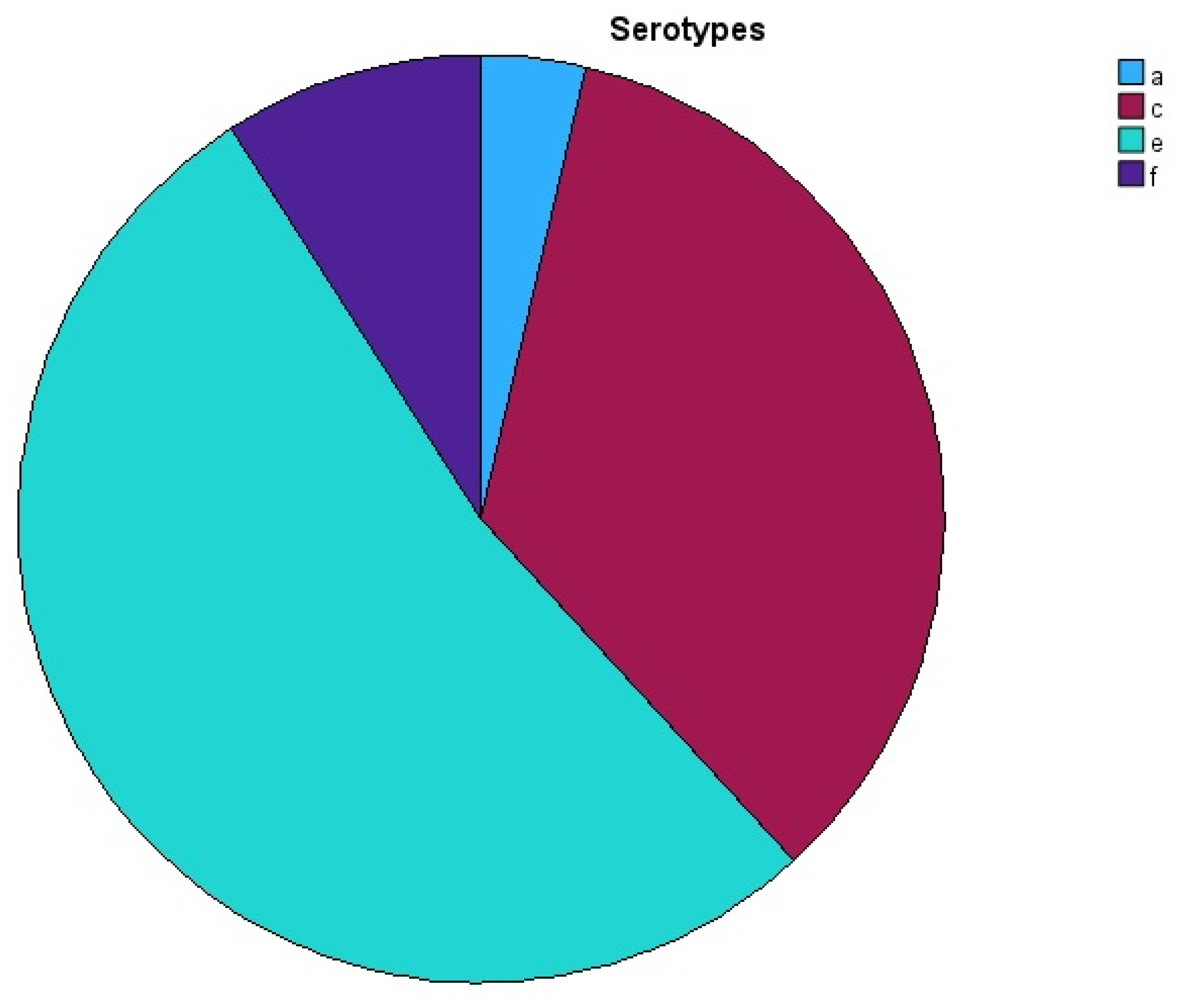
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abusleme, L.; Hoare, A.; Hong, B.Y.; Diaz, P.I. Microbial signatures of health, gingivitis, and periodontitis. Periodontol. 2000 2021, 86, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Monsarrat, P.; Blaizot, A.; Kémoun, P.; Ravaud, P.; Nabet, C.; Sixou, M.; Vergnes, J.N. Clinical research activity in periodontal medicine: A systematic mapping of trial registers. J. Clin. Periodontol. 2016, 43, 390–400. [Google Scholar] [CrossRef]
- Villoria, G.E.M.; Fischer, R.G.; Tinoco, E.M.B.; Meyle, J.; Loos, B.G. Periodontal disease: A systemic condition. Periodontology 2000 2024, 96, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Curtis, M.A.; Darveau, R.P.; Curtis, M.A. Microbial transitions from health to disease. Periodontology 2000 2021, 86, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.; Ward, J.M.; Ready, D. Aggregatibacter (Actinobacillus) actinomycetemcomitans: A triple A* periodontopathogen? Periodontol. 2000 2010, 54, 78–105. [Google Scholar] [CrossRef]
- Zambon, J.J. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 1985, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.C.; McArthur, W.P.; Baehni, P.C.; Hammond, B.F.; Taichman, N.S. Extraction and partial characterization of a leukotoxin from a plaque-derived Gram-negative microorganism. Infect. Immun. 1979, 25, 427–439. [Google Scholar] [CrossRef]
- Zambon, J.J.; Slots, J.; Genco, R.J. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect. Immun. 1983, 41, 19–27. [Google Scholar] [CrossRef]
- Huang, Y.; Kittichotirat, W.; Mayer, M.P.; Hall, R.; Bumgarner, R.; Chen, C. Comparative genomic hybridization and transcriptome analysis with a pan-genome microarray reveal distinctions between JP2 and non-JP2 genotypes of Aggregatibacter actinomycetemcomitans. Mol. Oral. Microbiol. 2013, 28, 1–17. [Google Scholar] [CrossRef]
- Kaplan, J.B.; Schreiner, H.C.; Furgang, D.; Fine, D.H. Population Structure and Genetic Diversity of Actinobacillus actinomycetemcomitans Strains Isolated from Localized Juvenile Periodontitis Patients. J. Clin. Microbiol. 2002, 40, 1181–1187. [Google Scholar] [CrossRef]
- Kilian, M.; Frandsen, E.V.; Haubek, D.; Poulsen, K. The etiology of periodontal disease revisited by population genetic analysis. Periodontology 2000 2006, 42, 158–179. [Google Scholar] [CrossRef]
- Bajanca, P.; Caniça, M. Emergence of nonencapsulated and encapsulated non-b-type invasive Haemophilus influenzae isolates in Portugal (1989–2001). J. Clin. Microbiol. 2004, 42, 807–810. [Google Scholar] [CrossRef]
- Ashton, F.E.; Ryan, J.A.; Borczyk, A.; Caugant, D.A.; Mancino, L.; Huang, D. Emergence of a virulent clone of Neisseria meningitidis serotype 2a that is associated with meningococcal group C disease in Canada. J. Clin. Microbiol. 1991, 29, 2489–2493. [Google Scholar] [CrossRef]
- van Winkelhoff, A.J.; Slots, J. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in nonoral infections. Periodontology 2000 1999, 20, 122–135. [Google Scholar] [CrossRef]
- Kim, T.S.; Frank, P.; Eickholz, P.; Eick, S.; Kim, C.K. Serotypes of Aggregatibacter actinomycetemcomitans in patients with different ethnic backgrounds. J. Periodontol. 2009, 80, 2020–2027. [Google Scholar] [CrossRef] [PubMed]
- Brogan, J.M.; Lally, E.T.; Poulsen, K.; Kilian, M.; Demuth, D.R. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: Analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect. Immun. 1994, 62, 501–508. [Google Scholar] [CrossRef]
- Haubek, D.; Ennibi, O.K.; Poulsen, K.; Poulsen, S.; Benzarti, N.; Kilian, M. Early-onset periodontitis in Morocco is associated with the highly leukotoxic clone of Actinobacillus actinomycetemcomitans. J. Dent. Res. 2001, 80, 1580–1583. [Google Scholar] [CrossRef]
- Brígido, J.A.; da Silveira, V.R.; Rego, R.O.; Nogueira, N.A. Serotypes of Aggregatibacter actinomycetemcomitans in relation to periodontal status and geographic origin of individuals-a review of the literature. Med. Oral. Patol. Oral. Cir. Bucal 2014, 19, e184–e191. [Google Scholar] [CrossRef]
- Yamamoto, M.; Nishihara, T.; Koseki, T.; He, T.; Yamato, K.; Zhang, Y.J.; Nakashima, K.; Oda, S.; Ishikawa, I. Prevalence of Actinobacillus actinomycetemcomitans serotypes in Japanese patients with periodontitis. J. Periodontal Res. 1997, 32, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Celenligil, H.; Ebersole, J.L. Analysis of serum antibody responses to periodontopathogens in early-onset periodontitis patients from different geographical locations. J. Clin. Periodontol. 1998, 25, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, L.; Rebeis, E.S.; Martins, E.D.S.; Sekiguchi, R.T.; Ando-Suguimoto, E.S.; Mafra, C.E.S.; Holzhausen, M.; Romito, G.A.; Mayer, M.P.A. IgG sera levels against a subset of periodontopathogens and severity of disease in aggressive periodontitis patients: A cross-sectional study of selected pocket sites. J. Clin. Periodontol. 2014, 41, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Saarela, M.; Asikainen, S.; Alaluusua, S.; Pyhälä, L.; Lai, C.H.; Jousimies-Somer, H. Frequency and stability of mono- or poly-infection by Actinobacillus actinomycetemcomitans serotypes a, b, c, d or e. Oral. Microbiol. Immunol. 1992, 7, 277–279. [Google Scholar] [CrossRef]
- Thiha, K.; Takeuchi, Y.; Umeda, M.; Huang, Y.; Ohnishi, M.; Ishikawa, I. Identification of periodontopathic bacteria in gingival tissue of Japanese periodontitis patients. Oral. Microbiol. Immunol. 2007, 22, 201–207. [Google Scholar] [CrossRef]
- Rylev, M.; Kilian, M. Prevalence and distribution of principal periodontal pathogens worldwide. J. Clin. Periodontol. 2008, 35, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Bandhaya, P.; Saraithong, P.; Likittanasombat, K.; Hengprasith, B.; Torrungruang, K. Aggregatibacter actinomycetemcomitans serotypes, the JP2 clone and cytolethal distending toxin genes in a Thai population. J. Clin. Periodontol. 2012, 39, 519–525. [Google Scholar] [CrossRef]
- Claesson, R.; Höglund-Åberg, C.; Haubek, D.; Johansson, A. Age-related prevalence and characteristics of Aggregatibacter actinomycetemcomitans in periodontitis patients living in Sweden. J. Oral. Microbiol. 2017, 9, 1334504. [Google Scholar] [CrossRef]
- Khzam, N.; Kujan, O.; Haubek, D.; Arslan, A.; Johansson, A.; Oscarsson, J.; Razooqi, Z.; Miranda, L.A. Prevalence of Subgingival Aggregatibacter actinomycetemcomitans: Descriptive Cross-Sectional Study. Pathogens 2024, 13, 531. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89 (Suppl. S1), S159–S172. [Google Scholar] [CrossRef]
- Razooqi, Z.; Höglund Åberg, C.; Kwamin, F.; Claesson, R.; Haubek, D.; Oscarsson, J.; Johansson, A. Aggregatibacter actinomycetemcomitans and Filifactor alocis as Associated with Periodontal Attachment Loss in a Cohort of Ghanaian Adolescents. Microorganisms 2022, 10, 2511. [Google Scholar] [CrossRef] [PubMed]
- Kirakodu, S.S.; Govindaswami, M.; Novak, M.J.; Ebersole, J.L.; Novak, K.F. Optimizing qPCR for the Quantification of Periodontal Pathogens in a Complex Plaque Biofilm. Open Dent. J. 2008, 2, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.B.; Perry, M.B.; MacLean, L.L.; Furgang, D.; Wilson, M.E.; Fine, D.H. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect. Immun. 2001, 69, 5375–5384. [Google Scholar] [CrossRef]
- Mombelli, A.; Gmür, R.; Lang, N.P.; Corbet, E.; Frey, J. Actinobacillus actinomycetemcomitans in Chinese adults: Serotype distribution and analysis of the leukotoxin gene promoter locus. J. Clin. Periodontol. 1999, 26, 505–510. [Google Scholar] [CrossRef]
- Yoshida, Y.; Suzuki, N.; Nakano, Y.; Shibuya, K.; Ogawa, Y.; Koga, T. Distribution of Actinobacillus actinomycetemcomitans serotypes and Porphyromonas gingivalis in Japanese adults. Oral. Microbiol. Immunol. 2003, 18, 135–139. [Google Scholar] [CrossRef]
- Zambon, J.J.; DeLuca, C.; Slots, J.; Genco, R.J. Studies of leukotoxin from Actinobacillus actinomycetemcomitans using the promyelocytic HL-60 cell line. Infect. Immun. 1983, 40, 205–212. [Google Scholar] [CrossRef]
- Asikainen, S.; Lai, C.H.; Alaluusua, S.; Slots, J. Distribution of Actinobacillus actinomycetemcomitans serotypes in periodontal health and disease. Oral. Microbiol. Immunol. 1991, 6, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-W.; Huang, Y.-F.; Chan, Y.; Chou, M.-Y. Relationship of Actinobacillus actinomycetemcomitans serotypes to periodontal condition: Prevalence and proportions in subgingival plaque. Eur. J. Oral Sci. 2005, 113, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Sakellari, D.; Katsikari, A.; Slini, T.; Ioannidis, I.; Konstantinidis, A.; Arsenakis, M. Prevalence and distribution of Aggregatibacter actinomycetemcomitans serotypes and the JP2 clone in a Greek population. J. Clin. Periodontol. 2011, 38, 108–114. [Google Scholar] [CrossRef] [PubMed]
- van der Reijden, W.A.; Bosch-Tijhof, C.J.; van der Velden, U.; van Winkelhoff, A.J. Java project on periodontal diseases: Serotype distribution of Aggregatibacter actinomycetemcomitans and serotype dynamics over an 8-year period. J. Clin. Periodontol. 2008, 35, 487–492. [Google Scholar] [CrossRef]
- Poulsen, K.; Theilade, E.; Lally, E.T.; Demuth, D.R.; Kilian, M. Population structure of Actinobacillus actinomycetemcomitans: A framework for studies of disease-associated properties. Microbiology 1994, 140 Pt 8, 2049–2060. [Google Scholar] [CrossRef][Green Version]
- Haubek, D.; Dirienzo, J.M.; Tinoco, E.M.B.; Westergaard, J.; López, N.J.; Chung, C.-P.; Poulsen, K.; Kilian, M. Racial tropism of a highly toxic clone of Actinobacillus actinomycetemcomitans associated with juvenile periodontitis. J. Clin. Microbiol. 1997, 35, 3037–3042. [Google Scholar] [CrossRef]
- Lakio, L.; Kuula, H.; Dogan, B.; Asikainen, S. Actinobacillus actinomycetemcomitans proportion of subgingival bacterial flora in relation to its clonal type. Eur. J. Oral. Sci. 2002, 110, 212–217. [Google Scholar] [CrossRef]
- Saito, A.; Hosaka, Y.; Nakagawa, T.; Seida, K.; Yamada, S.; Takazoe, I.; Okuda, K. Significance of serum antibody against surface antigens of Actinobacillus actinomycetemcomitans in patients with adult periodontitis. Oral. Microbiol. Immunol. 1993, 8, 146–153. [Google Scholar] [CrossRef]
- Riggio, M.P.; Macfarlane, T.W.; Mackenzie, D.; Lennon, A.; Smith, A.J.; Kinane, D. Comparison of polymerase chain reaction and culture methods for detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in subgingival plaque samples. J. Periodontal Res. 1996, 31, 496–501. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Dzink, J.L.; Hillman, J.D. Associations between microbial species in subgingival plaque samples. Oral. Microbiol. Immunol. 1988, 3, 1–7. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef]
- Chung, H.J.; Chung, C.P.; Son, S.H.; Nisengard, R.J. Actinobacillus actinomycetemcomitans serotypes and leukotoxicity in Korean localized juvenile periodontitis. J. Periodontol. 1989, 60, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Doğan, B.; Saarela, M.H.; Jousimies-Somer, H.; Alaluusua, S.; Asikainen, S. Actinobacillus actinomycetemcomitans serotype e—Biotypes, genetic diversity and distribution in relation to periodontal status. Oral. Microbiol. Immunol. 1999, 14, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Fine, D.H.; Markowitz, K.; Furgang, D.; Fairlie, K.; Ferrandiz, J.; Nasri, C.; McKiernan, M.; Gunsolley, J. Aggregatibacter actinomycetemcomitans and Its Relationship to Initiation of Localized Aggressive Periodontitis: Longitudinal Cohort Study of Initially Healthy Adolescents. J. Clin. Microbiol. 2007, 45, 3859–3869. [Google Scholar] [CrossRef] [PubMed]
- Dahlén, G.; Widar, F.; Teanpaisan, R.; Papapanou, P.N.; Baelum, V.; Fejerskov, O. Actinobacillus actinomycetemcomitans in a rural adult population in southern Thailand. Oral. Microbiol. Immunol. 2002, 17, 137–142. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Primer Sequence 5′→3′ | Size |
|---|---|---|
| Serotype b and c: forward | ARAAYTTYTCWTCGGGAATG (R = A/G; Y = C/T; W = A/T) | 333 bp |
| Serotype b: reverse | TCTCCACCATTTTTGAGTGG | 268 bp |
| Serotype c: reverse | GAAACCACTTCTATTTCTCC | |
| Serotype a: forward | GGACAAAGTGGTGTTGTTTGG | 362 bp |
| Serotype a: reverse | GCAAGCCAACTTCTACACAATG | |
| Serotype e: forward | CCTTCGACCAAACGGTAAAA | 283 bp |
| Serotype e: reverse | TTAAAAATAGCGTGCGTGAGC | |
| Serotype d: forward | TCCCAGAGGTTGGTTATTTTT | 300 bp |
| Serotype d: reverse | TTCTTTCCCAAAAACCAAGTTTA | |
| Serotype f: forward | TTGATTTTGCAGAGGTCAATG | 250 bp |
| Serotype f: reverse | TGGCAGAGAGTTTTCACTTGC | |
| Primers Annealing temperature.: 55 °C | ||
| Components | Volume/Reaction |
|---|---|
| 2x QIAGEN Multiplex PCR Master Mix | 12.5 µL |
| Forward and reverse primers | 5 µL |
| 5x Q-Solution | 5 µL |
| DNA | 2.5 µL |
| Total volume | 25 µL |
| Step | Time | Temperature |
|---|---|---|
| Initial heat activation | 15 min | 95 °C |
| 3-step cycling: denaturation | 30 s | 94 °C |
| Annealing | 90 s | 55 °C |
| Extension | 60 s | 72 °C |
| Number of cycles | 40 Cycles | |
| Final extension | 30 s | 72 °C |
| No. | ID | Serotype | No. | ID | Serotype | No. | ID | Serotype |
|---|---|---|---|---|---|---|---|---|
| 1 | 1S | c, e | 13 | 1C | c, e | 25 | 1P | c, e |
| 2 | 7S | a, c, e | 14 | 7C | - | 26 | 7P | a, e |
| 3 | 8S | c, e | 15 | 8C | e | 27 | 8P | c, e |
| 4 | 13S | c, e | 16 | 13C | c | 28 | 13P | c, e |
| 5 | 21S | e, f | 17 | 21C | e, f | 29 | 21P | e, f |
| 6 | 22S | e | 18 | 22C | - | 30 | 22P | c, e |
| 7 | 24S | e | 19 | 24C | c | 31 | 24P | e |
| 8 | 27S | e | 20 | 27C | c | 32 | 27P | c, e |
| 9 | 30S | e | 21 | 30C | - | 33 | 30P | e |
| 10 | 31S | e | 22 | 31C | - | 34 | 31P | e |
| 11 | 37S | c, e, f | 23 | 37C | c, e | 35 | 37P | c, e |
| 12 | 44S | c, e, f | 24 | 44C | c, e | 36 | 44P | c, e |
| Patient/Variables | Gender | Age in Years | Country of Origin | Stage/Grade |
|---|---|---|---|---|
| Pt 1 | M | 40 | China | IV/C |
| Pt 2 | F | 38 | Australia | II/B |
| Pt 3 | M | 38 | New Zealand (Māori) | III/B |
| Pt 4 | M | 33 | Vietnam | IV/C |
| Pt 5 | F | 36 | Libya | III/B |
| Pt 6 | M | 40 | Australia | III/B |
| Pt 7 | F | 39 | Poland | III/B |
| Pt 8 | F | 39 | Australia | III/B |
| Pt 9 | F | 32 | New Zealand (Māori) | IV/C |
| Pt 10 | F | 36 | Italy | IV/C |
| Pt 11 | M | 38 | New Zealand | III/B |
| Pt 12 | F | 32 | Italy | III/B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khzam, N.; Kujan, O.; Haubek, D.; Miranda, L.A. Serotype Distribution of Aggregatibacter actinomycetemcomitans in Periodontitis Patients. Pathogens 2025, 14, 805. https://doi.org/10.3390/pathogens14080805
Khzam N, Kujan O, Haubek D, Miranda LA. Serotype Distribution of Aggregatibacter actinomycetemcomitans in Periodontitis Patients. Pathogens. 2025; 14(8):805. https://doi.org/10.3390/pathogens14080805
Chicago/Turabian StyleKhzam, Nabil, Omar Kujan, Dorte Haubek, and Leticia Algarves Miranda. 2025. "Serotype Distribution of Aggregatibacter actinomycetemcomitans in Periodontitis Patients" Pathogens 14, no. 8: 805. https://doi.org/10.3390/pathogens14080805
APA StyleKhzam, N., Kujan, O., Haubek, D., & Miranda, L. A. (2025). Serotype Distribution of Aggregatibacter actinomycetemcomitans in Periodontitis Patients. Pathogens, 14(8), 805. https://doi.org/10.3390/pathogens14080805






